Comprehensive genomic characterization of squamous cell lung cancers (original) (raw)
Main
Lung cancer is the leading cause of cancer-related mortality worldwide, leading to an estimated 1.4 million deaths in 2010 (ref. [1](/articles/nature11404#ref-CR1 "World Health Organization Cancer, fact sheet no. 297 〈 http://www.who.int/mediacentre/factsheets/fs297/en/
〉 (accessed, February 2012)")). The discovery of recurrent mutations in the epidermal growth factor receptor (_EGFR_) kinase, as well as fusions involving anaplastic lymphoma kinase (_ALK_), has led to a marked change in the treatment of patients with lung adenocarcinoma, the most common type of lung cancer[2](/articles/nature11404#ref-CR2 "Soda, M. et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007)"),[3](/articles/nature11404#ref-CR3 "Paez, J. G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004)"),[4](/articles/nature11404#ref-CR4 "Lynch, T. J. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139 (2004)"),[5](/articles/nature11404#ref-CR5 "Pao, W. et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl Acad. Sci. USA 101, 13306–13311 (2004)"). More recent data have suggested that targeting mutations in _BRAF_, _AKT1_, _ERBB2_ and _PIK3CA_ and fusions that involve _ROS1_ and _RET_ may also be successful[6](/articles/nature11404#ref-CR6 "Felip, E., Gridelli, C., Baas, P., Rosell, R. & Stahel, R. Metastatic non-small-cell lung cancer: consensus on pathology and molecular tests, first-line, second-line, and third-line therapy. Ann. Oncol. 22, 1507–1519 (2011)"),[7](/articles/nature11404#ref-CR7 "Ju, Y. S. et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 22, 436–445 (2012)"). Unfortunately, activating mutations in _EGFR_ and _ALK_ fusions are typically not present in the second most common type of lung cancer, lung squamous cell carcinoma (SQCC)[8](/articles/nature11404#ref-CR8 "Rekhtman, N. et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin. Cancer. Res. 18, 1167–1176 (2012)"), and targeted agents developed for lung adenocarcinoma are largely ineffective against lung SQCC.Although no comprehensive genomic analysis of lung SQCCs has been reported, single-platform studies have identified regions of somatic copy number alterations in lung SQCCs, including amplification of SOX2, PDGFRA and FGFR1 and/or WHSC1L1 and deletion of CDKN2A9,10. DNA sequencing studies of lung SQCCs have reported recurrent mutations in several genes, including TP53, NFE2L2, KEAP1, BAI3, FBXW7, GRM8, MUC16, RUNX1T1, STK11 and ERBB4 (refs 11, 12). DDR2 mutations and FGFR1 amplification have been nominated as therapeutic targets13,14,15.
We have conducted a comprehensive study of lung SQCCs from a large cohort of patients as part of The Cancer Genome Atlas (TCGA) project. The twin aims are to characterize the genomic and epigenomic landscape of lung SQCC and to identify potential opportunities for therapy. We report an integrated analysis based on DNA copy number, somatic exonic mutations, messenger RNA sequencing, mRNA expression and promoter methylation for 178 histopathologically reviewed lung SQCCs, in addition to whole genome sequencing (WGS) of 19 samples and microRNA sequencing of 159 samples (Supplementary Table 1.1). Demographic and clinical data and results of the genomic analyses can be downloaded from the TCGA data portal (https://tcga-data.nci.nih.gov/docs/publications/lusc_2012/).
Samples and clinical data
Tumour samples were obtained from 178 patients with previously untreated stage I–IV lung SQCC. Germline DNA was obtained from adjacent, histologically normal tissues resected at the time of surgery (n = 137) or from peripheral blood (n = 41). All patients provided written informed consent to conduct genomic studies in accordance with local Institutional Review Boards. The demographic characteristics are described in Supplementary Table 1.2. The median follow-up for the cohort was 15.8 months, and 60% of patients were alive at the time of the last follow-up (data updated in November 2011). Ninety-six per cent of the patients had a history of tobacco use, similar to previous reports for North American patients with lung SQCC16. DNA and RNA were extracted from patient specimens and measured by several genomic assays, which included standard quality-control assessments (Supplementary Methods, sections 2–8). A committee of experts in lung cancer pathology performed a further review of all samples to confirm the histological subtype (Supplementary Fig. 1.1 and Supplementary Methods, section 1).
Somatic DNA alterations
The lung SQCCs analysed in this study display a large number and variety of DNA alterations, with a mean of 360 exonic mutations, 323 altered copy number segments and 165 genomic rearrangements per tumour.
Copy number alterations were analysed using several platforms. Analysis of single nucleotide polymorphism (SNP) 6.0 array data across the set of 178 lung SQCCs identified a high rate of copy number alteration (mean of 323 segments) when compared with other TCGA projects (as of 1 February 2012), including ovarian cancer (477 segments)17, glioblastoma multiforme (282 segments)18, colorectal carcinoma (213 segments), breast carcinoma (282 segments) and renal cell carcinoma (156 segments) (P < 1 × 10−15 by Fisher’s exact test). These segments gave rise to regions of both focal and broad somatic copy number alterations (SCNAs), with a mean of 47 focal and 23 broad events per tumour (broad events defined as ≥50% of the length of the chromosome arm). There was strong concordance between the three independent copy number assays for all regions of SCNA (Supplementary Figs 2.1–2.4).
At the level of whole chromosome arm SCNAs, lung SQCCs exhibit many similarities to 205 cases of lung adenocarcinoma analysed by TCGA (Supplementary Fig. 2.1a). The most notable difference between these cancers is selective amplification of chromosome 3q in lung SQCC, as has been reported9,19. Using the SNP 6.0 array platform and GISTIC 2.0 (refs 20, 21), we identified regions of significant copy number alteration (Supplementary Methods, section 2). There were 50 peaks of significant amplification or deletion (Q < 0.05), several of which included SCNAs previously seen in lung SQCCs including SOX2, PDGFRA and/or KIT, EGFR, FGFR1 and/or WHSC1L1, CCND1 and CDKN2A9,10,19 (Supplementary Fig. 2.1b and Supplementary Data 2.1 and 2.2). Other peaks defined regions of SCNA reported for the first time, including amplifications of chromosomal segments containing NFE2L2, MYC, CDK6, MDM2, BCL2L1 and EYS and deletions of FOXP1, PTEN and NF1 (Supplementary Fig. 2.1b).
Whole exome sequencing of 178 lung SQCCs and matched germline DNA targeted 193,094 exons from 18,863 genes. The mean sequencing coverage across targeted bases was 121×, with 83% of target bases above 30× coverage. We identified a total of 48,690 non-silent mutations with a mean of 228 non-silent and 360 total exonic mutations per tumour, corresponding to a mean somatic mutation rate of 8.1 mutations per megabase (Mb) and median of 8.4 per Mb. That rate is higher than rates observed in other TCGA projects including acute myelogenous leukaemia (0.56 per Mb), breast carcinoma (1.0 per Mb), ovarian cancer17 (2.1 per Mb), glioblastoma multiforme18 (2.3 per Mb) and colorectal carcinoma (3.2 per Mb) (data as of 1 February 2012, P < 2.2 × 10−16 by _t_-test or Wilcoxon’s rank sum test for lung SQCC versus all others). In lung SQCC, CpG transitions and transversions were the most commonly observed mutation types, with mean rates of 9.9 and 10.7 per sequenced megabase of CpG context, respectively, for a total mutation rate of 20.6 per Mb. At non-CpG sites, transversions at C:G sites were more common than transitions (7.3 versus 2.9 per Mb; total = 10.2 per Mb) and more common than transversions or transitions at A:T sites (1.5 versus 1.3 per Mb; total = 2.8 per Mb).
Significantly mutated genes were identified using a modified version of the MutSig algorithm (Supplementary Methods, section 3)22,23. We identified 10 genes with a false discovery rate (FDR) Q value < 0.1 (Supplementary Table 3.1): _TP53_, _CDKN2A_, _PTEN_, _PIK3CA_, _KEAP1_, _MLL2_, _HLA-A_, _NFE2L2_, _NOTCH1_ and _RB1_, all of which demonstrated robust evidence of gene expression as defined by reads per kilobase of exon model per million mapped reads (RPKM) > 1 (Fig. 1). TP53 mutation was observed in 81% of samples by automated analysis; visual review of sequencing reads identified a further 9% of samples with potential mutations in regions of sub-optimal coverage or in samples with low purity. Most observed mutations in NOTCH1 (8 out of 17) were truncating alterations, suggesting loss-of-function, as has recently been reported for head and neck SQCCs22,24. Mutations in HLA-A were also almost exclusively nonsense or splice site events (7 out of 8).
Figure 1: Significantly mutated genes in lung SQCC.
Significantly mutated genes (Q value < 0.1) identified by exome sequencing are listed vertically by Q value. The percentage of lung SQCC samples with a mutation detected by automated calling is noted at the left. Samples displayed as columns, with the overall number of mutations plotted at the top, and samples are arranged to emphasize mutual exclusivity among mutations. Syn., synonymous.
To increase our statistical power to detect mutated genes in the setting of the observed high background mutation rate, we performed a secondary MutSig analysis only considering genes previously observed to be mutated in cancer according to the COSMIC database. This yielded 12 other genes with FDR < 0.1: FAM123B (also known as WTX), HRAS, FBXW7, SMARCA4, NF1, SMAD4, EGFR, APC, TSC1, BRAF, TNFAIP3 and CREBBP (Supplementary Table 3.1). Both the spectrum and the frequency of EGFR mutations differed from those seen in lung adenocarcinomas. The two most common alterations in lung adenocarcinoma, Leu858Arg and inframe deletions in exon 19, were absent, whereas two Leu861Gln mutations were detected in EGFR.
As described in Supplementary Fig. 3.1, we verified somatic mutations by performing an independent hybrid-recapture of 76 genes in all samples. A total of 1,289 mutations were assayed, and we achieved satisfactory coverage to have power to verify at 1,283 positions. We validated 1,235 mutations (96.2%) (Supplementary Fig. 3.1 and Supplementary Methods, section 3). We also verified mutation calls using WGS and RNA sequencing data with similar results (Supplementary Figs 3.1, 4.3 and Supplementary Methods, sections 3 and 4).
WGS was performed for 19 tumour/normal pairs with a mean computed coverage of 54×. A mean of 165 somatic rearrangements was found per lung SQCC tumour pair (Supplementary Fig. 3.2), a value in excess of that reported for WGS studies of other tumour types including colorectal carcinoma (75)25, prostate carcinoma (108)26, multiple myeloma (21)23 and breast cancer (90)27. Although most inframe coding fusions detected in WGS were validated by RNA sequencing, no recurrent rearrangements predicted to generate fusion proteins were identified (Supplementary Data 3.1 and 4.1).
Somatically altered pathways
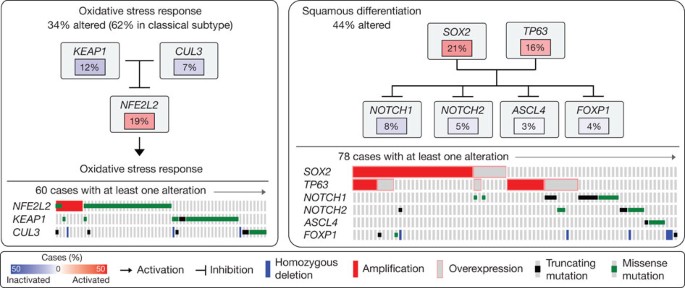
Many of the somatic alterations we have identified in lung SQCCs seem to be drivers of pathways important to the initiation or progression of the cancer. Specifically, genes involved in the oxidative stress response and squamous differentiation were frequently altered by mutation or SCNA. We observed mutations and copy number alterations of NFE2L2 and KEAP1 and/or deletion or mutation of CUL3 in 34% of cases (Fig. 2). NFE2L2 and KEAP1 code for proteins that bind to each other, have been shown to regulate the cell response to oxidative damage, chemo- and radiotherapy, and are somatically altered in a variety of cancer types28,29. We found mutations in NFE2L2 almost exclusively in one of two KEAP1 interaction motifs, DLG or ETGE. Mutations in KEAP1 and CUL3 showed a pattern consistent with loss-of-function and were mutually exclusive with mutations in NFE2L2 (Figs 1c and 2). PARADIGM SHIFT30 analysis predicts that mutations in NFE2L2 and KEAP1 exert a considerable functional effect (Supplementary Fig. 7.C.1, 7.C.2 and Supplementary Methods, section 7).
Figure 2: Somatically altered pathways in squamous cell lung cancer.
Left, alterations in oxidative stress response pathway genes as defined by somatic mutation, copy number alteration or up- or downregulation. Frequencies of alteration are expressed as a percentage of all cases, with background in red for activated genes and blue for inactivated genes. Right, alterations in genes that regulate squamous differentiation, as defined in the left panel.
We also found alterations in genes with known roles in squamous cell differentiation in 44% of samples, including overexpression and amplification of SOX2 and TP63, loss-of-function mutations in NOTCH1, NOTCH2 and ASCL4 and focal deletions in FOXP1 (Fig. 2). Although NOTCH1 has been well characterized as an oncogene in haematological cancers31, NOTCH1 and NOTCH2 truncating mutations have been reported in cutaneous SQCCs and lung SQCCs32. Truncating mutations in ASCL4 are the first to be reported in human cancer and may have a lineage role given the requirement for ASCL1 for survival of small-cell lung cancer cells33. Alterations in NOTCH1, NOTCH2 and ASCL4 were mutually exclusive and exhibited minimal overlap with amplification of TP63 and/or SOX2 (Fig. 2), suggesting that aberrations in those modulators of squamous cell differentiation have overlapping functional consequences.
mRNA expression profiling and subtype classification
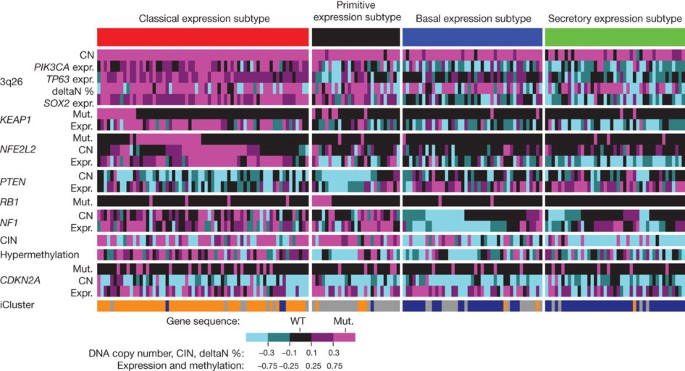
Whole-transcriptome expression profiles were generated by RNA sequencing for the entire cohort and by microarrays for a 121-sample subset. Of 20,502 genes analysed, the mean RNA coverage indices were 19× and 6,420 RPKM (Supplementary Fig. 4.1 and Supplementary Methods, section 4). Previously reported lung SQCC gene expression-subtype signatures34 were applied to both of the expression platforms, yielding four subtypes designated as classical (36%), basal (25%), secretory (24%) and primitive (15%). The concordance of subtypes between the two platforms was high (94% agreement) (Supplementary Fig. 4.2). Considerable correlations were found between the expression subtypes and genomic alterations in copy number, mutation and methylation (Fig. 3). The classical subtype was characterized by alterations in KEAP1, NFE2L2 and PTEN, as well as pronounced hypermethylation and chromosomal instability. The 3q26 amplicon was present in all of the subtypes, but it was most characteristic of the classical subtype, which also showed the greatest overexpression of three known oncogenes on 3q: SOX2, TP63 and PIK3CA. RNA sequencing data suggested that high expression levels of TP63, in samples with and without amplification of TP63, were associated with dominant expression of the deltaN isoform (also called p40), which lacks the amino-terminal transactivation domain, compared with the longer isoform, called tap63 (89% of tumours overexpressed deltaN compared with tap63; P < 2.2 × 10−16). The short deltaN isoform is thought to function as an oncogene35,36, and its expression was most enriched in the classical subtype. By contrast, the primitive expression subtype more commonly exhibited RB1 and PTEN alterations, and the basal expression subtype showed NF1 alterations (Fig. 3). Amplification of FGFR1 and WHSC1L1 was anticorrelated with the classical subtype and specifically with NFE2L2 or KEAP1 mutated samples. Although CDKN2A alterations are common in lung SQCCs, they are not associated with any particular expression subtype (Fig. 3).
Figure 3: Gene expression subtypes integrated with genomic alterations.
Tumours are displayed as columns, grouped by gene expression subtype. Subtypes were compared by Kruskal–Wallis tests for continuous features and by Fisher’s exact tests for categorical features. Displayed features showed significant association with gene expression subtype (P < 0.05), except for CDKN2A alterations. deltaN percentage represents transcript isoform usage between the TP63 isoforms, deltaN and tap63, as determined by RNA sequencing. Chromosomal instability (CIN) is defined by the mean of the absolute values of chromosome arm copy numbers (CN) from the GISTIC23,24 output. Absolute values are used so that amplification and deletion alterations are counted equally. Hypermethylation scores and iCluster assignments are described in Supplementary Figs 6.1 and 7.A1, respectively. CIN, methylation, gene expression and deltaN values were standardized for display using _z_-score transformation. Expr., expression; mut., mutation; WT, wild type.
Independent clustering of miRNA and methylation data indicated association with expression subtypes. The highest overall methylation was seen in the classical subtype (Fig. 3, Supplementary Figs 5.1 and 6.1, Supplementary Methods, sections 5 and 6, Supplementary Data 6.1 and 6.2 and Supplementary Table 5.1). Integrative clustering (iCluster)37 of mRNA, miRNA, methylation, SCNA and mutation data demonstrated concordance with the mRNA expression subtypes and associated alterations (Fig. 3, Supplementary Fig. 7.A.1 and Supplementary Methods, section 7). Independent correlation of somatic mutations, copy number alterations and gene expression signatures revealed notable subtype associations with alterations in the TP53, PI3K, RB1 and NFE2L2/KEAP1 pathways (Supplementary Fig. 7.B.1 and Supplementary Methods, section 7).
Analysis of the CDKN2A locus
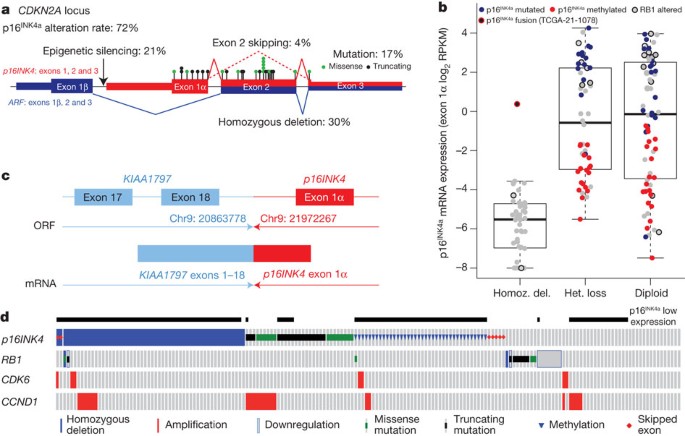
Integrated multiplatform analyses showed that CDKN2A, a known tumour suppressor gene in lung SQCC38 that encodes the p16INK4A and p14ARF proteins, is inactivated in 72% of cases of lung SQCC (Fig. 4a and Supplementary Data 7.1)—by epigenetic silencing by methylation (21%), inactivating mutation (18%), exon 1β skipping (4%) and homozygous deletion (29%).
Figure 4: Multi-faceted characterization of mechanisms of CDKN2A loss.
a, Schematic view of the exon structure of CDKN2A demonstrating the types of alterations identified in the study. The locations of point mutation are denoted by black and green circles. b, CDKN2A expression (y axis) versus CDKN2A copy number (x axis). Samples are represented by circles and colour-coded by specific type of CDKN2A alteration. Del., deletion; het., heterozygous; homoz., homozygous. c, Diagram of the KIAA1797-p16INK4 fusion identified by WGS. ORF, open reading frame. d, CDKN2A alterations and expression levels (binary) in each sample.
Analysis of mRNA expression across the CDKN2A locus revealed four distinct patterns of expression: complete absence of both p16INK4 and ARF (33%); expression of high levels of both p16INK4 and ARF (31%); high expression of ARF and absence of p16INK4 (31%); or expression of a transcript that represents a splicing of exon 1β from ARF with the shared exon 3 of ARF and p16INK4, generating a premature stop codon (4%) (Supplementary Fig. 4.4). Almost all of the cases completely lacking p16INK4 and ARF expression showed homozygous deletion (Fig. 4b and Supplementary Data 7.1). In one case, p16INK4 expression was detected but analysis of WGS data demonstrated an intergenic fusion event that resulted in detectable transcription between exon 1α p16INK4 and exon 18 of KIAA1797 (Fig. 4b, c). Interestingly, combined analysis of WGS and RNA sequencing data identified tumour suppressor gene inactivation by intra- or interchromosomal rearrangement in PTEN, NOTCH1, ARID1A, CTNNA2, VHL and NF1, in eight further cases (Supplementary Data 3.1 and 4.1).
In addition to homozygous deletion, there are frequent mutational events in CDKN2A (Fig. 4b and Supplementary Data 7.1). These account for 45% of the 56 cases with high p16INK4 and ARF expression. Furthermore, methylation of the exon 1α promoter accounts for many other cases of CDKN2A inactivation (70% of lung SQCCs with ARF expression in the absence of detectable p16INK4). Seven other tumours in the high-ARF/low-INK4A group had documented mutations of INK4A, primarily nonsense mutations, suggesting nonsense-mediated decay as a mechanism. Of the 28% of tumours without CDKN2A alterations, RB1 mutations were identified in eight cases and CDK6 amplification in one case (Fig. 4d).
Therapeutic targets
Molecularly targeted agents are now commonly used in patients with adenocarcinoma of the lung, whereas no effective targeted agents have been developed specifically for lung SQCCs13. We analysed our genomic data for evidence of the two common genomic alterations in adenocarcinomas of the lung: EGFR and KRAS mutations. Only one sample had a KRAS codon 61 mutation, and there were no exon 19 deletions or Leu858Arg mutations in EGFR. However, amplifications of EGFR were found in 7% of cases, as were two instances of the Leu861Gln EGFR mutation, which confers sensitivity to erlotinib and gefitinib39.
The presence of new potential therapeutic targets in lung SQCC was suggested by the observation that 96% (171 out of 178) of tumours contain one or more mutations in tyrosine kinases, serine/threonine kinases, phosphatidylinositol-3-OH kinase (PI(3)K) catalytic and regulatory subunits, nuclear hormone receptors, G-protein-coupled receptors, proteases and tyrosine phosphatases (Supplementary Fig. 7.D.1a and Supplementary Data 7.2 and 7.3). From 50 to 77% of the mutations were predicted to have a medium or high functional effect as determined by the mutation assessor score40 (Supplementary Fig. 7.D.1a), and 39% of tyrosine and 42% of serine/threonine kinase mutations were located in the kinase domain. Many of the alterations were in known oncogenes and tumour suppressors, as defined in the COSMIC database (Supplementary Data 7.3).
We selected potential therapeutic targets based on several features, including (1) availability of a US Food and Drug Administration (FDA)-approved targeted therapeutic agent or one under study in current clinical trials (Supplementary Data 7.2); (2) confirmation of the altered allele in RNA sequencing; and (3) the mutation assessor score40. Using those criteria, we identified 114 cases with somatic alteration of a potentially targetable gene (64%) (Supplementary Fig. 7.D.1b and Supplementary Data 7.4). Among these, we identified three families of tyrosine kinases, the erythroblastic leukaemia viral oncogene homologues (ERBBs), fibroblast growth factor receptors (FGFRs) and Janus kinases (JAKs), all of which were found to be mutated and/or amplified41. As discussed for EGFR, the mutational spectra in these potential therapeutic targets differed from those in lung adenocarcinoma (Supplementary Fig. 7.D.2)42.
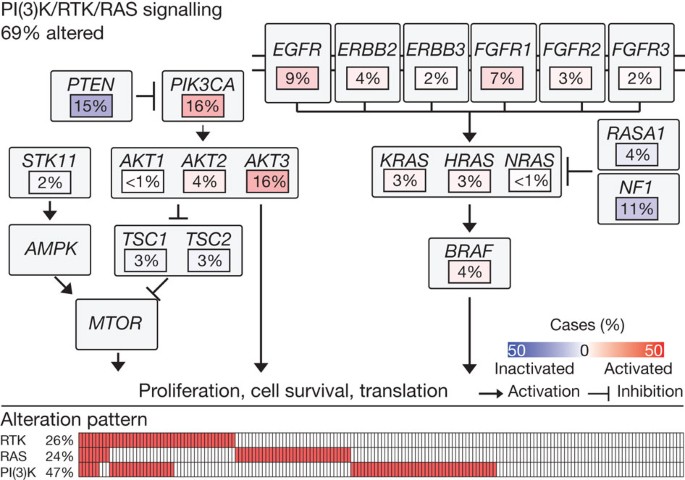
To complement a gene-centred search for potential therapeutic targets, we analysed core cellular pathways known to represent potential therapeutic vulnerabilities: PI(3)K/AKT, receptor tyrosine kinase (RTK) and RAS. Analysis of the 178 lung SQCCs revealed alteration in at least one of those pathways in 69% of samples after restriction of the analysis to mutations confirmed by RNA sequencing and to amplifications associated with overexpression of the target gene (Fig. 5). Mutational events that have been curated in COSMIC are also shown in Supplementary Fig. 7D.2, as is the distribution of mutations, amplifications and overexpression of the genes depicted in Fig. 5. (A summary of all samples and their significant mutations and copy number alterations, including alterations in Fig. 5, is shown in Supplementary Data 7.5.) Specifically, one of the components of the PI(3)K/AKT pathway was altered in 47% of tumours and RTK signalling probably affected by events such as EGFR amplification, BRAF mutation or FGFR amplification or mutation in 26% of tumours (Fig. 5 and Supplementary Fig. 7.D.3). Alterations in the PI(3)K/AKT pathway genes were mutually exclusive with EGFR alterations as identified by MEMo43 (Supplementary Fig. 7.D.4.). Although the dependence of lung SQCC on many of these individual alterations remains to be defined functionally, this analysis suggests new areas for potential therapeutic development in this cancer.
Figure 5: Alterations in targetable oncogenic pathways in lung SQCCs.
Pathway diagram showing the percentage of samples with alterations in the PI(3)K/RTK/RAS pathways. Alterations are defined by somatic mutations, homozygous deletions, high-level, focal amplifications, and, in some cases, by significant up- or downregulation of gene expression (AKT3, FGFR1, PTEN).
Discussion
Lung SQCCs are characterized by a high overall mutation rate of 8.1 mutations per megabase and marked genomic complexity. Similar to high-grade serous ovarian carcinoma17, almost all lung SQCCs display somatic mutation of TP53. There were also frequent alterations in the following pathways: CDKN2A/RB1, NFE2L2/KEAP1/CUL3, PI3K/AKT and SOX2/TP63/NOTCH1 pathways, providing evidence of common dysfunction in cell cycle control, response to oxidative stress, apoptotic signalling and/or squamous cell differentiation. Pathway alterations clustered according to expression-subtype in many cases, suggesting that those subtypes have a biological basis.
A role for somatic mutation in the cancer hallmark of avoiding immune destruction44 is suggested by the presence of inactivating mutations in the HLA-A gene. Somatic loss-of-function alterations of HLA-A have not been reported previously in genomic studies of lung cancer. Given the recently reported efficacy of anti-programmed death 1 (PD1)45 and anti-cytotoxic T-lymphocyte antigen 4 (CTLA4) antibodies in non-small-cell lung cancer46, these HLA-A mutations suggest a possible role for genotypic selection of patients for immunotherapies.
Targeted kinase inhibitors have been successfully used for the treatment of lung adenocarcinoma but minimally so in lung SQCC. The observations reported here suggest that a detailed understanding of the possible targets in lung SQCCs may identify targeted therapeutic approaches. Whereas EGFR and KRAS mutations, the two most common oncogenic aberrations in lung adenocarcinoma, are extremely rare in lung SQCC, alterations in the FGFR kinase family are common. Lung SQCCs also share many alterations in common with head and neck squamous cell carcinomas without evidence of human papilloma virus infection, including mutation in PIK3CA, PTEN, TP53, CDKN2A, NOTCH1 and HRAS22,24, suggesting that the biology of these two diseases may be similar.
The current study has identified a potentially targetable gene or pathway alteration in most lung SQCC samples studied. The data presented here can help to organize efforts to analyse lung SQCC clinical tumour specimens for a panel of specific, actionable mutations to select patients for appropriately targeted clinical trials. These data could thereby help to facilitate effective personalized therapy for this deadly disease.
Methods Summary
All specimens were obtained from patients with appropriate consent from the relevant Institutional Review Board. DNA and RNA were collected from samples using the Allprep kit (Qiagen). We used commercial technology for capture and sequencing of exomes from tumour DNA and normal DNA and whole-genome shotgun sequencing. Significantly mutated genes were identified by comparing them with expectation models based on the exact measured rates of specific sequence lesions. GISTIC23,24 analysis of the circular-binary-segmented Affymetrix SNP 6.0 copy number data was used to identify recurrent amplification and deletion peaks. Consensus clustering approaches were used to analyse mRNA, miRNA and methylation subtypes using previous approaches20,21,34,38,41,44.
References
- World Health Organization Cancer, fact sheet no. 297 〈http://www.who.int/mediacentre/factsheets/fs297/en/〉 (accessed, February 2012)
- Soda, M. et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007)
Article ADS CAS PubMed Google Scholar - Paez, J. G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004)
Article ADS CAS PubMed Google Scholar - Lynch, T. J. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139 (2004)
Article CAS PubMed Google Scholar - Pao, W. et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl Acad. Sci. USA 101, 13306–13311 (2004)
Article ADS CAS PubMed PubMed Central Google Scholar - Felip, E., Gridelli, C., Baas, P., Rosell, R. & Stahel, R. Metastatic non-small-cell lung cancer: consensus on pathology and molecular tests, first-line, second-line, and third-line therapy. Ann. Oncol. 22, 1507–1519 (2011)
Article CAS PubMed Google Scholar - Ju, Y. S. et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 22, 436–445 (2012)
Article CAS PubMed PubMed Central Google Scholar - Rekhtman, N. et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin. Cancer. Res. 18, 1167–1176 (2012)
Article CAS PubMed PubMed Central Google Scholar - Bass, A. J. et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nature Genet. 41, 1238–1242 (2009)
Article CAS PubMed Google Scholar - Ramos, A. H. et al. Amplification of chromosomal segment 4q12 in non-small cell lung cancer. Cancer Biol. Ther. 8, 2042–2050 (2009)
Article CAS PubMed Google Scholar - Shibata, T. et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl Acad. Sci. USA 105, 13568–13573 (2008)
Article ADS CAS PubMed PubMed Central Google Scholar - Kan, Z. et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466, 869–873 (2010)
Article ADS CAS PubMed Google Scholar - Hammerman, P. S., Sos, M. L., Ramos, A. & Xu, C. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discovery 1, 78 (2011)
Article CAS PubMed PubMed Central Google Scholar - Weiss, J. et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci. Transl. Med. 2, 62ra93 (2010)
Article CAS PubMed PubMed Central Google Scholar - Dutt, A. et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One 6, e20351 (2011)
Article ADS CAS PubMed PubMed Central Google Scholar - Kenfield, S. A., Wei, E. K., Stampfer, M. J., Rosner, B. A. & Colditz, G. A. Comparison of aspects of smoking among the four histological types of lung cancer. Tob. Control 17, 198–204 (2008)
Article CAS PubMed Google Scholar - The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011)
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008)
- Tonon, G. et al. High-resolution genomic profiles of human lung cancer. Proc. Natl Acad. Sci. USA 102, 9625–9630 (2005)
Article ADS CAS PubMed PubMed Central Google Scholar - Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010)
Article ADS CAS PubMed PubMed Central Google Scholar - Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011)
Article PubMed PubMed Central Google Scholar - Stransky, N. et al. The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160 (2011)
Article ADS CAS PubMed PubMed Central Google Scholar - Chapman, M. A. et al. Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 (2011)
Article ADS CAS PubMed PubMed Central Google Scholar - Agrawal, N. et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1 . Science 333, 1154–1157 (2011)
Article ADS CAS PubMed PubMed Central Google Scholar - Bass, A. J. et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nature Genet. 43, 964–968 (2011)
Article CAS PubMed Google Scholar - Berger, M. F. et al. The genomic complexity of primary human prostate cancer. Nature 470, 214–220 (2011)
Article ADS CAS PubMed PubMed Central Google Scholar - Stephens, P. J. et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462, 1005–1010 (2009)
Article ADS CAS PubMed PubMed Central Google Scholar - Singh, A. et al. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS Med. 3, e420 (2006)
Article PubMed PubMed Central Google Scholar - Singh, A., Bodas, M., Wakabayashi, N., Bunz, F. & Biswal, S. Gain of Nrf2 function in non-small-cell lung cancer cells confers radioresistance. Antioxid. Redox Signal. 13, 1627–1637 (2010)
Article CAS PubMed PubMed Central Google Scholar - Vaske, C. J. et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics 26, i237–i245 (2010)
Article CAS PubMed PubMed Central Google Scholar - Aster, J. C., Blacklow, S. C. & Pear, W. S. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J. Pathol. 223, 263–274 (2011)
Article Google Scholar - Wang, N. J. et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc. Natl Acad. Sci. USA 108, 17761–17766 (2011)
Article ADS CAS PubMed PubMed Central Google Scholar - Osada, H., Tatematsu, Y., Yatabe, Y., Horio, Y. & Takahashi, T. ASH1 gene is a specific therapeutic target for lung cancers with neuroendocrine features. Cancer Res. 65, 10680–10685 (2005)
Article CAS PubMed Google Scholar - Wilkerson, M. D. et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin. Cancer Res. 16, 4864–4875 (2010)
Article CAS PubMed PubMed Central Google Scholar - Bishop, J. A. et al. p40 (ΔNp63) is superior to p63 for the diagnosis of pulmonary squamous cell carcinoma. Mod. Pathol. 25, 405–415 (2011)
Article PubMed Google Scholar - Massion, P. P. et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res. 63, 7113–7121 (2003)
CAS PubMed Google Scholar - Shen, R., Olshen, A. B. & Ladanyi, M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics 25, 2906–2912 (2009)
Article CAS PubMed PubMed Central Google Scholar - Wikman, H. & Kettunen, E. Regulation of the G1/S phase of the cell cycle and alterations in the RB pathway in human lung cancer. Expert Rev. Anticancer Ther. 6, 515–530 (2006)
Article CAS PubMed Google Scholar - Kancha, R. K., Peschel, C. & Duyster, J. The epidermal growth factor receptor-L861Q mutation increases kinase activity without leading to enhanced sensitivity toward epidermal growth factor receptor kinase inhibitors. J. Thorac. Oncol. 6, 387–392 (2011)
Article PubMed Google Scholar - Reva, B., Antipin, Y. & Sander, C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39, e118 (2011)
Article CAS PubMed PubMed Central Google Scholar - Govindan, R. Summary of the proceedings from the 10th annual meeting of molecularly targeted therapy in non-small cell lung cancer. J. Thorac. Oncol. 5, S433 (2010)
Article PubMed Google Scholar - Ding, L. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075 (2008)
Article ADS CAS PubMed PubMed Central Google Scholar - Ciriello, G., Cerami, E., Sander, C. & Schultz, N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 22, 398–406 (2012)
Article CAS PubMed PubMed Central Google Scholar - Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011)
Article CAS PubMed Google Scholar - Brahmer, J. R. et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J. Clin. Oncol. 28, 3167–3175 (2010)
Article CAS PubMed PubMed Central Google Scholar - Lynch, T. J. et al. Phase II trial of ipilimumab (IPI) and paclitaxel/carboplatin (P/C) in first-line stage IIIb/IV non-small cell lung cancer (NSCLC). J. Clin. Oncol. 28, 7531 (2010)
Article Google Scholar
Acknowledgements
This study was supported by NIH grants U24 CA126561, U24 CA126551, U24 CA126554, U24 CA126543, U24 CA126546, U24 CA126563, U24 CA126544, U24 CA143845, U24 CA143858, U24 CA144025, U24 CA143882, U24 CA143866, U24 CA143867, U24 CA143848, U24 CA143840, U24 CA143835, U24 CA143799, U24 CA143883, U24 CA143843, U54 HG003067, U54 HG003079 and U54 HG003273.
Author information
Author notes
- Human Genome Sequencing Center, Baylor College of Medcine, Houston, Texas 77030, USA.
Authors and Affiliations
- The Eli and Edythe L. Broad Institute of Massachusetts Institute of Technology and Harvard University Cambridge, 02142, Massachusetts, USA
Peter S. Hammerman, Michael S. Lawrence, Douglas Voet, Rui Jing, Kristian Cibulskis, Andrey Sivachenko, Petar Stojanov, Aaron McKenna, Eric S. Lander, Gad Getz, Marcin Imielinski, Elena Helman, Bryan Hernandez, Nam H. Pho, Matthew Meyerson, Gordon Saksena, Andrew D. Cherniack, Stephen E. Schumacher, Barbara Tabak, Scott L. Carter, Nam H. Pho, Huy Nguyen, Andrew Crenshaw, Rameen Beroukhim, Wendy Winckler, Peter S. Hammerman, Gad Getz, Matthew Meyerson, Hailei Zhang, Sachet Shukla, Lynda Chin, Gad Getz, Michael Noble, Doug Voet, Gordon Saksena, Nils Gehlenborg, Daniel DiCara, Hailei Zhang, Spring Yingchun Liu, Michael S. Lawrence, Lihua Zou, Andrey Sivachenko, Pei Lin, Petar Stojanov, Rui Jing, Juok Cho, Marc-Danie Nazaire, Jim Robinson, Helga Thorvaldsdottir, Jill Mesirov, Lynda Chin, Matthew Meyerson, Gad Getz, Peter S. Hammerman, Bryan Hernandez, Marcin Imielinski, Michael S. Lawrence, Andrey Sivachenko, Peter S. Hammerman, Gad Getz, Andrey Sivachenko & Matthew Meyerson - Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, Massachusetts 02215, USA.,
Peter S. Hammerman, Matthew Meyerson, Stephen E. Schumacher, Barbara Tabak, Rameen Beroukhim, Peter S. Hammerman, Matthew Meyerson, Lynda Chin, Chang-Jiun Wu, Lynda Chin, Matthew Meyerson, Peter S. Hammerman, Bruce Johnson, David Kwiatkowski, Peter S. Hammerman, Bruce E. Johnson & Matthew Meyerson - Department of Biology, Massachusetts Institute of Technology, Cambridge, 02142, Massachusetts, USA
Eric S. Lander - Department of Systems Biology, Harvard University, Boston, Massachusetts 02115, USA.,
Eric S. Lander & Carrie Sougnez - Genetic Analysis Platform, The Eli and Edythe L. Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, 02142, Massachusetts, USA
Stacey Gabriel, Gad Getz, Carrie Sougnez, Robert C. Onofrio, Kristin Ardlie, Wendy Winckler, Gad Getz, Gad Getz, Gad Getz & Gad Getz - Department of Pathology, Harvard Medical School, Boston, 02115, Massachusetts, USA
Marcin Imielinski, Matthew Meyerson, Matthew Meyerson, Matthew Meyerson, Marcin Imielinski & Matthew Meyerson - Canada’s Michael Smith Genome Sciences Centre, BC Cancer Agency, Vancouver, British Columbia V5Z, Canada.,
Andy Chu, Hye-Jung E. Chun, Andrew J. Mungall, Erin Pleasance, A. Gordon Robertson, Payal Sipahimalani, Dominik Stoll, Miruna Balasundaram, Inanc Birol, Yaron S. N. Butterfield, Eric Chuah, Robin J. N. Coope, Richard Corbett, Noreen Dhalla, Ranabir Guin, An He, Carrie Hirst, Martin Hirst, Robert A. Holt, Darlene Lee, Haiyan I. Li, Michael Mayo, Richard A. Moore, Karen Mungall, Ka Ming Nip, Jacqueline E. Schein, Jared R. Slobodan, Angela Tam, Nina Thiessen, Richard Varhol, Thomas Zeng, Yongjun Zhao, Steven J. M. Jones, Marco A. Marra, Andy Chu, Elizabeth Chun, Andy Mungall, Gordon Robertson, Dominik Stoll, Andy Chu & Gordon Robertson - Department of Epidemiology and Biostatistics, University of California, San Francisco, San Francisco, California 94143, USA.,
Adam Olshen - Department of Medical Oncology, Belfer Institute for Applied Cancer Science, Dana-Farber Cancer Institute, Boston, Massachusetts 02115, USA.,
Alexei Protopopov, Jianhua Zhang, Xiaojia Ren, Hailei Zhang, Sachet Shukla, Lynda Chin, Jinhua Zhang, Lynda Chin & Jianjua John Zhang - Department of Genomic Medicine, Institute for Applied Cancer Science, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.,
Alexei Protopopov, Jianhua Zhang, Lynda Chin, Jinhua Zhang, Chang-Jiun Wu, Lynda Chin & Jianjua John Zhang - Department of Genetics, Harvard Medical School, Boston, 02115, Massachusetts, USA
Angela Hadjipanayis, Xiaojia Ren, Peng-Chieh Chen, Raju Kucherlapati, Raju Kucherlapati & Raju Kucherlapati - Division of Genetics, Brigham and Women’s Hospital, Boston, 02115, Massachusetts, USA
Angela Hadjipanayis, Xiaojia Ren, Peng-Chieh Chen, Psalm Haseley, Eunjung Lee, Peter J. Park, Raju Kucherlapati, Peter J. Park, Raju Kucherlapati & Raju Kucherlapati - The Center for Biomedical Informatics, Harvard Medical School, Boston, 02115, Massachusetts, USA
Semin Lee, Ruibin Xi, Lixing Yang, Psalm Haseley, Eunjung Lee, Peter J. Park, Nils Gehlenborg & Peter J. Park - Department of Dermatology, Harvard Medical School, Boston, 02115, Massachusetts, USA
Lynda Chin & Lynda Chin - Informatics Program, Children's Hospital, Boston, 02115, Massachusetts, USA
Peter J. Park & Peter J. Park - Computational Biology Center, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, USA.,
Nicholas D. Socci, Yupu Liang, Nikolaus Schultz, Laetitia Borsu, Alex E. Lash, Agnes Viale, Chris Sander, Nikolaus Schultz, Rileen Sinha, Giovanni Ciriello, Ethan Cerami, Benjamin Gross, Anders Jacobsen, Jianjiong Gao, B. Arman Aksoy, Nils Weinhold, Ricardo Ramirez, Barry S. Taylor, Yevgeniy Antipin, Boris Reva, Chris Sander, Chris Sander, Nikolaus Schultz, Rileen Sinha, Nikolaus Schultz, Chris Sander, Ronglai Shen & Rileen Sinha - Department of Molecular Oncology, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, USA.,
Marc Ladanyi, Marc Ladanyi, Marc Ladanyi & Marc Ladanyi - Department of Pathology and Human Oncology & Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, USA.,
Marc Ladanyi, Marc Ladanyi, Marc Ladanyi & Marc Ladanyi - Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.,
J. Todd Auman & J. Todd Auman - Institute for Pharmacogenetics and Individualized Therapy, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.,
J. Todd Auman & J. Todd Auman - Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.,
Katherine A. Hoadley, Piotr A. Mieczkowski, Derek Y. Chiang, Charles M. Perou, Katherine A. Hoadley, Charles M. Perou & Derek Chiang - Department of Pathology and Laboratory Medicine, University of North Carolina at Chapel Hill, Chapel Hill, Chapel Hill, North Carolina 27599, USA.,
Katherine A. Hoadley, Michael D. Topal, Lisle E. Mose, Stuart R. Jefferys, Charles M. Perou, Katherine A. Hoadley, Lisle E. Mose, Stuart R. Jefferys & Charles M. Perou - Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.,
Katherine A. Hoadley, Matthew D. Wilkerson, Yan Shi, Christina Liquori, Shaowu Meng, Ling Li, Yidi J. Turman, Michael D. Topal, Scot Waring, Elizabeth Buda, Jesse Walsh, Darshan Singh, Junyuan Wu, Anisha Gulabani, Peter Dolina, Tom Bodenheimer, Alan P. Hoyle, Janae V. Simons, Matthew G. Soloway, Saianand Balu, Brian D. O’Connor, Derek Y. Chiang, D. Neil Hayes, Charles M. Perou, Matthew D. Wilkerson, Katherine A. Hoadley, Ying Du, Christopher Cabanski, Vonn Walter, Darshan Singh, Junyuan Wu, Anisha Gulabani, Tom Bodenheimer, Alan P. Hoyle, Janae V. Simons, Matthew G. Soloway, Saianand Balu, Jan F. Prins, D. Neil Hayes, Charles M. Perou, Derek Chiang, D. Neil Hayes, W. Kimryn Rathmell, Ashley Hill, D. Neil Hayes & Matthew D. Wilkerson - Carolina Center for Genome Sciences, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.,
Donghui Tan & Yufeng Liu - Department of Biology, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.,
Corbin D. Jones - Department of Computer Science, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.,
Jan F. Prins - Department of Computer Science, University of Kentucky, Lexington, Kentucky 40506, USA.,
Jinze Liu, Kai Wang & Jinze Liu - Department of Internal Medicine, Division of Medical Oncology, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.,
D. Neil Hayes, D. Neil Hayes, D. Neil Hayes & D. Neil Hayes - Cancer Biology Division, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University, Baltimore, Maryland 21231, USA.,
Leslie Cope, Ludmila Danilova, James G. Herman, Stephen B. Baylin, Stephen B. Baylin, Leslie Cope, Ludmila Danilova, James G. Herman, Leslie Cope, James G. Herman & Stephen B. Baylin - University of Southern California Epigenome Center, University of Southern California, Los Angeles, California 90033, USA.,
Daniel J. Weisenberger, Dennis T. Maglinte, Fei Pan, David J. Van Den Berg, Timothy Triche Jr, Peter W. Laird, Daniel Weisenberger & Christopher Wilks - Department of Epidemiology and Biostatistics, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, USA.,
Ronglai Shen, Qianxing Mo, Venkatraman Seshan & Ronglai Shen - Department of Medicine, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, USA.,
Paul K. Paik - Department of Bioinformatics and Computational Biology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.,
Rehan Akbani, Nianxiang Zhang, Bradley M. Broom, Tod Casasent, Anna Unruh, Chris Wakefield, Keith A. Baggerly, John N. Weinstein, Rehan Akbani, John N. Weinstein & John N. Weinstein - Division of Pathology and Laboratory Medicine, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.,
R. Craig Cason - Department of Systems Biology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.,
John N. Weinstein, John N. Weinstein & John N. Weinstein - Department of Biomolecular Engineering and Center for Biomolecular Science and Engineering, University of California Santa Cruz, Santa Cruz, California 95064, USA.,
David Haussler, Joshua M. Stuart, Jingchun Zhu, Christopher Szeto, Sam Ng, Ted Goldstein, Peter Waltman, Artem Sokolov, Kyle Ellrott, Daniel Zerbino, Christopher Wilks, Singer Ma, Brian Craft, Sam Ng, Joshua Stuart & Charles Vaske - Howard Hughes Medical Institute, University of California Santa Cruz, Santa Cruz, California 95064, USA.,
David Haussler - Buck Institute for Age Research, Novato, California 94945, USA.,
Christopher C. Benz, Gary K. Scott & Christina Yau - Division of Hematology/Oncology, University of California San Francisco, San Francisco, 94143, California, USA
Eric A. Collisson, Eric Collisson & Eric A. Collisson - Department of Statistics and Operations Research, University of North Carolina Medical Center, Chapel Hill, North Carolina 27599, USA.,
J. S. Marron - Human Genome Sequencing Center and Dan L. Duncan Cancer Center Division of Biostatistics, Baylor College of Medicine, Houston, Texas 77030, USA.,
Chad J. Creighton, Yiqun Zhang, Chad J. Creighton & Chad J. Creighton - Department of Pathology, Memorial Sloan Kettering Cancer Center, New York, 10065, New York, USA
William D. Travis, Natasha Rekhtman, William D. Travis & William D. Travis - Department of Pathology, Mayo Clinic, Rochester, Minnesota 55905, USA.,
Joanne Yi, Marie C. Aubry & Cristiane Ida - Department of Pathology, Roswell Park Cancer Institute, Buffalo, New York 14263, USA.,
Richard Cheney, Carl Morrison & Carmelo Gaudioso - Department of Pathology, University of Pittsburgh Cancer Center, Pittsburgh, Pennsylvania 15213, USA.,
Sanja Dacic - Department of Pathology, Fox Chase Cancer Center, Philadelphia, Pennsylvania 19111, USA.,
Douglas Flieder, Jeff Boyd & JoEllen Weaver - Department of Pathology, University of North Carolina Medical Center, Chapel Hill, North Carolina 27599, USA.,
William Funkhouser - Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland 21287, USA.,
Peter Illei - Department of Pathology, Penrose-St. Francis Health System, Colorado Springs, Colorado 80907, USA.,
Jerome Myers, John Eckman & Jerome Myers - Department of Pathology and Medical Biophysics, Ontario Cancer Institute and Princess Margaret Hospital, Toronto, Ontario M5G 2MY, Canada.,
Ming-Sound Tsao, Ming-Sound Tsao, Ming-Sound Tsao, Bizhan Bandarchi-Chamkhaleh & Ming-Sound Tsao - International Genomics Consortium, Phoenix, Arizona 85004, USA.,
Robert Penny, David Mallery, Troy Shelton, Martha Hatfield, Scott Morris, Peggy Yena, Candace Shelton, Mark Sherman, Joseph Paulauskis, Robert Penny, Robert Penny, David Mallery, Erin Curley, Candace Shelton & Peggy Yena - Division of Oncology, Department of Medicine and The Genome Institute, Washington University School of Medicine, St. Louis, Missouri 63110, USA.,
Ramaswamy Govindan, Ron Bose, Li-Wei Chang, Li Ding, Christopher A. Maher, Ron Bose, Christopher A. Maher & Ramaswamy Govindan - Center for Individualized Medicine, Mayo Clinic, Rochester, Minnesota 55905, USA.,
Ijeoma Azodo, Farhad Kosari, Sandra Tomaszek, Dennis A. Wigle, Ping Yang, Dennis A. Wigle, Ijeoma A. Azodo, Sandra C. Tomaszek, Farhad Kosari & Dennis A. Wigle - Department of Surgery, University of Michigan, Ann Arbor, Michigan 48109, USA.,
David Beer - The University of Texas MD Anderson Cancer Center, Houston, 77030, Texas, USA
Lauren A. Byers & John Heymach - Departments of Hematology/Oncology and Cancer Biology, Vanderbilt University School of Medicine, Nashville, Tennessee 37232, USA.,
David Carbone, Jacob Kaufman & William Pao - Ontario Cancer Institute, IBM Life Sciences Discovery Centre, Toronto, Ontario M5G 1L7, Canada.,
Igor Jurisica - Department of Translational Genomics, University of Cologne, Cologne D-50931, Germany.,
Martin Peifer, Roman K. Thomas & Roman K. Thomas - Max Planck Institute for Neurological Research, Cologne D-50866, Germany.,
Martin Peifer, Roman K. Thomas & Roman K. Thomas - Department of Surgery, Memorial Sloan Kettering Cancer Center, New York, New York 10065, USA.,
Valerie Rusch & Valerie Rusch - Department of Pharmacology and Chemical Biology, University of Pittsburgh Medical Center, Pittsburgh, Pennsylvania 15232, USA.,
Jill Siegfried & Jill M. Siegfried - Department of Translational Cancer Genomics, Center of Integrated Oncology, University of Cologne, Cologne D-50924, Germany.,
Roman K. Thomas & Roman K. Thomas - SRA International, Fairfax, Virginia 22033, USA.,
Mark A. Jensen, Robert Sfeir, Ari B. Kahn, Anna L. Chu, Prachi Kothiyal, Zhining Wang, Eric E. Snyder, Joan Pontius, Todd D. Pihl, Brenda Ayala, Mark Backus, Jessica Walton, Julien Baboud, Dominique L. Berton, Matthew C. Nicholls, Deepak Srinivasan, Rohini Raman, Stanley Girshik, Peter A. Kigonya, Shelley Alonso, Rashmi N. Sanbhadti, Sean P. Barletta, John M. Greene & David A. Pot - Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota 55905, USA.,
Marie Christine Aubry & Christiane M. Ida - Department of Health Sciences Research, Mayo Clinic, Rochester, Minnesota 55905, USA.,
Ping Yang - Department of Surgery, Johns Hopkins School of Medicine, 600 North Wolfe Street, Baltimore, Maryland 21287, USA.,
Malcolm V. Brock, Kristen Rodgers & Travis Brown - Department of Oncology, Johns Hopkins School of Medicine, 600 North Wolfe Street, Baltimore, Maryland 21287, USA.,
Marian Rutledge & Beverly Lee - Department of Pathology, Johns Hopkins School of Medicine, 600 North Wolfe Street, Baltimore, Maryland 21287, USA.,
James Shin & Dante Trusty - Department of Pathology, University of Pittsburgh, Pittsburgh, Pennsylvania 15213, USA.,
Rajiv Dhir - Cureline, South San Francisco, 94080, California, USA
Olga Potapova & Elena Nemirovich-Danchenko - City Clinical Oncology Dispensary, St Petersburg 197022, Russia.,
Konstantin V. Fedosenko - Department of Pathology, Memorial Sloan Kettering Cancer Center, New York, New York 10065, USA.,
Maureen Zakowski - Helen F. Graham Cancer Center, Newark, Delaware 19713, USA.,
Mary V. Iacocca, Jennifer Brown, Brenda Rabeno, Christine Czerwinski & Nicholas Petrelli - St Joseph Medical Center, Towson, Maryland 21204, USA.,
Zhen Fan & Nicole Todaro - Department of Pathology, UNC Tissue Procurement Facility, UNC Lineberger Cancer Center, Chapel Hill, North Carolina 27599, USA.,
Leigh B. Thorne, Mei Huang & Lori Boice - Ontario Tumour Bank, Ontario Institute for Cancer Research, Toronto, Ontario M5G 0A3, Canada.,
John M. S. Bartlett, Sugy Kodeeswaran & Brent Zanke - Ontario Tumour Bank – Ottawa site, The Ottawa Hospital, Ottawa, Ontario K1H 8L6, Canada.,
Harman Sekhon - Indivumed GmbH, Hamburg, Falkenried 88, Haus D D-20251, Germany.,
Kerstin David - Indivumed Inc, Kensington, Maryland 20895, USA.,
Hartmut Juhl - ILSBio, LLC, Chestertown, Maryland 21620, USA.,
Xuan Van Le, Bernard Kohl & Richard Thorp - Ministry of Health, 138A Giang Vo Street, Hanoi, Vietnam.,
Nguyen Viet Tien - Hue Central Hospital, Hue City, 16 Le Loi, Hue, Vietnam.,
Nguyen Van Bang & Bui Duc Phu - Stanford University Medical Center, Stanford, 94305, California, USA
Howard Sussman - Center for Minority Health Research, University of Texas, M.D. Anderson Cancer Center, Houston, Texas 77030, USA.,
Richard Hajek - National Cancer Institute, 43 Quan Su Street, Hanoi, Vietnam.,
Nguyen Phi Hung - ILSBio LLC, Chestertown, Maryland 21620, USA.,
Khurram Z. Khan - ThoraxKlinik, Heidelberg University Hospital, Heidelberg 69126, Germany.,
Thomas Muley - The Cancer Genome Atlas Program Office, National Cancer Institute, National Institutes of Health, Bethesda, Maryland 20892, USA.,
Kenna R. Mills Shaw, Margi Sheth, Liming Yang, John A. Demchok & Laura A. L. Dillon - Center for Biomedical Informatics and Information Technology (CBIIT), National Cancer Institute, National Institutes of Health, Rockville, Maryland 20852, USA.,
Ken Buetow, Tanja Davidsen, Greg Eley & Carl Schaefer - MLF Consulting, Arlington, 02474, Maryland, USA
Martin Ferguson - National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland 20892, USA.,
Mark S. Guyer, Bradley A. Ozenberger, Jacqueline D. Palchik, Jane Peterson, Heidi J. Sofia & Elizabeth Thomson
Consortia
The Cancer Genome Atlas Research Network
Genome sequencing centres: Broad Institute
- Peter S. Hammerman
- , Michael S. Lawrence
- , Douglas Voet
- , Rui Jing
- , Kristian Cibulskis
- , Andrey Sivachenko
- , Petar Stojanov
- , Aaron McKenna
- , Eric S. Lander
- , Stacey Gabriel
- , Gad Getz
- , Carrie Sougnez
- , Marcin Imielinski
- , Elena Helman
- , Bryan Hernandez
- , Nam H. Pho
- & Matthew Meyerson
Genome characterization centres: BC Cancer Agency
- Andy Chu
- , Hye-Jung E. Chun
- , Andrew J. Mungall
- , Erin Pleasance
- , A. Gordon Robertson
- , Payal Sipahimalani
- , Dominik Stoll
- , Miruna Balasundaram
- , Inanc Birol
- , Yaron S. N. Butterfield
- , Eric Chuah
- , Robin J. N. Coope
- , Richard Corbett
- , Noreen Dhalla
- , Ranabir Guin
- , An He
- , Carrie Hirst
- , Martin Hirst
- , Robert A. Holt
- , Darlene Lee
- , Haiyan I. Li
- , Michael Mayo
- , Richard A. Moore
- , Karen Mungall
- , Ka Ming Nip
- , Adam Olshen
- , Jacqueline E. Schein
- , Jared R. Slobodan
- , Angela Tam
- , Nina Thiessen
- , Richard Varhol
- , Thomas Zeng
- , Yongjun Zhao
- , Steven J. M. Jones
- & Marco A. Marra
Broad Institute
- Gordon Saksena
- , Andrew D. Cherniack
- , Stephen E. Schumacher
- , Barbara Tabak
- , Scott L. Carter
- , Nam H. Pho
- , Huy Nguyen
- , Robert C. Onofrio
- , Andrew Crenshaw
- , Kristin Ardlie
- , Rameen Beroukhim
- , Wendy Winckler
- , Peter S. Hammerman
- , Gad Getz
- & Matthew Meyerson
Brigham & Women’s Hospital/Harvard Medical School
- Alexei Protopopov
- , Jianhua Zhang
- , Angela Hadjipanayis
- , Semin Lee
- , Ruibin Xi
- , Lixing Yang
- , Xiaojia Ren
- , Hailei Zhang
- , Sachet Shukla
- , Peng-Chieh Chen
- , Psalm Haseley
- , Eunjung Lee
- , Lynda Chin
- , Peter J. Park
- & Raju Kucherlapati
Memorial Sloan-Kettering Cancer Center (TCGA pilot phase only)
- Nicholas D. Socci
- , Yupu Liang
- , Nikolaus Schultz
- , Laetitia Borsu
- , Alex E. Lash
- , Agnes Viale
- , Chris Sander
- & Marc Ladanyi
University of North Carolina at Chapel Hill
- J. Todd Auman
- , Katherine A. Hoadley
- , Matthew D. Wilkerson
- , Yan Shi
- , Christina Liquori
- , Shaowu Meng
- , Ling Li
- , Yidi J. Turman
- , Michael D. Topal
- , Donghui Tan
- , Scot Waring
- , Elizabeth Buda
- , Jesse Walsh
- , Corbin D. Jones
- , Piotr A. Mieczkowski
- , Darshan Singh
- , Junyuan Wu
- , Anisha Gulabani
- , Peter Dolina
- , Tom Bodenheimer
- , Alan P. Hoyle
- , Janae V. Simons
- , Matthew G. Soloway
- , Lisle E. Mose
- , Stuart R. Jefferys
- , Saianand Balu
- , Brian D. O’Connor
- , Jan F. Prins
- , Jinze Liu
- , Derek Y. Chiang
- , D. Neil Hayes
- & Charles M. Perou
University of Southern California/Johns Hopkins
- Leslie Cope
- , Ludmila Danilova
- , Daniel J. Weisenberger
- , Dennis T. Maglinte
- , Fei Pan
- , David J. Van Den Berg
- , Timothy Triche Jr
- , James G. Herman
- , Stephen B. Baylin
- & Peter W. Laird
Genome data analysis centres: Broad Institute
- Gad Getz
- , Michael Noble
- , Doug Voet
- , Gordon Saksena
- , Nils Gehlenborg
- , Daniel DiCara
- , Jinhua Zhang
- , Hailei Zhang
- , Chang-Jiun Wu
- , Spring Yingchun Liu
- , Michael S. Lawrence
- , Lihua Zou
- , Andrey Sivachenko
- , Pei Lin
- , Petar Stojanov
- , Rui Jing
- , Juok Cho
- , Marc-Danie Nazaire
- , Jim Robinson
- , Helga Thorvaldsdottir
- , Jill Mesirov
- , Peter J. Park
- & Lynda Chin
Memorial Sloan-Kettering Cancer Center
- Nikolaus Schultz
- , Rileen Sinha
- , Giovanni Ciriello
- , Ethan Cerami
- , Benjamin Gross
- , Anders Jacobsen
- , Jianjiong Gao
- , B. Arman Aksoy
- , Nils Weinhold
- , Ricardo Ramirez
- , Barry S. Taylor
- , Yevgeniy Antipin
- , Boris Reva
- , Ronglai Shen
- , Qianxing Mo
- , Venkatraman Seshan
- , Paul K. Paik
- , Marc Ladanyi
- & Chris Sander
The University of Texas MD Anderson Cancer Center
- Rehan Akbani
- , Nianxiang Zhang
- , Bradley M. Broom
- , Tod Casasent
- , Anna Unruh
- , Chris Wakefield
- , R. Craig Cason
- , Keith A. Baggerly
- & John N. Weinstein
University of California Santa Cruz/Buck Institute
- David Haussler
- , Christopher C. Benz
- , Joshua M. Stuart
- , Jingchun Zhu
- , Christopher Szeto
- , Gary K. Scott
- , Christina Yau
- , Sam Ng
- , Ted Goldstein
- , Peter Waltman
- , Artem Sokolov
- , Kyle Ellrott
- , Eric A. Collisson
- , Daniel Zerbino
- , Christopher Wilks
- , Singer Ma
- & Brian Craft
University of North Carolina at Chapel Hill
- Matthew D. Wilkerson
- , J. Todd Auman
- , Katherine A. Hoadley
- , Ying Du
- , Christopher Cabanski
- , Vonn Walter
- , Darshan Singh
- , Junyuan Wu
- , Anisha Gulabani
- , Tom Bodenheimer
- , Alan P. Hoyle
- , Janae V. Simons
- , Matthew G. Soloway
- , Lisle E. Mose
- , Stuart R. Jefferys
- , Saianand Balu
- , J. S. Marron
- , Yufeng Liu
- , Kai Wang
- , Jinze Liu
- , Jan F. Prins
- , D. Neil Hayes
- & Charles M. Perou
Baylor College of Medicine
- Chad J. Creighton
- & Yiqun Zhang
Pathology committee
- William D. Travis
- , Natasha Rekhtman
- , Joanne Yi
- , Marie C. Aubry
- , Richard Cheney
- , Sanja Dacic
- , Douglas Flieder
- , William Funkhouser
- , Peter Illei
- , Jerome Myers
- & Ming-Sound Tsao
Biospecimen core resources: International Genomics Consortium
- Robert Penny
- , David Mallery
- , Troy Shelton
- , Martha Hatfield
- , Scott Morris
- , Peggy Yena
- , Candace Shelton
- , Mark Sherman
- & Joseph Paulauskis
Disease working group
- Matthew Meyerson
- , Stephen B. Baylin
- , Ramaswamy Govindan
- , Rehan Akbani
- , Ijeoma Azodo
- , David Beer
- , Ron Bose
- , Lauren A. Byers
- , David Carbone
- , Li-Wei Chang
- , Derek Chiang
- , Andy Chu
- , Elizabeth Chun
- , Eric Collisson
- , Leslie Cope
- , Chad J. Creighton
- , Ludmila Danilova
- , Li Ding
- , Gad Getz
- , Peter S. Hammerman
- , D. Neil Hayes
- , Bryan Hernandez
- , James G. Herman
- , John Heymach
- , Cristiane Ida
- , Marcin Imielinski
- , Bruce Johnson
- , Igor Jurisica
- , Jacob Kaufman
- , Farhad Kosari
- , Raju Kucherlapati
- , David Kwiatkowski
- , Marc Ladanyi
- , Michael S. Lawrence
- , Christopher A. Maher
- , Andy Mungall
- , Sam Ng
- , William Pao
- , Martin Peifer
- , Robert Penny
- , Gordon Robertson
- , Valerie Rusch
- , Chris Sander
- , Nikolaus Schultz
- , Ronglai Shen
- , Jill Siegfried
- , Rileen Sinha
- , Andrey Sivachenko
- , Carrie Sougnez
- , Dominik Stoll
- , Joshua Stuart
- , Roman K. Thomas
- , Sandra Tomaszek
- , Ming-Sound Tsao
- , William D. Travis
- , Charles Vaske
- , John N. Weinstein
- , Daniel Weisenberger
- , Dennis A. Wigle
- , Matthew D. Wilkerson
- , Christopher Wilks
- , Ping Yang
- & Jianjua John Zhang
Data coordination centre
- Mark A. Jensen
- , Robert Sfeir
- , Ari B. Kahn
- , Anna L. Chu
- , Prachi Kothiyal
- , Zhining Wang
- , Eric E. Snyder
- , Joan Pontius
- , Todd D. Pihl
- , Brenda Ayala
- , Mark Backus
- , Jessica Walton
- , Julien Baboud
- , Dominique L. Berton
- , Matthew C. Nicholls
- , Deepak Srinivasan
- , Rohini Raman
- , Stanley Girshik
- , Peter A. Kigonya
- , Shelley Alonso
- , Rashmi N. Sanbhadti
- , Sean P. Barletta
- , John M. Greene
- & David A. Pot
Tissue source sites
- Ming-Sound Tsao
- , Bizhan Bandarchi-Chamkhaleh
- , Jeff Boyd
- , JoEllen Weaver
- , Dennis A. Wigle
- , Ijeoma A. Azodo
- , Sandra C. Tomaszek
- , Marie Christine Aubry
- , Christiane M. Ida
- , Ping Yang
- , Farhad Kosari
- , Malcolm V. Brock
- , Kristen Rodgers
- , Marian Rutledge
- , Travis Brown
- , Beverly Lee
- , James Shin
- , Dante Trusty
- , Rajiv Dhir
- , Jill M. Siegfried
- , Olga Potapova
- , Konstantin V. Fedosenko
- , Elena Nemirovich-Danchenko
- , Valerie Rusch
- , Maureen Zakowski
- , Mary V. Iacocca
- , Jennifer Brown
- , Brenda Rabeno
- , Christine Czerwinski
- , Nicholas Petrelli
- , Zhen Fan
- , Nicole Todaro
- , John Eckman
- , Jerome Myers
- , W. Kimryn Rathmell
- , Leigh B. Thorne
- , Mei Huang
- , Lori Boice
- , Ashley Hill
- , Robert Penny
- , David Mallery
- , Erin Curley
- , Candace Shelton
- , Peggy Yena
- , Carl Morrison
- , Carmelo Gaudioso
- , John M. S. Bartlett
- , Sugy Kodeeswaran
- , Brent Zanke
- , Harman Sekhon
- , Kerstin David
- , Hartmut Juhl
- , Xuan Van Le
- , Bernard Kohl
- , Richard Thorp
- , Nguyen Viet Tien
- , Nguyen Van Bang
- , Howard Sussman
- , Bui Duc Phu
- , Richard Hajek
- , Nguyen Phi Hung
- , Khurram Z. Khan
- & Thomas Muley
Project team: National Cancer Institute
- Kenna R. Mills Shaw
- , Margi Sheth
- , Liming Yang
- , Ken Buetow
- , Tanja Davidsen
- , John A. Demchok
- , Greg Eley
- , Martin Ferguson
- , Laura A. L. Dillon
- & Carl Schaefer
National Human Genome Research Institute
- Mark S. Guyer
- , Bradley A. Ozenberger
- , Jacqueline D. Palchik
- , Jane Peterson
- , Heidi J. Sofia
- & Elizabeth Thomson
Writing committee
- Peter S. Hammerman
- , D. Neil Hayes
- , Matthew D. Wilkerson
- , Nikolaus Schultz
- , Ron Bose
- , Andy Chu
- , Eric A. Collisson
- , Leslie Cope
- , Chad J. Creighton
- , Gad Getz
- , James G. Herman
- , Bruce E. Johnson
- , Raju Kucherlapati
- , Marc Ladanyi
- , Christopher A. Maher
- , Gordon Robertson
- , Chris Sander
- , Ronglai Shen
- , Rileen Sinha
- , Andrey Sivachenko
- , Roman K. Thomas
- , William D. Travis
- , Ming-Sound Tsao
- , John N. Weinstein
- , Dennis A. Wigle
- , Stephen B. Baylin
- , Ramaswamy Govindan
- & Matthew Meyerson
Contributions
The TCGA research network contributed collectively to this study. Biospecimens were provided by the tissue source sites and processed by the biospecimen core resource. Data generation and analyses were performed by the genome sequencing centres, cancer genome characterization centres and genome data analysis centres. All data were released through the data coordinating centre. Project activities were coordinated by the National Cancer Institute and National Human Genome Research Institute project teams. We also acknowledge the following TCGA investigators who made substantial contributions to the project: P.S.H. and D.N.H. (manuscript coordinators); M.D.W. (data coordinator); P.S.H. and N.S. (analysis coordinators); P.S.H., M.S.L., A. Sivachecnko, B.H. and G.G. (DNA sequence analysis); M.D.W., J.L. and D.N.H. (mRNA sequence analysis); L. Cope, J.G.H. and L. Danilova (DNA methylation analysis); A.C., G.S., N.H.P., R.K. and M.L. (copy number analysis); N.S., R. Bose, C.J.C., R. Sinha, C.M., S.N., E.A.C., R. Shen, J.N.W. and C. Sander (pathway analysis); A.C. and G.R. (miRNA sequence analysis); W.D.T., B.E.J., D.A.W. and M.-S.T. (pathology and clinical expertise); S.B.B., R. Govindan and M. Meyerson (project chairs).
Corresponding author
Correspondence toMatthew Meyerson.
Ethics declarations
Competing interests
The author declare no competing financial interests.
Additional information
The primary and processed data used to generate the analyses presented here can be downloaded by registered users fromThe Cancer Genome Atlas (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp, https://cghub.ucsc.edu/ and https://tcga-data.nci.nih.gov/docs/publications/lusc_2012/).
(Participants are arranged by area of contribution and then by institution.)
Supplementary information
Supplementary Information
This file contains Supplementary Methods 1-8, which includes Supplementary Figures and Tables and additional references (see Supplementary Information file pages 1-2 for details). (PDF 16829 kb)
Supplementary Data
This zipped file contains Supplementary Data files 2.1, 2.2, 3.1, 3.2, 4.1, 4.2 , 6.1, 6.2, 7.1, 7.2, 7.3 (Supplementary table to support figure 5A_Bose_02 23 2012), 7.4 and 7.5 (see Supplementary Information file for details). (ZIP 9075 kb)
PowerPoint slides
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence (http://creativecommons.org/licenses/by-nc-sa/3.0/).
About this article
Cite this article
The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers.Nature 489, 519–525 (2012). https://doi.org/10.1038/nature11404
- Received: 09 March 2012
- Accepted: 09 July 2012
- Published: 09 September 2012
- Issue Date: 27 September 2012
- DOI: https://doi.org/10.1038/nature11404