Genome-scale loss-of-function screening with a lentiviral RNAi library (original) (raw)
- Perspective
- Published: 23 August 2006
Nature Methods volume 3, pages 715–719 (2006)Cite this article
- 5261 Accesses
- 9 Altmetric
- Metrics details
Abstract
The discovery that RNA interference (RNAi) is functional in mammalian cells led us to form The RNAi Consortium (TRC) with the goal of enabling large-scale loss-of-function screens through the development of genome-scale RNAi libraries and methodologies for their use. These resources form the basis of a loss-of-function screening platform created at the Broad Institute. Our human and mouse libraries currently contain >135,000 lentiviral clones targeting 27,000 genes. Initial screening efforts have demonstrated that these libraries and methods are practical and powerful tools for high-throughput lentivirus RNAi screens.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Additional access options:
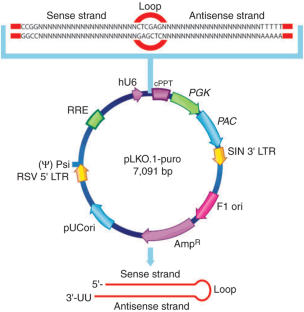
Figure 1: Vector map for the pLKO.1 lentiviral vector.
Similar content being viewed by others
References
- Elbashir, S.M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001).
Article CAS Google Scholar - Brummelkamp, T.R. & Bernards, R. New tools for functional mammalian cancer genetics. Nat. Rev. Cancer 3, 781–789 (2003).
Article CAS Google Scholar - Hannon, G.J. & Rossi, J.J. Unlocking the potential of the human genome with RNA interference. Nature 431, 371–378 (2004).
Article CAS Google Scholar - Moffat, J. & Sabatini, D.M. Building mammalian signalling pathways with RNAi screens. Nat. Rev. Mol. Cell Biol. 7, 177–187 (2006).
Article CAS Google Scholar - Berns, K. et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428, 431–437 (2004).
Article CAS Google Scholar - Brummelkamp, T.R. et al. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat. Chem. Biol. 2, 202–206 (2006).
Article CAS Google Scholar - Kolfschoten, I.G. et al. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell 121, 849–858 (2005).
Article CAS Google Scholar - Moffat, J. et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 (2006).
Article CAS Google Scholar - Ngo, V.N. et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441, 106–110 (2006).
Article CAS Google Scholar - Paddison, P.J. et al. A resource for large-scale RNA-interference-based screens in mammals. Nature 428, 427–431 (2004).
Article CAS Google Scholar - Pelkmans, L. et al. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 436, 78–86 (2005).
Article CAS Google Scholar - Westbrook, T.F. et al. A genetic screen for candidate tumor suppressors identifies REST. Cell 121, 837–848 (2005).
Article CAS Google Scholar - Dull, T. et al. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72, 8463–8471 (1998).
CAS PubMed PubMed Central Google Scholar - Follenzi, A., Ailles, L.E., Bakovic, S., Geuna, M. & Naldini, L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25, 217–222 (2000).
Article CAS Google Scholar - Zufferey, R. et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 72, 9873–9880 (1998).
CAS PubMed PubMed Central Google Scholar - Zeng, Y., Wagner, E.J. & Cullen, B.R. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9, 1327–1333 (2002).
Article CAS Google Scholar - Silva, J.M. et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 37, 1281–1288 (2005).
Article CAS Google Scholar - Khvorova, A., Reynolds, A. & Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216 (2003).
Article CAS Google Scholar - Schwarz, D.S. et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208 (2003).
Article CAS Google Scholar - Birmingham, A. et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3, 199–204 (2006).
Article CAS Google Scholar - Jackson, A.L. et al. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21, 635–637 (2003).
Article CAS Google Scholar - Chen, W., Arroyo, J.D., Timmons, J.C., Possemato, R. & Hahn, W.C. Cancer-associated PP2A Aalpha subunits induce functional haploinsufficiency and tumorigenicity. Cancer Res. 65, 8183–8192 (2005).
Article CAS Google Scholar - Masutomi, K. et al. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl. Acad. Sci. USA 102, 8222–8227 (2005).
Article CAS Google Scholar
Acknowledgements
The authors thank J. Grenier, G. Hinkle, B. Luo, J. Moffat and S. Silver for their contributions to the work reviewed here and for their comments on the manuscript. The TRC library is the product of The RNAi Consortium (TRC). We thank the members of TRC (Academia Sinica, Bristol-Meyers Squibb, Eli Lilly, Novartis, Sigma-Aldrich, the Broad Institute of MIT and Harvard, Dana-Farber Cancer Institute, Massachusetts General Hospital, Harvard Medical School, the Massachusetts Institute of Technology, and the Whitehead Institute for Biomedical Research) for their scientific contributions and financial support. The Broad Institute and TRC do not profit from the distribution of the TRC library. Sigma-Aldrich, one of five sponsoring members of The RNAi Consortium who contributed equally to funding the work described here, is a commercial distributor of TRC shRNA library clones.
Author information
Authors and Affiliations
- Broad Institute of MIT and Harvard, Cambridge, 02142, Massachusetts, USA
David E Root, Nir Hacohen, William C Hahn, Eric S Lander & David M Sabatini - Center for Immunology and Inflammatory Diseases, Massachusetts General Hospital, Boston, 02114, Massachusetts, USA
Nir Hacohen - Harvard Medical School, Boston, 02115, Massachusetts, USA
Nir Hacohen, William C Hahn & Eric S Lander - Department of Medical Oncology, Center for Cancer Systems Biology, Center for Cancer Genome Discovery, Dana-Farber Cancer Institute, Boston, 02115, Massachusetts, USA
William C Hahn - Whitehead Institute for Biomedical Research, Cambridge, 02142, Massachusetts, USA
Eric S Lander & David M Sabatini - Department of Biology, Massachusetts Institute of Technology, Cambridge, 02139, Massachusetts, USA
Eric S Lander & David M Sabatini
Authors
- David E Root
You can also search for this author inPubMed Google Scholar - Nir Hacohen
You can also search for this author inPubMed Google Scholar - William C Hahn
You can also search for this author inPubMed Google Scholar - Eric S Lander
You can also search for this author inPubMed Google Scholar - David M Sabatini
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toDavid E Root.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Root, D., Hacohen, N., Hahn, W. et al. Genome-scale loss-of-function screening with a lentiviral RNAi library.Nat Methods 3, 715–719 (2006). https://doi.org/10.1038/nmeth924
- Published: 23 August 2006
- Issue Date: 01 September 2006
- DOI: https://doi.org/10.1038/nmeth924