Autophagy: molecular machinery for self-eating (original) (raw)
Introduction
Normal cellular development and growth require a well-regulated balance between protein synthesis and degradation. Eukaryotic cells have two major avenues for degradation, the proteasome and autophagy. Autophagy, which literally means ‘self-eating’, is involved in the bulk degradation of long-lived cytosolic proteins and organelles, whereas the ubiquitin–proteasome system degrades specific short-lived proteins. Chaperone-mediated autophagy is a mechanism that allows the degradation of cytosolic proteins that contain a particular pentapeptide consensus motif.1, 2 Accordingly, it has a limited degradative capacity and will not be discussed in this review. The two other types of autophagy are macroautophagy and microautophagy.3, 4 Both of these pathways involve dynamic membrane rearrangements and, unlike proteasomal degradation, terminate at the lysosome/vacuole. Microautophagy involves the direct engulfment of cytoplasm at the surface of the degradative organelle by protrusion, septation, and/or invagination of the limiting membrane. On the other hand, macroautophagy involves the sequestration of cytoplasm into a double-membrane cytosolic vesicle, termed an autophagosome (Figure 1). Formation of the autophagosome is thought to take place at the perivacuolar, pre-autophagosomal structure (PAS).5, 6 Completed autophagosomes, which are 300–900 nm in diameter, fuse with the lysosome/vacuole, and the inner single-membrane vesicle is released into the lumen. The resulting autophagic body is lysed, the contents are degraded, and the resulting macromolecules recycled. In this review, we focus on macroautophagy, hereafter referred to as autophagy.
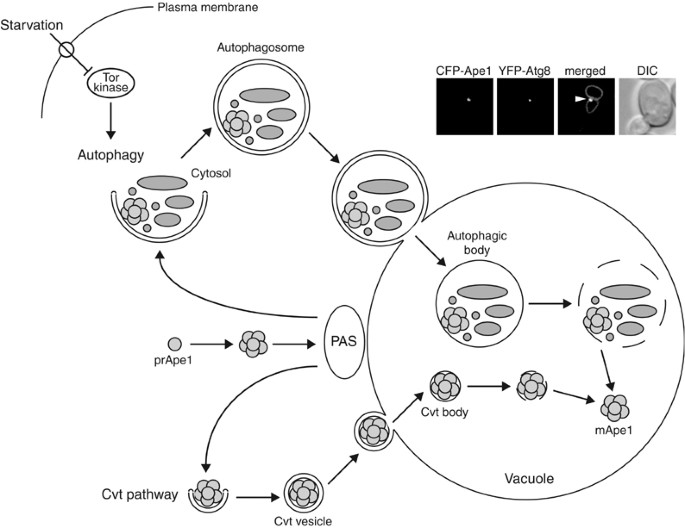
Figure 1
Autophagy and the Cvt pathway in yeast. Morphological steps are schematically shown in autophagy and the Cvt pathway. Both pathways require the sequestration of cargo components within distinct double-membrane vesicles, whose formation is thought to occur at the PAS. Autophagy is induced by inactivation of Tor kinase under starvation conditions such as nitrogen depletion. Organelles and bulk cytoplasm are sequestered into a double-membrane autophagosome, which is 300–900 nm in diameter. The Cvt pathway is a biosynthetic process that delivers resident hydrolases including Ape1 to the vacuole in vegetative conditions. The Cvt vesicle (140–160 nm in diameter) enwraps specific cargos such as the prApe1 oligomer (Ape1 complex). The Ape1 complex is also sequestered specifically by autophagosomes under starvation conditions. Once completed, the double-membrane vesicles target to and fuse with the vacuole, and the single-membrane vesicles (autophagic or Cvt body) are released into the lumen. Subsequently, these vesicles are degraded, allowing maturation of prApe1 into the mature form (mApe1) by removal of its propeptide, and the degradation of the cytoplasm. Fluorescent images show that the cargo protein CFP-Ape1 localizes at the perivacuolar PAS (white arrowhead) marked by YFP-Atg8; the vacuole is labeled with FM 4-64 in the merged image. DIC, differential interference contrast
Autophagy is evolutionarily conserved in eukaryotes from yeast to mammal, and has important roles in various cellular functions.3, 7, 8 In yeast, for example, nutrient starvation induces a high level of autophagy, which allows unneeded proteins to be degraded and the amino acids recycled for the synthesis of proteins that are essential for survival. In higher eukaryotes, autophagy is also induced in response to the nutrient depletion that occurs in animals at birth after severing of the trans-placental food supply, as well as that of nutrient-starved cultured cells and tissues.9, 10 Moreover, autophagy is involved in physiological cellular processes such as lifespan extension and cellular development and differentiation.8 Autophagy may play a protective role against the progression of some human diseases, including cancer, muscular disorders, and neurodegeneration, such as Huntington's, Alzheimer's, and Parkinson's diseases,8, 11, 12 and acts as a cellular defense mechanism to prevent infection by certain pathogenic bacteria and viruses;12, 13, 14 conversely, there is evidence that autophagy is involved in type II programmed cell death and might contribute to the pathology of some diseases.4, 15
Autophagy was morphologically identified first in the 1960s in mammalian cells; however, the molecular mechanism for this process has only recently begun to be elucidated. One breakthrough for studying the molecular basis of autophagy was achieved by identifying the genes involved in this process,16, 17 which in yeast are termed autophagy-related (ATG) genes.18 ATG genes were also discovered from studies of the biosynthetic cytoplasm to vacuole targeting (Cvt) pathway and pexophagy (peroxisome degradation) in baker's and methylotrophic yeasts, respectively.19, 20 Currently, there are 27 ATG genes whose products appear to be specific to Atg processes that have been identified in yeast.18 In addition, components required for the early secretory pathway as well as those involved in fusion with the vacuole are required for autophagy.21, 22, 23 It is known that the biosynthetic Cvt and degradative autophagy pathways are topologically and morphologically similar and share most of the Atg components (Figure 1).24, 25, 26 In contrast to autophagy, however, the Cvt pathway is a highly selective process that involves the sequestration of at least two specific cargos that are resident vacuolar hydrolases, aminopeptidase I (Ape1) and _α_-mannosidase (Ams1).27, 28 These proteins are sequestered within a double-membrane vesicle, termed a Cvt vesicle. In addition to differences in cargo selectivity – the Cvt vesicle appears to exclude bulk cytosol – there is a significant difference in the vesicle size between autophagy and the Cvt pathway; the Cvt vesicle is fairly consistent in size, and is much smaller than the autophagosome, being 140–160 nm in diameter.26 Similarly, the sequestering vesicle that forms during pexophagy, the pexophagosome, appears to enwrap only peroxisomes, although it may be substantially larger than a Cvt vesicle in order to accommodate its cargo.29
Studies with yeasts have advanced our understanding of the molecular mechanism required for autophagy, pexophagy, and the Cvt pathway. Additionally, orthologs of the ATG gene products have recently been found and functionally characterized in higher eukaryotes such as mammal, insect, worm, and plant, and have been shown to play important roles in Atg cellular functions like those noted above.3, 8 These findings have revealed that the molecular machineries of autophagy found in yeast are also commonly utilized in various other eukaryotic cells. This review summarizes our current understanding about the molecular mechanism of autophagy, with an emphasis on yeast.
Regulation and Induction of Autophagy
Autophagy occurs at a basal level in normal growing conditions; however, certain types of environmental stress result in a dramatic induction. For example, yeast autophagy is induced by nutrient starvation, including nitrogen and carbon depletion. The protein kinase target of rapamycin (Tor) is a negative regulator that specifically responds to nitrogen levels although the nutrient sensor upstream of Tor is unknown.30 Under nutrient-rich conditions, Tor kinase is active and autophagy is inhibited, whereas Tor is inactivated under nutrient-deficient conditions and autophagic activity is enhanced. The Tor protein (mTor) is conserved in mammalian cells and also senses environmental change to modulate autophagy, but the mechanism of regulation may be more complicated than that in yeast.31, 32 In mammalian cells, the negative regulatory cascade upstream of mTor includes a class I phosphatidylinositol (PtdIns) 3-kinase, PDK1, and Akt/PKB, while the phosphatase PTEN acts antagonistically to the PtdIns 3-kinase to induce autophagy.
In yeast, Tor controls the phosphorylation state of Atg13, one of the proteins required for autophagy (Figure 2).33, 34 In nutrient-rich conditions, Atg13 is highly phosphorylated and has a lower affinity for Atg1 kinase; in this situation autophagy is repressed. In contrast, under starvation conditions or following treatment with the Tor inhibitor rapamycin, Atg13 is rapidly and partially dephosphorylated and interacts with Atg1 with a higher affinity. These changes correlate with an increase in autophagic activity. Atg1 is the sole serine/threonine protein kinase identified among the Atg proteins,35 and is essential for both the Cvt pathway and autophagy;24 however, the requirement for Atg1 kinase activity relative to these two pathways is not clear. One report shows that Atg1 kinase activity, regulated by Atg13, is required for autophagy.34 According to this model, inhibiting Tor with rapamycin or through starvation allows hypophosphorylated Atg13 to interact with Atg1, resulting in increased Atg1 kinase activity and autophagy, although the downstream target of Atg1 is still not known. In contrast, another study has shown that the Cvt pathway requires higher levels of Atg1 kinase activity.36 In this case, Atg1 is proposed to have a primarily structural role during autophagy, although a lower level of kinase activity is still required. The role of Atg1 kinase activity still remains arguable; however, the Atg1 kinase complex may have an important role in induction and regulation, possibly switching the mode between the Cvt pathway and autophagy in response to environmental changes.
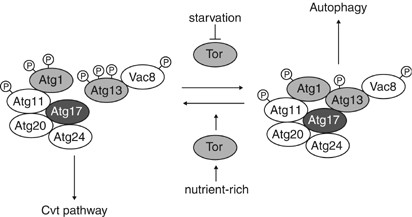
Figure 2
Regulatory complex for autophagy induction. Tor kinase regulates the induction of autophagy upon sensing the nutrient conditions. Atg1 kinase, which is essential for both autophagy and the Cvt pathway, forms a complex with several proteins that are characterized as being preferentially involved in the Cvt pathway (in white circles) or autophagy (in dark gray circle). Under nutrient-rich conditions, Tor kinase is active, and Atg13 is hyperphosphorylated. This highly phosphorylated Atg13 has a lower affinity for Atg1 and Atg17, and autophagy is downregulated. Under starvation conditions, Tor is inactive and Atg13 is hypophosphorylated. Dephosphorylated Atg13 interacts with Atg1 and Atg17 with a higher affinity. The enhancement of the formation of the Atg1-Atg13-Atg17 complex mediates the induction of autophagy. The Atg20-Atg24 complex, Atg11, and Vac8 might also belong to this complex, but a complete holo-complex as depicted has not been identified
The Atg1-Atg13 complex also interacts with multiple proteins such as Vac8, Atg11, and Atg17.34, 37, 38 Atg11 and Vac8 are also phosphorylated, independently of the Atg1-Atg13 complex, although the role of their phosphorylation is unknown.37 Atg17 is required for autophagy under starvation conditions, but is not needed for the Cvt pathway under nutrient-rich conditions.34 Recent reports have shown the presence of smaller autophagosomes, which are nearly 200 nm in diameter, in _atg17_Δ cells.39, 40 These abnormal autophagosomes allow uptake of precursor Ape1 (prApe1), but are defective for pexophagy and cause a dramatic reduction in bulk autophagy. Atg17 interacts with Atg13 in the absence of Atg1 but not vice versa, suggesting that Atg17 is assembled with the Atg1-Atg13 complex through a direct interaction with Atg13. This Atg17 interaction seems to have an important role in the regulation of autophagosome biogenesis; Atg17 mutants that are not able to interact with the Atg1-Atg13 complex have reduced levels of autophagy. These results suggest that the Atg1-Atg13-Atg17 complex might play a role in autophagy separate from induction and may be involved in controlling autophagosome formation, specifically in determining the ultimate size of the vesicle. Although Atg1 is conserved in mammals, it is not known how the Atg1 homolog acts in the mammalian autophagy system. In contrast, Atg13 and Atg17 homologs are not found in mammals.
Selective Autophagy/Cvt Pathway
Autophagy is generally known as a process involved in the degradation of bulk cytoplasmic components that are nonspecifically sequestered into an autophagosome. However, there are some specific cargos for autophagy. In yeasts such as Saccharomyces cerevisiae, Pichia pastoris, Hansenula polymorpha, and Yarrowia lipolytica, peroxisomes are specifically sequestered and degraded by both micro- and macroautophagic processes.29, 41, 42, 43 When yeasts are grown on a carbon source that requires peroxisomes for their metabolism such as oleate or methanol, peroxisomes proliferate. Shifting to an alternate carbon source such as glucose renders the excess peroxisomes superfluous and they are rapidly eliminated. The specific degradation of peroxisomes under nutrient-rich conditions, as opposed to starvation, is termed pexophagy. Micro- and macropexophagy appear to involve the same machinery as that of nonspecific autophagy,29 but all of the components that render pexophagy specific have not been elucidated, although Atg11, a specificity factor for the Cvt pathway, plays a role in specific peroxisome degradation. Even in the case of bulk autophagy, it has been reported that at least one protein, cytosolic acetaldehyde dehydrogenase or Ald6, is preferentially sequestered into autophagosomes relative to other cytosolic proteins.44
The prApe1 is sequestered within either Cvt vesicles or autophagosomes, depending on the nutrient conditions, and delivered to the vacuole. Autophagy and the Cvt pathway are topologically and mechanistically similar and share most of the same machinery (Figure 1).24, 25, 26 Recently, the mechanism of the selective recognition and packaging of prApe1 has been relatively well clarified (Figure 3). preApe1 protein is synthesized in the cytosol.45 After synthesis, prApe1 forms a dodecamer and these dodecamers are further assembled into a larger oligomeric structure, called the Ape1 complex, in a process dependent on the prApe1 propeptide.46 The Ape1 complex is ultimately enwrapped within either Cvt vesicles or autophagosomes at the perivacuolar PAS.26, 47, 48
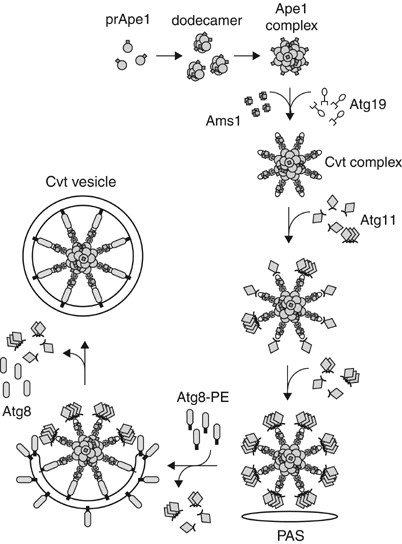
Figure 3
Selective recognition and packaging of cargo, and vesicle formation in the Cvt pathway. The Cvt pathway is an Atg process that allows the selective transport of the vacuolar hydrolases Ape1 and Ams1 into the vacuole. The preApe1 is synthesized in the cytosol, subsequently assembles into dodecamers and further forms a larger oligomer, termed the Ape1 complex. The receptor protein Atg19 binds to the Ape1 complex through the prApe1 propeptide to form the Cvt complex; the Ams1 oligomer is recruited to the complex via its binding to Atg19. Atg11 is assembled with the Cvt complex by interacting with Atg19 and guides the complex to the PAS. Atg11 forms a homodimer or homo-oligomer at the PAS, although it is not clear whether it occurs after or before the arrival at this site. Once the Cvt complex reaches the PAS, Atg components such as Atg8 conjugated to phosphatidylethanolamine (Atg8–PE) are recruited for vesicle formation. Atg19 further interacts with Atg8–PE, which might ensure the proper packaging of the cargo into the forming Cvt vesicle. Atg11 dissociates from the Cvt complex at the PAS prior to vesicle completion, although the exact timing is unknown
The receptor protein Atg19 binds to the Ape1 complex through the prApe1 propeptide to form the Cvt complex in the cytosol.49 In the absence of Atg19, prApe1 can form an Ape1 complex, but it does not localize at the PAS.47, 48 Another vacuole resident hydrolase, _α_-mannosidase (Ams1), also binds Atg19 at a separate site than that used for prApe1, and is sequestered within the vesicle along with the Cvt complex.48 Localization of the Cvt complex to the PAS requires at least one additional protein, Atg11;38 without Atg11, Atg19 and prApe1 can form the Cvt complex, but the complex does not target to the PAS.48 The C-terminus of Atg19 binds to Atg11 via a C-terminal coiled-coil region; movement to the PAS requires additional coiled-coil domains of Atg11.50 It remains unknown, however, whether another protein is needed to mediate the transport and targeting step. Once the cargo complex arrives at the PAS, Atg19 might mediate sequestration by the forming vesicle through an interaction with Atg8–PE.48 The Atg19-Atg8–PE interaction might have a role in guaranteeing that nonspecific cytosolic components are excluded from the forming Cvt vesicle, whereas the Cvt complex cargo is selectively packed into the vesicle. After the Cvt vesicle is completed, Atg19 remains incorporated within the vesicle and is transported to the vacuole along with the cargo, where it is degraded.49 In contrast, it is thought that Atg11 is not delivered to the vacuole, but is released from the cargo molecules and recycled before vesicle completion.38 Although the timing of Atg11 release from Atg19 is not clear, Atg1 kinase may be involved in this step; it was observed that Atg11 homo-oligomerization occurs more efficiently without the Atg1-Atg13 kinase complex, or in the presence of a kinase-defective Atg1 mutant.50
Under starvation conditions, neither Atg11 nor Atg19 is required for bulk nonselective autophagy. However, the efficiency of prApe1 transport to the vacuole is significantly reduced in the absence of Atg11 and Atg19, indicating that, under autophagic/starvation conditions, these proteins are required for selective autophagic transport of the Ape1 complex, functioning in a similar manner as in the Cvt pathway.51 It was observed that some Atg components that normally localize to the PAS do not show this localization without the Cvt complex or Atg11, or in the presence of certain Atg11 mutants that are unable to deliver the Cvt complex to the PAS.51, 50 This inability to correctly assemble the PAS is only seen under vegetative/Cvt pathway conditions, but not during starvation/autophagy. This observation may indicate that there are different ways to form the PAS depending on nutrient conditions, and further suggests a mechanism that allows the Cvt pathway to be highly selective for cargo.
Cargos for selective autophagy have not yet been documented in mammalian systems. For example, the biosynthetic Cvt pathway is not known to exist outside of yeast. Similarly, specific degradation of peroxisomes has not been shown to occur in higher eukaryotes. Along these lines, the Atg proteins that are known to play a role in cargo recognition do not have homologs in any of the characterized higher eukaryotic systems. It is possible that single-celled organisms have a greater demand for modulating autophagic processes; however, it is likely that at least degradative types of specific autophagy do take place in all organisms.
Conjugation Systems
Two unique ubiquitin-like conjugation systems, Atg8–phosphatidylethanolamine (Atg8–PE) and Atg12–Atg5, are involved in the biogenesis of autophagic vesicles (Figure 4).52 These conjugation systems are widely conserved in various eukaryotes and have an essential role in autophagy.3, 8
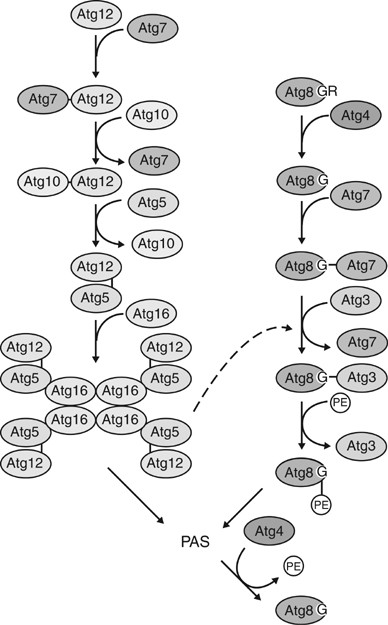
Figure 4
Two conjugation machineries. One conjugation begins with Atg7, which is homologous to the E1 ubiquitin-activating enzyme, linking to Atg12 through the formation of a thioester bond involving the C-terminal glycine residue of Atg12. Activated Atg12 is transferred to the E2-like protein Atg10, which catalyzes the conjugation with Atg5 through the formation of an isopeptide bond between the activated glycine of Atg12 and an internal lysine of Atg5. Finally, Atg16 is assembled with the Atg12–Atg5 conjugate. Atg16 can also form a homotetramer, resulting in a multimer of the Atg12–Atg5-Atg16 complex. The other system starts with the cysteine protease Atg4, which proteolytically removes the C-terminal arginine of Atg8 to allow access to a glycine residue. The same E1-like Atg7 activates this exposed glycine residue and then transfers activated Atg8 to Atg3. Finally, Atg3 mediates the conjugation of Atg8 with phosphatidylethanolamine to form Atg8–PE. The Atg12–Atg5-Atg16 conjugate is also required for stability of Atg8–PE, although the mechanism remains unclear. Atg12–Atg5-Atg16 and Atg8–PE both localize to the PAS and appear to be involved in vesicle formation. Atg8–PE is a structural component of the completed Cvt vesicle and autophagosome, but is enriched on the inner membrane. During or soon after vesicle formation, Atg8–PE on the vesicle outer membrane is cleaved off by Atg4 protease
In the Atg12–Atg5 conjugation system, an irreversible isopeptide bond is formed between a C-terminal glycine residue of Atg12 and a central lysine residue of Atg5.53 Two additional proteins are required to form the conjugate; one is Atg7, which is a homolog of the E1 ubiquitin-activating enzyme Uba1,54 the other is Atg10, which functions like an E2 ubiquitin-conjugating enzyme.55 As in the ubiquitin system, the E1-like Atg7 binds the C-terminal glycine of Atg12 through its active site cysteine to form an intermediate complex via a thioester bond. ATP hydrolysis results in Atg12 activation. Subsequently, activated Atg12 is transferred to the E2-like Atg10 and transiently linked through another thioester bond. Finally, Atg12 is covalently bonded to an internal lysine residue of Atg5 to form the final conjugate. The Atg12–Atg5 conjugate binds another protein, Atg16, noncovalently.56 Atg16 can form a homo-oligomer to mediate the formation of a higher multimeric structure of Atg12, Atg5, and Atg16 that has a molecular mass of 350 kDa in yeast and 800 kDa in mouse, which are predicted to be composed of a tetramer and octamer of the Atg12–Atg5-Atg16 complex, respectively.57, 58 Furthermore, experiments in which Atg16 oligomerization was regulated showed that the formation of the multimeric Atg12–Atg5-Atg16 complex is functionally important for auto-phagy.57
The other conjugation system involves Atg8 as a modifier of a lipid, phosphatidylethanolamine.59, 60 Atg8 was originally characterized as a microtubule-associated protein required for autophagy and the Cvt pathway,61 although a functional association with microtubules has not been substantiated. In the first step of Atg8 conjugation, the cysteine protease Atg4 proteolytically removes a C-terminal arginine residue of Atg8, exposing a glycine that is now accessible to the E1-like Atg7, the same enzyme used in the Atg12–Atg5 conjugation system. Atg7 activates Atg8, which is then transferred to another E2-like enzyme, Atg3, and eventually conjugated to phosphatidylethanolamine through an amide bond. Atg8 conjugated to PE behaves like a membrane protein. Unlike the Atg12–Atg5 conjugation, modification of PE with Atg8 is a reversible event, in which Atg4 can again cleave Atg8 after the glycine residue to remove it from the lipid.59 Recently, the Atg8–PE conjugation process was reconstituted using proteins purified from Escherichia coli.62 Only Atg7 and Atg3 are required to generate Atg8–PE in vitro if the Atg8 is genetically modified to remove the C-terminal arginine, bypassing the initial Atg4 cleavage event; however, Atg8–PE conjugation appears to be regulated in a more complex manner in vivo because depletion of the Atg12–Atg5 conjugate causes instability of Atg8–PE.5
Both the Atg12–Atg5-Atg16 complex and the Atg8–PE conjugate localize at the PAS and have some role in vesicle formation.5, 63 The Atg12–Atg5-Atg16 complex is not observed on the completed autophagosome. In mammalian cells, however, the complex is clearly associated with the forming autophagosome, termed the phagophore or isolation membrane.64, 58 These results lead us to suggest that during the membrane formation stage the Atg12–Atg5-Atg16 complex drives the expansion and/or curvature of the membrane envelope and finally dissociates from the vesicle just before, or immediately after, completion. On the other hand, Atg8–PE is detected on both the forming intermediate vesicle and the completed autophagosome.65 It is transported to the lysosome/vacuole and is degraded along with the cargo. Accordingly, Atg8–PE is the best candidate for a structural component of the autophagosome. The Atg8/Atg8–PE levels are significantly induced under starvation conditions. This may be an important factor allowing the increase in the size of the autophagosome relative to the Cvt vesicle. Along these lines, aberrant or small autophagosome-like structures are observed in _atg8_Δ cells under starvation conditions. Based on these observations, Atg8 might be another factor that is involved in controlling the size of the autophagosome.
There are at least three Atg8 homologs in mammalian cells, GATE-16, GABARAP, and MAP1LC3.66, 67 All three proteins appear to be modified with lipid in the same manner as occurs in yeast, and have been shown to localize to the autophagosome. Among them, LC3 has been best characterized and is used as a marker for the mammalian autophagosome, similar to yeast Atg8. Although yeast Atg8 is clearly induced during autophagy, this is not necessarily the case with the mammalian homologs. LC3 levels typically increase following autophagic induction, but this increase is cell line-dependent and the magnitude is not as great as seen with Atg8.
Phosphatidylinositol 3-Phosphate and Phosphatidylinositol 3-Kinase Complex
Phosphatidylinositol 3-phosphate (PtdIns(3)P) is known to have an important role in various cellular functions. There is only one PtdIns 3-kinase, Vps34, identified in yeast. Vps34 is involved in forming two distinct PtdIns 3-kinase complexes (Figure 5).68 Complex I is composed of Vps34, Vps15, Vps30/Atg6, and Atg14, while complex II contains the same proteins, except that Atg14 is replaced by Vps38. In the absence of Atg14, complex I is disrupted, resulting in the normal sorting of carboxypeptidase Y (Prc1), which is normally transported from the late Golgi to the vacuole through the CPY pathway, but the loss of autophagy and the Cvt pathway. On the other hand, Vps38 depletion causes a defect in Prc1 sorting, but allows normal progression of autophagy and the Cvt pathway.
Figure 5
Two PtdIns 3-kinase complexes in yeast. (a) Both complexes share Vps15, Vps30/Atg6 and Vps34. Complex I has Atg14 and functions in autophagy and the Cvt pathway. Complex II contains Vps38 and is involved in the CPY and Mvb pathways. (b) It is hypothesized that complex I might generate PtdIns(3)P for autophagy and the Cvt pathway at the PAS. The Atg20-Atg24 complex, Atg18, Atg21 and Atg27 are recruited to the PAS through their association with PtdIns(3)P
PtdIns(3)P is known to be bound by proteins that have PtdIns(3)P-binding sites such as the PX and FYVE domains. Two proteins, Atg20 and Atg24, have PX domains and bind to PtdIns(3)P.69, 70 Atg20 and Atg24 also belong to the sorting nexin family that functions in protein trafficking from the Golgi to the endosome, but both proteins are involved only in the Cvt pathway. These proteins interact with each other and show two types of subcellular localization, at the endosome and at the PAS. Their endosomal localization requires the PtdIns 3-kinase complex II, whereas the PAS localization requires complex I. The endosomal localization of the Atg20-Atg24 complex appears to be dispensable for the Cvt pathway. Conversely, this complex has a role in the Cvt pathway at the PAS. Moreover, Atg24 and possibly Atg20 interact with Atg17, and Atg20 also interacts with Atg11;69, 50 hence, the Atg20-Atg24 complex might function as part of the Atg1 kinase complex (Figure 2).
Atg18, Atg21, and Atg27 are also recruited to the PAS in a manner that is dependent on PtdIns 3-kinase complex I;71, 72, 73 all three proteins bind to PtdIns(3)P although none of them have known binding domains. Atg18 is required for both the Cvt and autophagy pathways, whereas Atg21 and Atg27 are primarily required for the Cvt pathway. The role of PtdIns(3)P in autophagy still remains to be investigated, but it might mediate the recruitment of some Atg components to the PAS to enable them to be involved in the vesicle-forming machinery; however, there is no direct evidence that PtdIns(3)P exists in the putative membrane of the PAS.
In contrast to yeast, mammalian cells regulate autophagy via two PtdIns 3-kinase complexes, class I and class III.32 The class III enzyme is similar to yeast Vps34, generates PtdIns(3)P, and plays a stimulatory role in autophagy. The class I enzyme converts PtdIns(4,5)P2 to PtdIns(3,4,5)P3. This enzyme complex functions at the plasma membrane and is part of a signaling cascade that activates mTor and other kinases, and thus has an inhibitory role. The function of PtdIns(3)P in mammalian cells is not clear, although it likely has a role similar to that in yeast; however, the Atg20, Atg21, Atg24, and Atg27 proteins, which appear to bind this phosphoinositide, do not have homologs in higher eukaryotes. Atg18 is the only PtdIns(3)P-binding protein with a mammalian homolog, but it has not yet been characterized in higher eukaryotes.
Retrieval
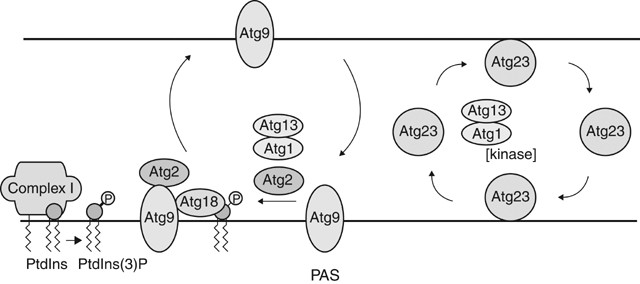
Most of the Atg components are either soluble or peripherally membrane-associated proteins, and display a single punctate localization at the PAS. Atg9, however, is predicted to be a multi-spanning transmembrane protein.74 In contrast to other Atg proteins, Atg9 localizes not only to the PAS but is also distributed to several other punctate structures.5 Similarly, Atg23, which is a peripheral membrane protein, also shows a multiple punctate localization including the PAS.75 Recently our laboratory has shown that, in the absence of the Atg1-Atg13 kinase complex, both Atg9 and Atg23 accumulate only at the PAS.76 This result suggests that Atg9 and Ag23 cycle between the PAS and the additional structures/organelles, and that the Atg1–Atg13 complex is involved in the retrieval of these proteins from the PAS. Additionally, only Atg23 retrieval requires a high level of Atg1 kinase activity, whereas Atg2, Atg18, and PtdIns 3-kinase complex I components are necessary for the retrieval of Atg9 (Figure 6).
Figure 6
Recycling of Atg9 and Atg23. Atg9 and Atg23 localize at the PAS and at multiple peripheral punctate structures (shown to be the mitochondria for Atg9). Retrieval of the integral membrane protein Atg9 from the PAS involves the Atg1-Atg13 complex, Atg2, and Atg18. The PtdIns 3-kinase complex I is proposed to generate PtdIns(3)P at the PAS, and Atg18 is likely localized to the PAS through its association with this phosphoinositide. The Atg1-Atg13 complex induces the interaction of Atg2, Atg9 and Atg18. Finally, this ternary complex allows Atg9 retrieval from the PAS. Atg23 is a peripheral membrane protein. It is not known whether Atg23 is recruited to the PAS from a cytosolic or membrane-associated pool. In contrast to Atg9, a high level of Atg1 kinase activity is needed to mediate the retrieval of Atg23 from the PAS
Atg9 interacts with Atg2 and can also bind Atg18.76, 77 Recruitment of the peripheral membrane protein Atg2 to the PAS requires Atg1, Atg13, PtdIns 3-kinase complex I, and Atg18,71, 78 whereas Atg9 interaction with Atg18 requires both Atg1 and Atg2.76 One model is that once Atg9 is recruited from the peripheral structures to the PAS, the Atg1-Atg13 kinase complex mediates the interaction of Atg9 with Atg2 and Atg18 and the formation of this ternary complex allows Atg9 to be retrieved from the PAS back to the peripheral sites.
The identity of the peripheral (non-PAS) structures or organelles where Atg23 is localized is still not known. A recent analysis indicates that the peripheral structures of Atg9 correspond to the mitochondria,79 although the signal and mechanism that target Atg9 to the mitochondria are not known. It is not understood how the Atg9 and Atg23 proteins, and their cycling, function in the autophagy pathway. However, both proteins have some role in vesicle formation because a sequestering vesicle is not formed in the absence of Atg9.74 In contrast, without Atg23, autophagosomes with essentially normal size are formed, but at greatly reduced numbers.75 Recently, two mammalian homologs of Atg9 were identified.80 Interestingly, in addition to the integral membrane regions, one of them has a putative signal sequence for mitochondrial localization; however, both proteins primarily localize to the perinuclear region and do not localize to mitochondria in cultured cells. A retrieval process for Atg9 has not been documented in higher eukaryotes, although it is likely that a similar mechanism exists. Along these lines, it is interesting that there is a homolog of Atg18 as noted earlier, suggesting that this protein may have a conserved role in the retrieval process.
Vesicle Biogenesis
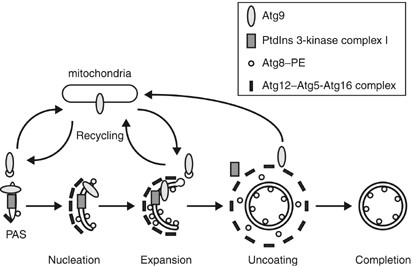
How is the vesicle formed in autophagy? This is a critical question, but one that has not been satisfactorily answered. The double-membrane vesicles for autophagy and the Cvt pathway are generated de novo in the sense that they do not appear to be derived by budding from a pre-existing organelle. Figure 7 shows a hypothetical model for vesicle formation. The PAS is the putative site of vesicle formation; most of the Atg components localize to this site, at least transiently.5, 63 The Atg proteins comprise the machinery for forming the vesicle and almost all of the Atg proteins may somehow be involved in the vesicle formation step. In contrast to vesicles that form by budding, an initial membrane must be formed in autophagy; this is the nucleation step of vesicle formation. The origin of this membrane is unknown. There is a general, although not universal, consensus that the endoplasmic reticulum (ER) is a source of the membrane for forming the vesicle. It is reported that ER function and machinery of the early secretory pathway are required for autophagy and the Cvt pathway in yeast;21, 22, 23 however, further study is necessary for understanding this relationship. The absence of appropriate conditional mutants in higher eukaryotes has prevented a similar analysis concerning the origin of the sequestering membrane, and the corresponding studies have been morphological and cytological in nature.
Figure 7
Schematic model for the step of vesicle formation. First, membrane nucleation is dependent on the PtdIns 3-kinase complex I, and Atg9. Atg proteins, including the Atg12–Atg5-Atg16 complex and Atg8–PE conjugate, components of the vesicle-forming machinery, are recruited to the PAS. The Atg12–Atg5-Atg16 complex mediates the expansion of the vesicle possibly by acting as a coat. Atg8–PE is recruited into the forming vesicle dependent on the Atg12–Atg5-Atg16 complex, and also acts in the elongation of the vesicle as a structural component. Atg9 cycles between the PAS and mitochondria; Atg9 cycling may supply the lipid for the expanding membrane. Upon, or just prior to, vesicle completion, the coat proteins and the Atg proteins, except for the Atg8–PE that is oriented toward the lumen (and Atg19), are dissociated from the vesicle. Finally, the completed vesicle can fuse with the vacuole
Among the Atg components, the PtdIns 3-kinase complex I is proposed to play some role in membrane nucleation. Complex I might generate PtdIns(3)P at the PAS and facilitate the recruitment of certain Atg components, such as the vesicle-forming machinery. The Atg12–Atg5-Atg16 complex and Atg8–PE conjugate are included in this machinery, and their localization to the PAS also requires Atg9.5 The PtdIns 3-kinase complex I and Atg9 may function before Atg12–Atg5-Atg16 and Atg8–PE; although Atg9 and complex I do not affect the formation of the conjugates, they are required for their proper localization at the PAS. In their absence, the conjugates are diffuse in the cytosol without any punctate localization. The Atg12–Atg5-Atg16 complex and Atg8–PE conjugate are proposed to be first located at the initial nucleation membrane and then function in vesicle expansion. Without the Atg12–Atg5-Atg16 complex, Atg8–PE does not localize at the PAS, resulting in a block in vesicle formation. In contrast, neither the localization of Atg5 at the PAS nor Atg12–Atg5 conjugate formation requires Atg8–PE.5, 57 Thus, the Atg12–Atg5-Atg16 complex is formed and may function prior to Atg8–PE at the PAS. Subsequently, Atg8–PE can localize to the PAS, or is stabilized at this site, and becomes incorporated into the membrane of the forming vesicle, possibly playing a role in determining vesicle size. During vesicle expansion, the Atg12–Atg5-Atg16 complex might transiently coat the forming membrane vesicle and control the vesicle curvature.
Expansion of the vesicle presumably requires the delivery of lipids, possibly in the form of lipid bilayers present in transport vesicles; however, the origin of these lipids is not entirely known. Atg9 localizes at the PAS and mitochondria as multiple dots, and may cycle between these structures.76, 79 It is not clear how Atg9 cycling functions; however, one hypothesis is that Atg9 marks the membrane source of the autophagosome. Atg9 may mediate the delivery of lipids/membrane by recycling from the donor (the mitochondria) to the acceptor membrane (autophagosome). This proposed lipid flow mediated by Atg9 cycling could be regulated via the Atg1-Atg13 complex and the PtdIns 3-kinase complex I in a manner that includes Atg2 and Atg18 (Figure 6). The Atg9 protein is required for the recruitment of the Atg12–Atg5-Atg16 complex and Atg8–PE to the PAS.5 In contrast, Atg1 and Atg2 are not needed for the PAS localization of the conjugates,5 suggesting that Atg9 retrieval from the PAS, which depends on Atg1 and Atg2, is also not important in this regard. Atg9 cycling might supply the membrane to drive vesicle expansion in concert with the Atg12–Atg5-Atg16 complex and Atg8–PE.
The sequestering vesicle might be uncoated before, or immediately after, the vesicle is completed. In fact, transient association of the Atg5 protein with the forming autophagosome has been shown by the real-time imaging of GFP-tagged Atg5 in mammalian cells.64 GFP-Atg5 initially appears as a small dot, becomes elongated into a spherical structure and finally dissociates when the autophagosome is completed. Upon vesicle completion, other Atg proteins comprising the vesicle-forming machinery are also dissociated from the vesicle. After uncoating, presumed fusion machinery present on the vesicle would become exposed to drive the fusion of the autophagosome with the vacuole. Thus, coating could act in part to prevent premature fusion of the forming intermediate vesicle with the vacuole.
Fusion with the Vacuole and Breakdown of the Vesicle
Once completed, the double-membrane autophagosome is transported to the vacuole and the outer membrane of the vesicle fuses with the vacuolar membrane, in a process that is dependent upon Ccz1 and Mon1.81 Ccz1 and Mon1 form a complex and also function in homotypic vacuole fusion.82 The Ccz1-Mon1 complex form part of a common fusion machinery consisting of the SNARE proteins Vam3, Vti1 and Vam7, the NSF Sec18, the _α_-SNAP Sec17, the Rab GTPase Ypt7 and the class C Vps/HOPS complex.4, 31 After fusion, the autophagosome inner single-membrane vesicle is released inside the vacuole lumen, and is now termed the autophagic body (Figure 1).
Subsequently, the membrane of the autophagic body is broken down in the vacuole lumen. The degradation of the autophagic body is dependent on two resident vacuole proteases, Pep4 and Prb1, and acidification of the vacuole lumen.83, 84 In addition to these factors, the transmembrane protein Atg15 is also required for lysis.85, 86 Atg15 has a conserved functional motif in esterases and lipases. This protein is transported to the vacuole through the multivesicular body (Mvb) pathway.87 Breakdown of the autophagic body results in release of the cargo into the vacuole lumen, where the cytoplasmic components are degraded. Finally, autophagy can be completed by allowing the resulting macromolecules to be recycled for reuse in the synthesis of proteins necessary for survival under nutrient-depleted conditions.
In mammalian cells, autophagosomes fuse with lysosomes, the analog of the yeast vacuole, to generate the autophagolysosome. It has been suggested that the Rab GTPase Rab7 is targeted to the autophagosome membrane and is involved in a late step, fusion with the lysosome, to allow maturation.88, 89 In contrast to mammalian Rab GTPase, it has not been demonstrated that the yeast Rab GTPase Ypt7 localizes at the autophagosome membrane. After fusion, lytic enzymes inside the lysosome lumen presumably degrade the autophagic body, as in the yeast vacuole.
Conclusion
Autophagy involves membrane rearrangement for the sequestration of cytoplasm and its delivery to the lysosome or vacuole. It is a process that is highly conserved in all eukaryotes and has important roles in cellular processes involved in normal growth, development and differentiation. Studies of autophagy, pexophagy and the Cvt pathway in yeasts have identified 27 ATG genes required for these two pathways and revealed the unique machineries that drive them. Most of the protein components identified in yeasts are also found in higher eukaryotes, indicating that there is a high degree of conservation from unicellular to multicellular organisms with regard to autophagy. Many questions, however, still remain to be answered, especially those related to the steps of vesicle formation, including how autophagosomes are generated, and the origin for the lipid membrane. The yeast model organism remains a powerful tool to examine these and other questions. Further studies of autophagy, pexophagy and the Cvt pathway will continue to provide answers and further our understanding of the molecular mechanism that directs these processes.
Abbreviations
Ape1:
aminopeptidase I
Atg:
autophagy-related
Cvt:
cytoplasm to vacuole targeting
PAS:
pre-autophagosomal structure
References
- Majeski AE and Dice JF (2004) Mechanisms of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 36: 2435–2444
Article CAS PubMed Google Scholar - Massey A, Kiffin R and Cuervo AM (2004) Pathophysiology of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 36: 2420–2434
Article CAS PubMed Google Scholar - Reggiori F and Klionsky DJ (2002) Autophagy in the eukaryotic cell. Eukaryot. Cell. 1: 11–21
Article CAS PubMed PubMed Central Google Scholar - Wang C-W and Klionsky DJ (2003) The molecular mechanism of autophagy. Mol. Med. 9: 65–76
Article PubMed PubMed Central Google Scholar - Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T and Ohsumi Y (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20: 5971–5981
Article CAS PubMed PubMed Central Google Scholar - Noda T, Suzuki K and Ohsumi Y (2002) Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 12: 231–235
Article CAS PubMed Google Scholar - Klionsky DJ and Emr SD (2000) Autophagy as a regulated pathway of cellular degradation. Science 290: 1717–1721
Article CAS PubMed PubMed Central Google Scholar - Levine B and Klionsky DJ (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6: 463–477
Article CAS PubMed Google Scholar - Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T and Mizushima N (2004) The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036
Article CAS PubMed Google Scholar - Mizushima N, Yamamoto A, Matsui M, Yoshimori T and Ohsumi Y (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15: 1101–1111
Article CAS PubMed PubMed Central Google Scholar - Cuervo AM (2004) Autophagy: in sickness and in health. Trends Cell Biol. 14: 70–77
Article CAS PubMed Google Scholar - Shintani T and Klionsky DJ (2004) Autophagy in health and disease: a double-edged sword. Science 306: 990–995
Article CAS PubMed PubMed Central Google Scholar - Kirkegaard K, Taylor MP and Jackson WT (2004) Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2: 301–314
Article CAS PubMed PubMed Central Google Scholar - Levine B (2005) Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 120: 159–162
CAS PubMed Google Scholar - Bursch W (2004) Multiple cell death programs: Charon's lifts to Hades. FEMS Yeast Res. 5: 101–110
Article CAS PubMed Google Scholar - Tsukada M and Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333: 169–174
Article CAS PubMed Google Scholar - Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M and Wolf DH (1994) Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349: 275–280
Article CAS PubMed Google Scholar - Klionsky DJ, Cregg JM, Dunn Jr WA, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M and Ohsumi Y (2003) A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5: 539–545
Article CAS PubMed Google Scholar - Harding TM, Morano KA, Scott SV and Klionsky DJ (1995) Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131: 591–602
Article CAS PubMed Google Scholar - Yuan W, Tuttle DL, Shi YJ, Ralph GS and Dunn Jr WA (1997) Glucose-induced microautophagy in Pichia pastoris requires the _α_-subunit of phosphofructokinase. J. Cell Sci. 110: 1935–1945
Article CAS PubMed Google Scholar - Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T and Ohsumi Y (2001) Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell 12: 3690–3702
Article CAS PubMed PubMed Central Google Scholar - Hamasaki M, Noda T and Ohsumi Y (2003) The early secretory pathway contributes to autophagy in yeast. Cell Struct. Funct. 28: 49–54
Article CAS PubMed Google Scholar - Reggiori F, Wang C-W, Nair U, Shintani T, Abeliovich H and Klionsky DJ (2004) Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 15: 2189–2204
Article CAS PubMed PubMed Central Google Scholar - Harding TM, Hefner-Gravink A, Thumm M and Klionsky DJ (1996) Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J. Biol. Chem. 271: 17621–17624
Article CAS PubMed Google Scholar - Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y and Klionsky DJ (1996) Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. USA 93: 12304–12308
Article CAS PubMed PubMed Central Google Scholar - Baba M, Osumi M, Scott SV, Klionsky DJ and Ohsumi Y (1997) Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 139: 1687–1695
Article CAS PubMed PubMed Central Google Scholar - Scott SV, Baba M, Ohsumi Y and Klionsky DJ (1997) Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J. Cell Biol. 138: 37–44
Article CAS PubMed PubMed Central Google Scholar - Hutchins MU and Klionsky DJ (2001) Vacuolar localization of oligomeric _α_-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J. Biol. Chem. 276: 20491–20498
Article CAS PubMed Google Scholar - Hutchins MU, Veenhuis M and Klionsky DJ (1999) Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 112: 4079–4087
Article CAS PubMed Google Scholar - Noda T and Ohsumi Y (1998) Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273: 3963–3936
Article CAS Google Scholar - Klionsky DJ (2005) The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118: 7–18
Article CAS PubMed Google Scholar - Codogno P and Meijer AC (2004) Signaling pathways in mammalian autophagy. In Autophagy, Klionsky DJ (ed) (Geogetown, TX: Landes Bioscience) pp. 26–47
Google Scholar - Funakoshi T, Matsuura A, Noda T and Ohsumi Y (1997) Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 192: 207–213
Article CAS PubMed Google Scholar - Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M and Ohsumi Y (2000) Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150: 1507–1513
Article CAS PubMed PubMed Central Google Scholar - Matsuura A, Tsukada M, Wada Y and Ohsumi Y (1997) Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192: 245–250
Article CAS PubMed Google Scholar - Abeliovich H, Zhang C, Dunn Jr WA, Shokat KM and Klionsky DJ (2003) Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell 14: 477–490
Article CAS PubMed PubMed Central Google Scholar - Scott SV, Nice III DC, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y and Klionsky DJ (2000) Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J. Biol. Chem. 275: 25840–25849
Article CAS PubMed Google Scholar - Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn Jr W and Klionsky DJ (2001) Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153: 381–396
Article CAS PubMed PubMed Central Google Scholar - Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M and Ohsumi Y (2005) Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell 16: 2544–2553
Article CAS PubMed PubMed Central Google Scholar - Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang C-W and Klionsky DJ (2005) Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell 16: 3438–3453
Article CAS PubMed PubMed Central Google Scholar - Tuttle DL, Lewin AS and Dunn Jr WA (1993) Selective autophagy of peroxisomes in methylotrophic yeasts. Eur. J. Cell Biol. 60: 283–290
CAS PubMed Google Scholar - Veenhuis M, Douma A, Harder W and Osumi M (1983) Degradation and turnover of peroxisomes in the yeast Hansenula polymorpha induced by selective inactivation of peroxisomal enzymes. Arch. Microbiol. 134: 193–203
Article CAS PubMed Google Scholar - Gunkel K, van der Klei IJ, Barth G and Veenhuis M (1999) Selective peroxisome degradation in Yarrowia lipolytica after a shift of cells from acetate/oleate/ethylamine into glucose/ammonium sulfate-containing media. FEBS Lett. 451: 1–4
Article CAS PubMed Google Scholar - Onodera J and Ohsumi Y (2004) Ald6p is a preferred target for autophagy in yeast, Saccharomyces cerevisiae. J. Biol. Chem. 279: 16071–16076
Article CAS PubMed Google Scholar - Klionsky DJ, Cueva R and Yaver DS (1992) Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 119: 287–299
Article CAS PubMed Google Scholar - Oda MN, Scott SV, Hefner-Gravink A, Caffarelli AD and Klionsky DJ (1996) Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J. Cell Biol. 132: 999–1010
Article CAS PubMed Google Scholar - Suzuki K, Kamada Y and Ohsumi Y (2002) Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae. Dev. Cell 3: 815–824
Article CAS PubMed Google Scholar - Shintani T, Huang W-P, Stromhaug PE and Klionsky DJ (2002) Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell 3: 825–837
Article CAS PubMed PubMed Central Google Scholar - Scott SV, Guan J, Hutchins MU, Kim J and Klionsky DJ (2001) Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell 7: 1131–1141
Article CAS PubMed PubMed Central Google Scholar - Yorimitsu T and Klionsky DJ (2005) Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell 16: 1593–1605
Article CAS PubMed PubMed Central Google Scholar - Shintani T and Klionsky DJ (2004) Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279: 29889–29894
Article CAS PubMed Google Scholar - Ohsumi Y (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell. Biol. 2: 211–216
Article CAS PubMed Google Scholar - Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M and Ohsumi Y (1998) A protein conjugation system essential for autophagy. Nature 395: 395–398
Article CAS PubMed Google Scholar - Kim J, Dalton VM, Eggerton KP, Scott SV and Klionsky DJ (1999) Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol. Biol. Cell 10: 1337–1351
Article CAS PubMed PubMed Central Google Scholar - Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T and Ohsumi Y (1999) Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 18: 5234–5241
Article CAS PubMed PubMed Central Google Scholar - Mizushima N, Noda T and Ohsumi Y (1999) Apg16p is required for the function of the Apg12p–Apg5p conjugate in the yeast autophagy pathway. EMBO J. 18: 3888–3896
Article CAS PubMed PubMed Central Google Scholar - Kuma A, Mizushima N, Ishihara N and Ohsumi Y (2002) Formation of the approximately 350-kDa Apg12–Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 277: 18619–18125
Article CAS Google Scholar - Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y and Yoshimori T (2003) Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12–Apg5 conjugate. J. Cell Sci. 116: 1679–1688
Article CAS PubMed Google Scholar - Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T and Ohsumi Y (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151: 263–276
Article CAS PubMed PubMed Central Google Scholar - Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T and Ohsumi Y (2000) A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492
Article CAS PubMed Google Scholar - Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf DH and Thumm M (1998) Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 17: 3597–3607
Article CAS PubMed PubMed Central Google Scholar - Ichimura Y, Imamura Y, Emoto K, Umeda M, Noda T and Ohsumi Y (2004) In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J. Biol. Chem. 279: 40584–40592
Article CAS PubMed Google Scholar - Kim J, Huang W-P, Stromhaug PE and Klionsky DJ (2002) Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 277: 763–773
Article CAS PubMed Google Scholar - Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y and Yoshimori T (2001) Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152: 657–668
Article CAS PubMed PubMed Central Google Scholar - Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T and Ohsumi Y (1999) Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147: 435–446
Article CAS PubMed PubMed Central Google Scholar - Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y and Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19: 5720–5728
Article CAS PubMed PubMed Central Google Scholar - Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y and Yoshimori T (2004) LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117: 2805–2812
Article CAS PubMed Google Scholar - Kihara A, Noda T, Ishihara N and Ohsumi Y (2001) Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152: 519–530
Article CAS PubMed PubMed Central Google Scholar - Nice DC, Sato TK, Stromhaug PE, Emr SD and Klionsky DJ (2002) Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 277: 30198–30207
Article CAS PubMed Google Scholar - Hettema EH, Lewis MJ, Black MW and Pelham HRB (2003) Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 22: 548–557
Article CAS PubMed PubMed Central Google Scholar - Guan J, Stromhaug PE, George MD, Habibzadegah-Tari P, Bevan A, Dunn Jr WA and Klionsky DJ (2001) Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol. Biol. Cell 12: 3821–3838
Article CAS PubMed PubMed Central Google Scholar - Wurmser AE and Emr SD (2002) Novel PtdIns(3)P-binding protein Etf1 functions as an effector of the Vps34 PtdIns 3-kinase in autophagy. J. Cell Biol. 158: 761–772
Article CAS PubMed PubMed Central Google Scholar - Stromhaug PE, Reggiori F, Guan J, Wang C-W and Klionsky DJ (2004) Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell 15: 3553–3566
Article CAS PubMed PubMed Central Google Scholar - Noda T, Kim J, Huang W-P, Baba M, Tokunaga C, Ohsumi Y and Klionsky DJ (2000) Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148: 465–480
Article CAS PubMed PubMed Central Google Scholar - Tucker KA, Reggiori F, Dunn Jr WA and Klionsky DJ (2003) Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy. J. Biol. Chem. 278: 48445–48452
Article CAS PubMed Google Scholar - Reggiori F, Tucker KA, Stromhaug PE and Klionsky DJ (2004) The Atg1–Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 6: 79–90
Article CAS PubMed Google Scholar - Wang C-W, Kim J, Huang W-P, Abeliovich H, Stromhaug PE, Dunn Jr WA and Klionsky DJ (2001) Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J. Biol. Chem. 276: 30442–30451
Article CAS PubMed Google Scholar - Shintani T, Suzuki K, Kamada Y, Noda T and Ohsumi Y (2001) Apg2p functions in autophagosome formation on the perivacuolar structure. J. Biol. Chem. 276: 30452–30460
Article CAS PubMed Google Scholar - Reggiori F, Shintani T, Nair U and Klionsky DJ (2005) Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy 1: 101–109
Article CAS PubMed Google Scholar - Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K and Scherer SW (2005) Endothelial nitric oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J. Biol. Chem. 280: 18283–18290
Article CAS PubMed Google Scholar - Wang C-W, Stromhaug PE, Shima J and Klionsky DJ (2002) The Ccz1–Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J. Biol. Chem. 277: 47917–47927
Article CAS PubMed Google Scholar - Wang C-W, Stromhaug PE, Kauffman EJ, Weisman LS and Klionsky DJ (2003) Yeast homotypic vacuole fusion requires the Ccz1–Mon1 complex during the tethering/docking stage. J. Cell Biol. 163: 973–985
Article CAS PubMed PubMed Central Google Scholar - Takeshige K, Baba M, Tsuboi S, Noda T and Ohsumi Y (1992) Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119: 301–311
Article CAS PubMed Google Scholar - Nakamura N, Matsuura A, Wada Y and Ohsumi Y (1997) Acidification of vacuoles is required for autophagic degradation in the yeast, Saccharomyces cerevisiae. J. Biochem. 121: 338–344
Article CAS PubMed Google Scholar - Teter SA, Eggerton KP, Scott SV, Kim J, Fischer AM and Klionsky DJ (2001) Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J. Biol. Chem. 276: 2083–2087
Article CAS PubMed Google Scholar - Epple UD, Suriapranata I, Eskelinen E-L and Thumm M (2001) Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J. Bacteriol. 183: 5942–5955
Article CAS PubMed PubMed Central Google Scholar - Epple UD, Eskelinen E-L and Thumm M (2003) Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J. Biol. Chem. 278: 7810–7821
Article CAS PubMed Google Scholar - Gutierrez MG, Munafo DB, Beron W and Colombo MI (2004) Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell. Sci. 117: 2687–2697
Article CAS PubMed Google Scholar - Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P and Eskelinen E-L (2004) Role for Rab7 in maturation of late autophagic vacuoles. J. Cell. Sci. 117: 4837–4848
Article PubMed CAS Google Scholar
Acknowledgements
We thank Drs. Julie E Legakis and Usha Nair for critically reading the manuscript. This work was supported by Public Health Service grant GM53396 from the National Institutes of Health (to DJK).
Author information
Authors and Affiliations
- Departments of Molecular, Cellular and Developmental Biology and Biological Chemistry, Life Sciences Institute, University of Michigan, Ann Arbor, 48109, MI, USA
T Yorimitsu & D J Klionsky
Authors
- T Yorimitsu
You can also search for this author inPubMed Google Scholar - D J Klionsky
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toD J Klionsky.
Additional information
Edited by G Melino
Rights and permissions
About this article
Cite this article
Yorimitsu, T., Klionsky, D. Autophagy: molecular machinery for self-eating.Cell Death Differ 12 (Suppl 2), 1542–1552 (2005). https://doi.org/10.1038/sj.cdd.4401765
- Received: 16 May 2005
- Revised: 26 July 2005
- Accepted: 02 August 2005
- Published: 25 October 2005
- Issue Date: 01 November 2005
- DOI: https://doi.org/10.1038/sj.cdd.4401765