Plaque-associated lipids in Alzheimer’s diseased brain tissue visualized by nonlinear microscopy (original) (raw)
- Article
- Open access
- Published: 27 August 2015
Scientific Reports volume 5, Article number: 13489 (2015)Cite this article
- 5018 Accesses
- 74 Citations
- 11 Altmetric
- Metrics details
Subjects
Abstract
By simultaneous coherent anti-Stokes Raman scattering (CARS) and 2-photon fluorescence microscopy of Thioflavin-S stained Alzheimer´s diseased human brain tissues, we show evidence of lipid deposits co-localizing with fibrillar β-amyloid (Aβ) plaques. Two lipid morphologies can be observed; lamellar structures and coalescing macro-aggregates of sub-micron sizes to ~25 μm. No significant lipid deposits were observed in non-fibrillar, diffuse plaques identified by Aβ immuno-staining. CARS microscopy of unlabeled samples confirms the lamellar and macro-aggregate lipid morphologies. The composition of the plaques was analyzed by CARS microspectroscopy and Raman microscopy; vibrational signatures of lipids with long acyl chains co-localize with the β-sheet vibrations. The lipid fluidity was evaluated from the CARS spectra, illustrating that the lipid composition/organization varies throughout the plaques. Altogether this indicates close amyloid-lipid interplay in fibrillar Aβ plaques, rendering them more dynamic compositions than previously believed and, hence, potential sources of toxic oligomers.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is estimated to be the fourth-leading cause of death in high income countries and the only lethal disease among the top ten that currently has no means for cure, prevention or even delay[1](/articles/srep13489#ref-CR1 "World health organization, The top 10 causes of death. Available at: http://www.who.int/mediacentre/factsheets/fs310/en/index1.html
(Date of access: 18/02/2015)"). Hence, the prevalence of AD continues to increase with 7.7 million new cases diagnosed world-wide every year and is expected to escalate further with the aging population[2](/articles/srep13489#ref-CR2 "World Health Organization, Executive summary in Dementia: a public health priority, 1-2 (WHO, 2012). Available at:
http://apps.who.int/iris/bitstream/10665/75263/1/9789241564458_eng.pdf?ua=1
(Date of access: 18/02/2015)"). Early pathological hallmarks are the formation of soluble β-amyloid (Aβ) oligomers and synaptic dysfunction, believed to cause mild cognitive impairment long before the accumulation of amyloid plaques and neurodegeneration[3](/articles/srep13489#ref-CR3 "Lue, L. F. et al. Soluble amyloid beta peptide concentration as a predictor of synaptic change in Alzheimer’s disease. Am. J. Pathol. 155, 853–862 (1999)."),[4](/articles/srep13489#ref-CR4 "Mucke, L. et al. High-level neuronal expression of A beta(1-42) in wild-type human amyloid protein precursor transgenic mice: Synaptotoxicity without plaque formation. J. Neurosci. 20, 4050–4058 (2000)."),[5](/articles/srep13489#ref-CR5 "Gong, Y. S. et al. Alzheimer’s disease-affected brain: Presence of oligomeric A beta ligands (ADDLs) suggests a molecular basis for reversible memory loss. Proc. Natl. Acad. Sci. U.S.A. 100, 10417–10422 (2003)."),[6](/articles/srep13489#ref-CR6 "Cleary, J. P. et al. Natural oligomers of the amyloid-protein specifically disrupt cognitive function. Nature Neurosci. 8, 79–84 (2005)."),[7](/articles/srep13489#ref-CR7 "Shankar, G. M. et al. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat. Med. 14, 837–842 (2008)."),[8](/articles/srep13489#ref-CR8 "Lesne, S. E. et al. Brain amyloid-beta oligomers in ageing and Alzheimer’s disease. Brain 136, 1383–1398 (2013)."). Aβ oligomers have been shown to bind to cellular membranes in human cortical neurons with high affinity and more specifically to synaptic contacts[9](/articles/srep13489#ref-CR9 "Deshpande, A., Mina, E., Glabe, C. & Busciglio, J. Different conformations of amyloid beta induce neurotoxicity by distinct mechanisms in human cortical neurons. J. Neurosci. 26, 6011–6018 (2006)."). Here they disrupt the membrane barrier function by _e.g._ altering the membrane structure, complete membrane rupture or forming pores, followed by Ca2+ influx and accumulation of reactive oxygen species (for a review, see Stefani[10](/articles/srep13489#ref-CR10 "Stefani, M. Structural features and cytotoxicity of amyloid oligomers: Implications in Alzheimer’s disease and other diseases with amyloid deposits. Prog. Neurobiol. 99, 226–245 (2012).")). Aβ oligomers have also been shown to specifically bind to post-synaptic receptors, in turn modulating down-stream signaling pathways causing alterations in the axonal transport of synaptic vesicles and mitochondria leading to synaptic impairment (for reviews, see Larson _et al._[11](/articles/srep13489#ref-CR11 "Larson, M. E. & Lesne, S. E. Soluble Aβ oligomer production and toxicity. J. Neurochem. 120, 125–139 (2012).") and Overk _et al._[12](/articles/srep13489#ref-CR12 "Overk, C. R. & Masliah, E. Pathogenesis of synaptic degeneration in Alzheimer’s disease and Lewy body disease. Biochem. Pharmacol. 88, 508–516 (2014).")). In the advanced stages of AD, Aβ oligomers accumulate to insoluble plaques, surrounded by a distinct halo of free Aβ oligomers[13](/articles/srep13489#ref-CR13 "Koffie, R. M., et al. Oligomeric amyloid beta associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc. Natl. Acad. Sci. U.S.A. 106, 4012–4017 (2009)."). Neuroinflammation and activation of the mitochondrial apoptotic pathway[14](/articles/srep13489#ref-CR14 "Canevari, L., Abramov, A. Y. & Duchen, M. R. Toxicity of amyloid beta peptide: Tales of calcium, mitochondria and oxidative stress. Neurochem. Res. 29, 637–650 (2004)."),[15](/articles/srep13489#ref-CR15 "Cha, M. Y. et al. Mitochondria-Specific Accumulation of Amyloid beta Induces Mitochondrial Dysfunction Leading to Apoptotic Cell Death. Plos One 7, e34929 (2012). 10.1371/journal.pone.0034929") result in manifest neurodegeneration, the degree of which correlates well with the gradient distribution of free oligomers in the halo and the surrounding neuropil[13](/articles/srep13489#ref-CR13 "Koffie, R. M., et al. Oligomeric amyloid beta associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc. Natl. Acad. Sci. U.S.A. 106, 4012–4017 (2009)."). It is proposed that the Aβ oligomers in the halo are released by the fibrils in the plaque[13](/articles/srep13489#ref-CR13 "Koffie, R. M., et al. Oligomeric amyloid beta associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc. Natl. Acad. Sci. U.S.A. 106, 4012–4017 (2009)."), which is supported by _in vitro_ studies showing that Aβ fibrils continuously dissolve and reform[16](/articles/srep13489#ref-CR16 "Sanchez, L. et al. A beta 40 and A beta 42 Amyloid Fibrils Exhibit Distinct Molecular Recycling Properties. J. Am. Chem. Soc. 133, 6505–6508 (2011)."), rather than being irreversibly trapped as inert fibrous species. Lipids interacting with amyloid fibrils have been shown to significantly promote the disintegration into toxic oligomers by acting as detergents[17](/articles/srep13489#ref-CR17 "Widenbrant, M. J. O., Rajadas, J., Sutardja, C. & Fuller, G. G. Lipid-induced beta-amyloid peptide assemblage fragmentation. Biophys. J. 91, 4071–4080 (2006)."),[18](/articles/srep13489#ref-CR18 "Martins, I. C. et al. Lipids revert inert A beta amyloid fibrils to neurotoxic protofibrils that affect learning in mice. Embo J. 27, 224–233 (2008)."). Interestingly, there is a line of evidence that fibrillar plaques contain lipids[19](/articles/srep13489#ref-CR19 "Rak, M., Del Bigio, M. R., Mai, S., Westaway, D. & Gough, K. Dense-core and diffuse Abeta plaques in TgCRND8 mice studied with synchrotron FTIR microspectroscopy. Biopolymers 87, 207–217 (2007)."),[20](/articles/srep13489#ref-CR20 "Kuzyk, A. et al. Association among Amyloid Plaque, Lipid and Creatine in Hippocampus of TgCRND8 Mouse Model for Alzheimer Disease. J. Biol. Chem. 285, 31202–31207 (2010)."),[21](/articles/srep13489#ref-CR21 "Panchal, M. et al. Enrichment of cholesterol in microdissected Alzheimer’s disease senile plaques as assessed by mass spectrometry. J. Lipid. Res. 51, 598–605 (2010)."),[22](/articles/srep13489#ref-CR22 "Kastyak-Ibrahim, M. Z. et al. Biochemical label-free tissue imaging with subcellular-resolution synchrotron FTIR with focal plane array detector. Neuroimage 60, 376–383 (2012)."),[23](/articles/srep13489#ref-CR23 "Liao, C. R. et al. Synchrotron FTIR reveals lipid around and within amyloid plaques in transgenic mice and Alzheimer’s disease brain. Analyst 138, 3991–3997 (2013)."),[24](/articles/srep13489#ref-CR24 "Sole-Domenech, S. et al. Localization of cholesterol, amyloid and glia in Alzheimer’s disease transgenic mouse brain tissue using time-of-flight secondary ion mass spectrometry (ToF-SIMS) and immunofluorescence imaging. Acta Neuropathol. 125, 145–157 (2013)."), hence, boosting the release of oligomers and rendering them a huge reservoir of toxic species. This lipid-promoted fibril fragmentation is further confirmed by molecular dynamics simulations[25](/articles/srep13489#ref-CR25 "Friedman, R., Pellarin, R. & Caflisch, A. Soluble Protofibrils as Metastable Intermediates in Simulations of Amyloid Fibril Degradation Induced by Lipid Vesicles. J. Phys. Chem. Lett. 1, 471–474 (2010)."). Altogether, these studies underline that Aβ oligomers are the active players in AD, however, also that their dynamics and distribution is strongly modulated by their interaction with lipids. While analytical methods and fluorescence microscopy[26](/articles/srep13489#ref-CR26 "Kaplan, B., Martin, B. M., Livneh, A., Pras, M. & Gallo, G. Biochemical micro-techniques in the diagnosis and classification of amyloidosis. Curr. Pharm. Anal. 2, 45–52 (2006)."),[27](/articles/srep13489#ref-CR27 "Nesterov, E. E. et al. In vivo optical Imaging of amyloid aggregates in brain: Design of fluorescent markers. Angew. Chem. Int. Ed. (English) 44, 5452–5456 (2005)."),[28](/articles/srep13489#ref-CR28 "Pedersen, J. T. & Heegaard, N. H. H. Analysis of Protein Aggregation in Neurodegenerative Disease. Anal. Chem. 85, 4215–4227 (2013).") allow us to obtain important quantitative and spatial information on Aβ species alone, a more holistic understanding of the underlying chain of mechanisms could be gained by taking into account their close interplay with lipids. Such information is currently almost non-existing, partly due to the lack of reliable methods to detect as fragile entities as lipids; their organization and distribution are highly influenced by changes in their physicochemical environment in conjunction with _e.g._ extraction procedures, sample preparation and labeling. Lipidomic analyses, involving chemical extraction of lipids from larger tissue samples and plasma, reveal elevated ceramide, cholesterol and triglyceride levels in conjunction with amyloid deposition[29](/articles/srep13489#ref-CR29 "Touboul, D. & Gaudin, M. Lipidomics of Alzheimer’s disease. Bioanalysis 6, 541–561 (2014)."), though without any information on the spatial distribution in the brain. Fluorescence microscopy suggests increased accumulation of cholesterol in senile plaques[30](/articles/srep13489#ref-CR30 "Mori T. et al. Cholesterol accumulates in senile plaques of Alzheimer disease patients and in transgenic APP(SW) mice. J. Neuropathol. Exp. 60, 778–785 (2001)."),[31](/articles/srep13489#ref-CR31 "Burns, M. P. et al. Co-localization of cholesterol, apolipoprotein E and fibrillar A beta in amyloid plaques. Mol. Brain Res. 110, 119–125 (2003)."), but these results have been shown to originate from false-positive staining[32](/articles/srep13489#ref-CR32 "Lebouvier, T., Perruchini, C., Panchal, M., Potier, M. C. & Duyckaerts, C. Cholesterol in the senile plaque: often mentioned, never seen. Acta Neuropathol. 117, 31–34 (2009)."). Time-of-flight secondary ion mass spectrometry (ToF-SIMS) imaging reports a slight increase in the overall cholesterol signal in the cortex of human AD brain tissue compared to controls[33](/articles/srep13489#ref-CR33 "Lazar, A. N. et al. Time-of-flight secondary ion mass spectrometry (TOF-SIMS) imaging reveals cholesterol overload in the cerebral cortex of Alzheimer disease patients. Acta Neuropathol. 125, 133–144 (2013)."). Minor depositions of cholesterol granules were observed immediately around the plaques in an AD mice model[24](/articles/srep13489#ref-CR24 "Sole-Domenech, S. et al. Localization of cholesterol, amyloid and glia in Alzheimer’s disease transgenic mouse brain tissue using time-of-flight secondary ion mass spectrometry (ToF-SIMS) and immunofluorescence imaging. Acta Neuropathol. 125, 145–157 (2013)."), however, the signals were comparatively weak and the granular distribution could in a follow-up study not be confirmed[34](/articles/srep13489#ref-CR34 "Carlred, L. et al. Simultaneous Imaging of Amyloid-beta and Lipids in Brain Tissue Using Antibody-Coupled Liposomes and Time-of-Flight Secondary Ion Mass Spectrometry. J. Am. Chem. Soc. 136, 9973–9981 (2014)."). Altogether, no significant lipid deposits are reported by ToF-SIMS, when studying a shallow (10 nm) two-dimensional plane of the surface (sensitive to influence of topographic variations) and to the low m/z region. No data in the high m/z region _e.g._ triglycerides seem to be available in the current literature to the best of our knowledge. Fourier transform infrared (FTIR) microscopy is capable of simultaneous imaging of lipids and amyloid proteins, revealing elevated lipid concentrations in regions identified as core plaques in transgenic mouse models and human AD brain tissues[19](/articles/srep13489#ref-CR19 "Rak, M., Del Bigio, M. R., Mai, S., Westaway, D. & Gough, K. Dense-core and diffuse Abeta plaques in TgCRND8 mice studied with synchrotron FTIR microspectroscopy. Biopolymers 87, 207–217 (2007)."),[22](/articles/srep13489#ref-CR22 "Kastyak-Ibrahim, M. Z. et al. Biochemical label-free tissue imaging with subcellular-resolution synchrotron FTIR with focal plane array detector. Neuroimage 60, 376–383 (2012)."),[23](/articles/srep13489#ref-CR23 "Liao, C. R. et al. Synchrotron FTIR reveals lipid around and within amyloid plaques in transgenic mice and Alzheimer’s disease brain. Analyst 138, 3991–3997 (2013)."). Due to the low number of pixels covering the plaque region, each pixel representing \~5 μm, it is difficult to conclude any detailed information on the lipid distribution and to what extent lipids co-localize with amyloid fibers. Altogether, there are convincing data that there are accumulations of lipid in the vicinity of fibrillar Aβ plaques, but no detailed information on the morphology of the lipid deposits, on their fluidity (_i.e._ stability) or degree of co-localization with the Aβ fibrils – all important information in order to better understand the underlying accumulation process(es) and their ability to release toxic oligomers.In this study we use the strengths of multimodal non-linear optical (NLO) microscopy, combining coherent anti-Stokes Raman scattering (CARS) and multiphoton excited fluorescence (MPEF), for simultaneous investigations of spatial distributions of lipids and fluorescently stained Aβ fibrils in human AD brain tissue sections in order to complement with more detailed information. CARS microscopy has been previously shown to be a valuable label-free tool to visualize lipids in other tissues and cells by exciting and probing CH stretching vibrations of fatty acids via a nonlinear optical wave-mixing process35,36,37,38,39. In the current study, images of the distributions of lipids in AD brain tissue are complemented by MPEF images of the Aβ fibrils labeled with Thioflavin-S40. Three-dimensional renderings of the lipid/Aβ fibril distributions are presented at high spatial resolution (~0.3 μm in x/y and ~1 μm in z), allowing us to resolve regions with distinct different contents of lipids and Aβ fibrils within and around core plaques and assess to what extent they co-localize. We complement the data by label-free imaging using CH-CARS only (no Thioflavin-S staining), as well as Raman microscopy in the CH vibrational region (lipids) and the amide vibrational region (Aβ species), in order to avoid possible labelling effects on the distribution/composition of the fragile lipids/amyloid co-arrangements. In contrast to previous FTIR and ToF-SIMS imaging studies, we present simultaneously collected images of lipids and Aβ species in soft, wet tissue samples, allowing us to directly relate the distributions without e.g. freeze-drying. Furthermore, by pixel-wise forming a ratio of the CARS signal at the symmetric and asymmetric CH vibrations, maps of the lipid fluidity (degree of unsaturation) throughout the plaque formations and surrounding regions are formed. This is of high interest, as the lipid environment controls the degree of fibril fragmentation; polyunsaturated lipids with a capability to form micelles are particularly efficient in preventing the aggregation of fibrils, leaving the Aβ species to a high degree as toxic protofibrils41. This kind of visual information can be used to assess to what extent the plaques are capable of releasing toxic oligomers, hence, their neurotoxicity.
Results
Tissue sections from the prefrontal cortex of AD diseased human brains were investigated. Fibrillar Aβ plaques were identified by MPEF from Thioflavin-S stained tissue sections (Fig. 1a–c). Simultaneously, the CARS signal was detected by tuning to the symmetric CH vibrational mode at 2840 cm−1, characteristic of long-chained triglycerides42. The CARS images revealed lipid accumulations primarily in the areas of fibrillar Aβ plaques (Fig. 1d–f). In general, core Aβ plaques contained higher Thioflavin-S and lipid concentrations on a macro scale (~100 μm) compared to the surrounding tissue (dashed outlines in Fig. 1a–f). However, within the plaque region, Thioflavin-S and lipid concentrations showed little correlation. Plaque areas with comparatively high fluorescence signals could be associated with both high and low lipid signals (Fig. 1g–i). Variations in the lipid distribution at different depths in the tissue could also be observed at the spatial resolution of 1 μm. A 3D rendering and a z-stack representation of the lipid distribution in a plaque is exemplified in Fig. 1j,k. Excitation beams could readily penetrate through the whole thickness of a tissue section (~40 μm).
Figure 1
Visualization of Aβ and lipid distributions in core plaques by fluorescence/CARS microscopy.
(a–c) Thioflavin-S fluorescence; (d–f) CARS images at 2840 cm−1; (g–i) profile plots along the lines indicated in (a,d), (b,e) and (c,f) respectively; (j) a 3D rendering of a CARS microscopy image (yellow) of lipids distributed in the AD plaque region, visualizing the different morphologies: crystalline needle-like structures (left) and more diffusely distributed (right); (k) z-stack representation of the CARS signal from the plaque in (d). Scale bars, 25 μm.
In contrast to fibrillar Aβ plaques (Thioflavin-S positive), diffuse plaques (non-fibrillar; Thioflavin-S negative, here identified by Aβ immuno-staining) did not exhibit any significant lipid deposits (Supplementary Fig. S1 online). Further systematic studies are needed to determine whether fibrillar Aβ plaque content is prerequisite for lipid accumulation, or whether there are also classes of Aβ oligomers that co-localize with lipids.
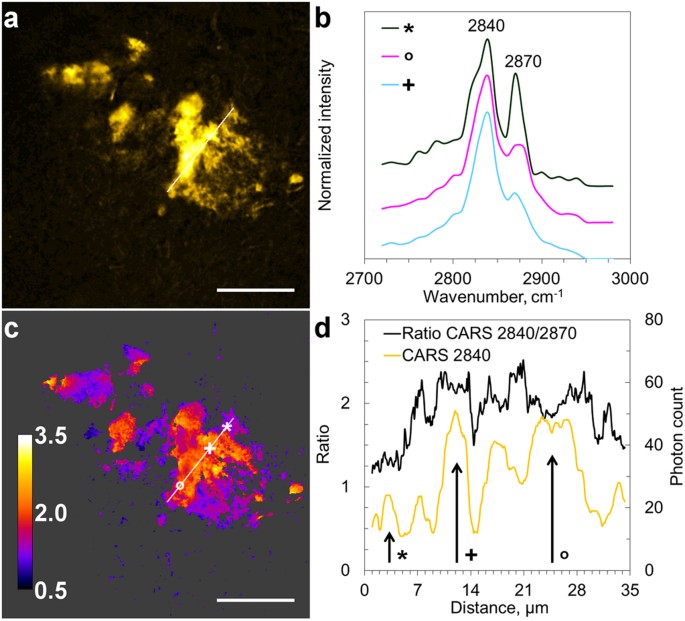
In order to avoid possible interference from the labeling on the lipid distribution, we further studied unstained AD diseased human brain tissue sections. These sections were adjacent to samples with core plaques identified by Thioflavin-S staining. Detailed information on the lipid composition in the unstained samples was acquired by scanning CARS spectra in the CH-stretching vibration region (2720–2980 cm−1). Spectra of lipid aggregates in AD brain tissue, shown in Fig. 2a, were dominated by two peaks at 2840 cm−1 and 2870 cm−1 (Fig. 2b), corresponding to the symmetric and asymmetric CH2 vibrations, respectively. The spectral shape is similar to that obtained for lipids with long acyl-chains, however, spatial variations were observed (Fig. 2b) and minor contributions from e.g. cholesterol43 cannot be excluded.
Figure 2
Visualization of the heterogeneity in lipid aggregates composition in AD brain tissue.
(a) CARS (2840 cm−1) image of lipid aggregate; (b) CARS spectra collected at positions marked by symbols in c, curves are displayed with an offset for clarity; (c) CARS ratio image, intensities at 2840 cm−1 vs. 2870 cm−1, representing variations in lipid fluidity; (d) profile plots along the lines indicated in (a,c). Scale bars, 25 μm.
Spectra collected at different spatial positions revealed notable differences in chemical composition, observed as different contributions from the two CH2 vibrations (Fig. 2b). In order to investigate the heterogeneity of the lipid composition within the lipid aggregates, ratios of CARS signals at 2840 cm−1 and 2870 cm−1 were calculated (Fig. 2c), providing an estimate of lipid fluidity44,45. Higher 2840 cm−1/2870 cm−1 ratios indicate lower packing of acyl chains, higher fluidity and/or more unsaturated lipids promoting Aβ fibril fragmentation41. Variations in the 2840 cm−1/2870 cm−1 ratio within the lipid aggregates suggest that they are complex and composed of areas with different lipid compositions with different propensity to release toxic Aβ oligomers.
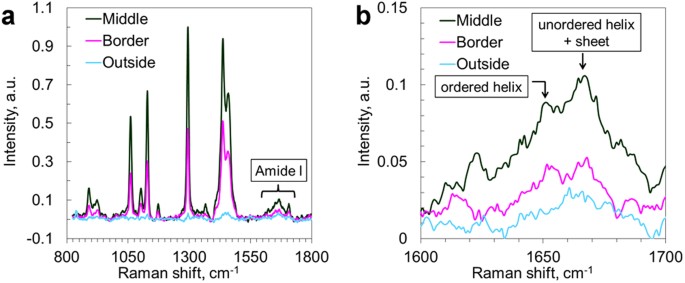
In order to investigate the contents of unlabeled samples, avoiding the potential impact of the labeling agent, we further complemented our data with Raman micro-spectroscopy (Fig. 3) in the fingerprint region, focusing in particular on the range 1620–1670 cm−1 - the amide I vibrations46. Spectra from the central part of the lipid aggregates (confirmed by strong CH vibrations) in AD brain tissue exhibited a distinct peak at 1667 cm−1 (β-sheet and unordered α-helix) and a small shoulder at 1655 cm−1 (ordered α-helix)47, in contrast to a featureless amide I band in the spectra from the surrounding tissue. In the plaque border region, the β-sheet peak could still be distinguished, but it was significantly weaker relative to the α-helix peak than in the core region. This indicates a higher amount of β-sheet formations in the lipid-rich areas compared to the surrounding brain tissues and confirms that the lipid aggregates indeed co-localize with Aβ species in AD brain tissues48. Furthermore, the overall Raman scattering intensities in the amide region collected in the center of the aggregate were significantly higher than at the border of and outside the aggregate, indicating that higher amounts of proteins could in general be found in the lipid deposit region. In order to identify which classes of lipids were present in the lipid aggregates, the fingerprint Raman micro-spectroscopy data were compared with spectra collected on different brain lipids as presented by Krafft et al.49. The shape of the spectra collected on the plaque-associated lipids (Fig. 3) was found to agree well with that of long-acyl chain fatty acids, hence, likely to be the main lipid component of the aggregates. The ester bond vibrations at 1729 and 1744 cm−1 are lacking as well as the phosphate group vibration at 860 cm−1, which suggests that triglycerides and phospholipids are minor components. Furthermore, the numerous sharp bands between 400 and 1200 cm−1, characteristic for cholesterol, could not be distinguished, which agrees with the observations made in the high-frequency CH vibrational region, i.e. minor contents of cholesterol.
Figure 3
Raman spectra from lipid aggregates in human AD brain tissue.
(a) Raman spectra in the finger print region highlighting the Amide I band; (b) Zoom-in on the Amide I band, from (a). Spectra were collected in the center and on the border of lipid aggregate, as well as from the tissue outside the lipid aggregate.
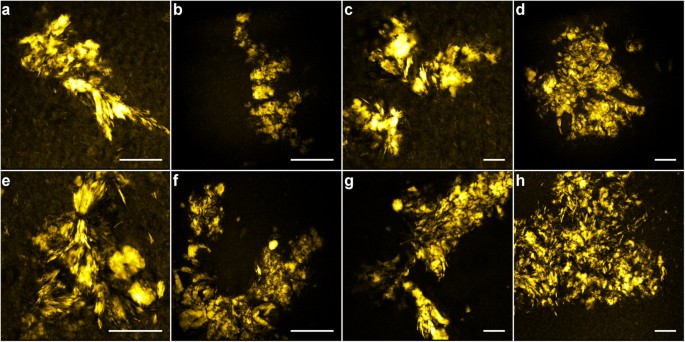
Characteristic morphologies of lipid aggregates are observed and can be recognized in CARS images of tissue samples at different locations and from different patients; Fig. 4(a–d) patient №1, Fig. 4(e–h) patient №2. Some lipid aggregates seem to form lamellar structures, suggesting that lipids organize along and possibly separate Aβ species (Fig. 4a,e,g); some are composed of larger, coalescent structures (Fig. 4b,c,e), while others are formed from multiple, smaller domains (Fig. 4d,f–h). Sizes range from sub-micron to ~25 μm.
Figure 4
Differences in lipid aggregate morphology found in human AD brain tissues investigated by CARS microscopy (2840 cm−1) including two main categories; lamellar structures (e.g. in a, e and g) and coalescent structures of different sizes, possibly originating from lipid micelles initially decorating the Aβ fibrils or ApoE particles, alternatively from lipid microvesicles shedded by microglia (see discussion).
(a–d) Patient №1; (e–h) patient №2. Scale bars, 25 μm. Note the different scales in the images.
Discussion
Biochemical analyses of lipid extracts from AD brain tissues and plasma29, as well as FTIR microscopy of brain tissue sections with fibrillar Aβ plaques19,20,22,23, report solid evidence for altered lipid composition and manifest lipid accumulations, respectively. However, more detailed information on the morphologies of the lipid deposits and their possible co-arrangements with Aβ species are needed to be able to find plausible underlying mechanisms. Information on the degree and character of Aβ/lipid co-arrangements in AD brain tissues are also important to sort out the conflicting reports on the toxicity of fibrillar plaques, taking into consideration the observations that lipid-decorated fibrils are readily destabilized and fragmented into toxic Aβ oligomers in contrast to pure, non-lipidated Aβ fibrils which are largely inert18. With the increased spatial resolution and three-dimensional imaging capability of NLO microscopy, our data reveal a morphologically and chemically more complex structure of Aβ fibrillar plaques than previously been reported. In the CARS microscopy images we note that lipids arrange as aggregates of primarily two different characters; (A) lamellar structures, as if templated by fibrillar Aβ species and (B) coalescent macro-aggregates of various sizes and shapes (see examples in Fig. 1 and Fig. 4). These two arrangements could be found in labelled (Fig. 1) as well as in unlabeled samples (Fig. 2 and Fig. 4), confirming that they were not caused or affected by the labelling. Several different mechanisms underlying the formation of lipid lamella and coalescent regions are plausible. Electron microscopy has shown that Aβ fibrils actively extract lipids from vesicle membranes through a detergent-like mechanism50. When multiple protein and membrane segments interact under amyloid promoting conditions, it has been shown that they form multi-lamellar suprastructures51. The protein assemblies line up with lipids in-between and the positively charged residues connect to the negatively charged lipids. These mechanisms could explain the formation of the lipid lamella. Hellstrand et al. also report that α-synuclein form fairly aligned bundles of fibrils at no or low concentrations of lipids, however, a random network is rather established at higher lipid concentrations with excess lipids being trapped as deformed vesicles in between the fibrils52. The latter organization with lipid vesicles could potentially result in the coalescence to lipid macro-aggregates of different sizes, as we observe in the CARS microscopy images. Hence, the formation of lipid aggregates of two different characters might be due to local differences in lipid concentrations; indeed, the lipid droplet regions typically generate a stronger CARS signal than the lamellar structures. Several other mechanisms could explain the formation of coalescing lipid deposits. Microvesicles budding off from microglia have been shown to adhere to fibrillar plaques and both feed them with and extract toxic Aβ oligomers53. Also the lipid transporter protein Apolipoprotein E (ApoE) and its lipid cargo have been shown to co-localize with fibrillar Aβ plaques31. Both microglial lipid vesicles and ApoE show no affinity to Aβ oligomers53,54, which could explain why no lipid deposits are observed in diffuse plaques consisting of Aβ oligomers.
The proposed mechanisms suggest that the lipids in the aggregates observed in the CARS microscopy images originate from cell/organelle membranes, alternatively lipids carried by the ApoE complex (primarily cholesterol but also phospholipids55). Analytical data indeed report elevated levels of cholesterol in senile plaques29. FTIR spectroscopy instead suggests that the many CH and CO-related absorption peaks can be attributed to phospholipids, however, due to overlapping features no conclusive interpretations of the contents could be made more than the presence of lipids with long acyl chains19. Our Raman and CARS spectra confirm the FTIR data that the primary content is lipids with long acyl chains, which could mean e.g. fatty acids or ceramides. We hardly see any signs of the phosphate-group vibration at 860 cm−1, indicating no/small amounts of phospholipids. Furthermore, no signs of the ester-vibrations at 1729 and 1744 cm−1, characteristic of triglycerides, can be noted. However, this may be due to the low number of phosphate- and ester groups relative to the number of CH bonds per lipid molecule. Furthermore, we cannot exclude the presence of cholesterol, as its spectral features in the high-wavenumber CH region partly overlap with that of other lipids. For more specific characterization of the lipid content in the plaque-associated lipid aggregates, it would be interesting to complement with ToF-SIMS measurements in the high m/zregion.
The CARS spectra and CARS CH ratio image in Fig. 2 reveal that the composition of the lipids varies throughout a plaque. Data indicate that the region characterized by larger coalescent lipid aggregates consist of lipids with higher fluidity, whereas the lamellar regions pinched by the protein-lipid interaction sites consist of more ordered lipids. More thorough investigations are needed in order to interpret these variations. Furthermore, it would be interesting to compare the degree of lipid fluidity with the distribution of different Aβ species by immunostaining and fluorescence microscopy, in order to confirm previous observations that lipids with higher fluidity and a capability of forming micelles have a higher tendency to destabilize the Aβ fibrils to toxic Aβ oligomers41. With this knowledge the CARS CH ratio images could be used as indirect toxicity maps, requiring no immunostaining.
In conclusion, we have shown that a sub-population of fibrillar plaques co-localizes with lipid deposits, which consist of lipids with long acyl chains organized as either multi-lamellar structures or as collections of coalescent lipid aggregates of sizes from sub-micron to ~25 μm. No lipid deposits have been observed in diffuse plaques, lacking fibrillar Aβ. We show that the lipid composition in the lipid deposits, here in terms of the lipid fluidity, varies throughout the plaque region; important information as it determines to what extent toxic Aβ oligomers could be released from the fibrillar plaques. This study demonstrates the strength of NLO microscopy for simultaneous characterization of three-dimensional distributions of lipids and Aβ species under biologically relevant conditions in human AD brain tissues, illustrating that the chemical build-up of AD plaques is more complex than previously reported. It further allows us to study the intricate interplay between lipids and amyloids, necessary to deepen our understanding of the toxic mechanisms of Aβ plaques.
Materials and Methods
AD brain tissue samples
Human brain samples from two different individuals were obtained with informed consent from the families and with the ethical approval of institutional Ethics Committees from New York Brain Bank at Columbia University (Alzheimer Disease Research Centre, Taub Institute), New York, USA and Lund University (Elisabet Englund, Dnr 286–2014), Lund, Sweden. Experiments were performed in accordance with the guidelines approved by Department of Experimental Medical Science at Lund University, Lund, Sweden and Department of Biology and Biological Engineering at Chalmers University of Technology, Göteborg, Sweden. All samples were taken from the prefrontal cortex, more specifically from gyrus rectus. The brain samples were fixed with formaldehyde solution, which has been shown not to interfere with the CARS signal56, sectioned into 40 μm thick slices and kept in phosphate buffered saline (PBS). Control samples were labeled with Thioflavin-S or with Cy2 conjugated antibodies against β-amyloid. For CARS microscopy measurements, human brain slices were mounted in a compartment, containing PBS, formed by a spacer between two coverslips secured with double adhesive tape.
Laser setup
The laser source consists of a Nd:YVO4 pump laser (picoTRAIN, HighQ GmbH) and an optical parametric oscillator (OPO), (Levante Emerald OPO, APE GmbH). The laser has two output beams; (i) the fundamental (1064 nm, used as Stokes beam) and (ii) the frequency doubled (532 nm), characterized by a pulse length of 7 ps and a repetition rate of 76 MHz. The 532 nm beam pumps the OPO, providing a variable output wavelength in the range of 690–990 nm (used as pump/probe beam). The OPO output was spatially overlapped with the 1064 nm beam using a dichroic mirror (DMSP1000, Thorlabs). A delay line in the 1064 nm beam path was used for temporal overlap of the excitation beams.
Microscope setup
Spatially and temporally combined excitation beams were coupled into the mirror scanning unit (Nikon C1) of the confocal microscope (Nikon Eclipse TE2000-E) and focused onto the sample by a high-NA objective (Nikon Plan Fluor 40x/1.30 oil). Forward propagating CARS emission was collected with a high-NA lens, passed the filter block and was focused on a single–photon-counting photomultiplier tube (PMC-100-20, Becker & Hickl GmbH). Epi-propagating fluorescence was collected by the excitation objective and directed by dichroic mirror to the back-port of the microscope. After passing the filter block the fluorescence emission was focused on a single-photon-counting photomultiplier tube (PMC-100-1, Becker & Hickl GmbH).
CARS and fluorescence microscopy
In the current study the Stokes beam was fixed at 1064 nm, while the pump/probe beam was tuned in the 807–825 nm range. When the energy difference between the Stokes and pump/probe beams matched the energy of a C-H vibrational mode, an enhanced coherent emission from the acyl chain in lipids was generated at the anti-Stokes wavelength of the pump/probe beam. The CH2 stretch vibration at 2840 cm−1 was probed by tuning the OPO to 817 nm. The forward propagating CARS signal was separated from the excitation light and the fluorescence by four band-pass filters (650/50 nm, Chroma Technologies). Simultaneously, the multi-photon excitation fluorescence (MPEF) from labeled samples was detected in the epi-direction, spectrally selected by optical filters: for Thioflavin S we used a combination of short-pass filters (600SP and 2 × 750SP, Ealing) and for Cy2 we used a band-pass filter (525/50 nm, Chroma). Z-stacks were acquired with a distance of 1 μm, forming 3D images of the AD brain tissues. Single images were obtained with acquisition times of 20 s. Images were formed in ImageJ57 by color-coding the number of detected photons in each pixel.
CARS spectroscopy
For spectral analysis of the CH-stretching vibrational region, the OPO was tuned from 825.2 nm to 807.9 nm, corresponding to the vibrational range from 2720 cm−1 to 2980 cm−1, in steps of 5 cm−1.
Raman microscopy
For Raman measurements, unstained human AD brain samples were mounted on a cover slip and dried in air. Raman micro-spectroscopy measurements were conducted using a confocal Raman microscope (inVia Raman, Renishaw) with a continuous wave laser source (532 nm). Single Raman spectra were acquired in extended spectral scanning mode (500–3000 cm−1) with a 2400 mm−1 grating, using a 100x objective, 25 mW excitation power at the sample position and a 10 s integration time.
Analysis of Raman spectra
Spectral data from Raman microscopy experiments was analyzed using in-house written Matlab routines. All spectra were interpolated to a uniform frequency separation of 1 cm−1, baseline corrected using the airPLS algorithm58 and smoothed with 7 points moving average filter.
Additional Information
How to cite this article: Kiskis, J. et al. Plaque-associated lipids in Alzheimer's diseased brain tissue visualized by nonlinear microscopy. Sci. Rep. 5, 13489; doi: 10.1038/srep13489 (2015).
References
- World health organization, The top 10 causes of death. Available at: http://www.who.int/mediacentre/factsheets/fs310/en/index1.html (Date of access: 18/02/2015)
- World Health Organization, Executive summary in Dementia: a public health priority, 1-2 (WHO, 2012). Available at: http://apps.who.int/iris/bitstream/10665/75263/1/9789241564458_eng.pdf?ua=1 (Date of access: 18/02/2015)
- Lue, L. F. et al. Soluble amyloid beta peptide concentration as a predictor of synaptic change in Alzheimer’s disease. Am. J. Pathol. 155, 853–862 (1999).
Article CAS ADS Google Scholar - Mucke, L. et al. High-level neuronal expression of A beta(1-42) in wild-type human amyloid protein precursor transgenic mice: Synaptotoxicity without plaque formation. J. Neurosci. 20, 4050–4058 (2000).
Article CAS Google Scholar - Gong, Y. S. et al. Alzheimer’s disease-affected brain: Presence of oligomeric A beta ligands (ADDLs) suggests a molecular basis for reversible memory loss. Proc. Natl. Acad. Sci. U.S.A. 100, 10417–10422 (2003).
Article CAS ADS Google Scholar - Cleary, J. P. et al. Natural oligomers of the amyloid-protein specifically disrupt cognitive function. Nature Neurosci. 8, 79–84 (2005).
Article CAS Google Scholar - Shankar, G. M. et al. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat. Med. 14, 837–842 (2008).
Article CAS Google Scholar - Lesne, S. E. et al. Brain amyloid-beta oligomers in ageing and Alzheimer’s disease. Brain 136, 1383–1398 (2013).
Article Google Scholar - Deshpande, A., Mina, E., Glabe, C. & Busciglio, J. Different conformations of amyloid beta induce neurotoxicity by distinct mechanisms in human cortical neurons. J. Neurosci. 26, 6011–6018 (2006).
Article CAS Google Scholar - Stefani, M. Structural features and cytotoxicity of amyloid oligomers: Implications in Alzheimer’s disease and other diseases with amyloid deposits. Prog. Neurobiol. 99, 226–245 (2012).
Article CAS Google Scholar - Larson, M. E. & Lesne, S. E. Soluble Aβ oligomer production and toxicity. J. Neurochem. 120, 125–139 (2012).
Article CAS Google Scholar - Overk, C. R. & Masliah, E. Pathogenesis of synaptic degeneration in Alzheimer’s disease and Lewy body disease. Biochem. Pharmacol. 88, 508–516 (2014).
Article CAS Google Scholar - Koffie, R. M., et al. Oligomeric amyloid beta associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc. Natl. Acad. Sci. U.S.A. 106, 4012–4017 (2009).
Article CAS ADS Google Scholar - Canevari, L., Abramov, A. Y. & Duchen, M. R. Toxicity of amyloid beta peptide: Tales of calcium, mitochondria and oxidative stress. Neurochem. Res. 29, 637–650 (2004).
Article CAS Google Scholar - Cha, M. Y. et al. Mitochondria-Specific Accumulation of Amyloid beta Induces Mitochondrial Dysfunction Leading to Apoptotic Cell Death. Plos One 7, e34929 (2012). 10.1371/journal.pone.0034929
Article CAS ADS PubMed PubMed Central Google Scholar - Sanchez, L. et al. A beta 40 and A beta 42 Amyloid Fibrils Exhibit Distinct Molecular Recycling Properties. J. Am. Chem. Soc. 133, 6505–6508 (2011).
Article CAS Google Scholar - Widenbrant, M. J. O., Rajadas, J., Sutardja, C. & Fuller, G. G. Lipid-induced beta-amyloid peptide assemblage fragmentation. Biophys. J. 91, 4071–4080 (2006).
Article CAS Google Scholar - Martins, I. C. et al. Lipids revert inert A beta amyloid fibrils to neurotoxic protofibrils that affect learning in mice. Embo J. 27, 224–233 (2008).
Article CAS Google Scholar - Rak, M., Del Bigio, M. R., Mai, S., Westaway, D. & Gough, K. Dense-core and diffuse Abeta plaques in TgCRND8 mice studied with synchrotron FTIR microspectroscopy. Biopolymers 87, 207–217 (2007).
Article CAS Google Scholar - Kuzyk, A. et al. Association among Amyloid Plaque, Lipid and Creatine in Hippocampus of TgCRND8 Mouse Model for Alzheimer Disease. J. Biol. Chem. 285, 31202–31207 (2010).
Article CAS Google Scholar - Panchal, M. et al. Enrichment of cholesterol in microdissected Alzheimer’s disease senile plaques as assessed by mass spectrometry. J. Lipid. Res. 51, 598–605 (2010).
Article CAS Google Scholar - Kastyak-Ibrahim, M. Z. et al. Biochemical label-free tissue imaging with subcellular-resolution synchrotron FTIR with focal plane array detector. Neuroimage 60, 376–383 (2012).
Article CAS Google Scholar - Liao, C. R. et al. Synchrotron FTIR reveals lipid around and within amyloid plaques in transgenic mice and Alzheimer’s disease brain. Analyst 138, 3991–3997 (2013).
Article CAS ADS Google Scholar - Sole-Domenech, S. et al. Localization of cholesterol, amyloid and glia in Alzheimer’s disease transgenic mouse brain tissue using time-of-flight secondary ion mass spectrometry (ToF-SIMS) and immunofluorescence imaging. Acta Neuropathol. 125, 145–157 (2013).
Article CAS Google Scholar - Friedman, R., Pellarin, R. & Caflisch, A. Soluble Protofibrils as Metastable Intermediates in Simulations of Amyloid Fibril Degradation Induced by Lipid Vesicles. J. Phys. Chem. Lett. 1, 471–474 (2010).
Article CAS Google Scholar - Kaplan, B., Martin, B. M., Livneh, A., Pras, M. & Gallo, G. Biochemical micro-techniques in the diagnosis and classification of amyloidosis. Curr. Pharm. Anal. 2, 45–52 (2006).
Article CAS Google Scholar - Nesterov, E. E. et al. In vivo optical Imaging of amyloid aggregates in brain: Design of fluorescent markers. Angew. Chem. Int. Ed. (English) 44, 5452–5456 (2005).
Article CAS Google Scholar - Pedersen, J. T. & Heegaard, N. H. H. Analysis of Protein Aggregation in Neurodegenerative Disease. Anal. Chem. 85, 4215–4227 (2013).
Article CAS Google Scholar - Touboul, D. & Gaudin, M. Lipidomics of Alzheimer’s disease. Bioanalysis 6, 541–561 (2014).
Article CAS Google Scholar - Mori T. et al. Cholesterol accumulates in senile plaques of Alzheimer disease patients and in transgenic APP(SW) mice. J. Neuropathol. Exp. 60, 778–785 (2001).
Article CAS Google Scholar - Burns, M. P. et al. Co-localization of cholesterol, apolipoprotein E and fibrillar A beta in amyloid plaques. Mol. Brain Res. 110, 119–125 (2003).
Article CAS Google Scholar - Lebouvier, T., Perruchini, C., Panchal, M., Potier, M. C. & Duyckaerts, C. Cholesterol in the senile plaque: often mentioned, never seen. Acta Neuropathol. 117, 31–34 (2009).
Article CAS Google Scholar - Lazar, A. N. et al. Time-of-flight secondary ion mass spectrometry (TOF-SIMS) imaging reveals cholesterol overload in the cerebral cortex of Alzheimer disease patients. Acta Neuropathol. 125, 133–144 (2013).
Article CAS Google Scholar - Carlred, L. et al. Simultaneous Imaging of Amyloid-beta and Lipids in Brain Tissue Using Antibody-Coupled Liposomes and Time-of-Flight Secondary Ion Mass Spectrometry. J. Am. Chem. Soc. 136, 9973–9981 (2014).
Article CAS Google Scholar - Le, T. T., Yue, S. & Cheng, J. X. Shedding new light on lipid biology with coherent anti-Stokes Raman scattering microscopy. J. Lipid Res. 51, 3091–3102 (2010).
Article CAS Google Scholar - Evans, C. L., Potma, E. O., Puoris’haag, M., Cote, D., Lin, C. P. & Xie, X. S. Chemical imaging of tissue in vivo with video-rate coherent anti-Stokes Raman scattering microscopy. Proc. Natl. Acad. Sci. U.S.A. 102, 16807–16812 (2005).
Article CAS ADS Google Scholar - Evans, C. L., Xu, X., Kesari, S., Xie, X. S., Wong, S. T. C. & Young, G. S. Chemically-selective imaging of brain structures with CARS microscopy. Opt Express 15, 12076–12087 (2007).
Article CAS ADS Google Scholar - Hellerer, T., Axang, C., Brackmann, C., Hillertz, P., Pilon, M. & Enejder, A. Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-Stokes Raman scattering (CARS) microscopy. Proc. Natl. Acad. Sci. U.S.A. 104, 14658–14663 (2007).
Article CAS ADS Google Scholar - Wang, H. F., Fu, Y., Zickmund, P., Shi, R. Y. & Cheng, J. X. Coherent anti-stokes Raman scattering imaging of axonal myelin in live spinal tissues. Biophys. J. 89, 581–591 (2005).
Article CAS Google Scholar - Kelenyi, G. Thioflavin S fluorescent and Congo Red anisotropic stainings in histologic demonstration of amyloid. Acta Neuro-pathol. 7, 336–348 (1967).
CAS Google Scholar - Johansson, A. S. et al. Docosahexaenoic acid stabilizes soluble amyloid-beta protofibrils and sustains amyloid-beta-induced neurotoxicity in vitro. Febs J. 274, 990–1000 (2007).
Article CAS Google Scholar - Nan, X., Cheng, J. X. & Xie, X. S. Vibrational imaging of lipid droplets in live fibroblast cells with coherent anti-Stokes Raman scattering microscopy. J. Lipid. Res. 44, 2202–2208 (2003).
Article CAS Google Scholar - Lim, R. S. et al. Identification of cholesterol crystals in plaques of atherosclerotic mice using hyperspectral CARS imaging. J. Lipid. Res. 52, 2177–2186 (2011).
Article CAS Google Scholar - Larsson, K. & Rand, R. P. Detection of changes in the environment of hydrocarbon chains by Raman spectroscopy and its application to lipid-protein systems. Biochim. Biophys. Acta 326, 245–255 (1973).
Article CAS Google Scholar - Bonn, M., Müller, M., Rinia, H. A. & Burger, K. N. J. Imaging of chemical and physical state of individual cellular lipid droplets using multiplex CARS microscopy. J. Raman Spectrosc. 40, 763–769 (2009).
Article CAS ADS Google Scholar - Krafft, C., Sobottka, S. B., Schackert, G. & Salzer, R. Near infrared Raman spectroscopic mapping of native brain tissue and intracranial tumors. Analyst 130, 1070–1077 (2005).
Article CAS ADS Google Scholar - Sane, S. U., Cramer, S. M. & Przybycien, T. M. A Holistic Approach to Protein Secondary Structure Characterization Using Amide I Band Raman Spectroscopy. Anal. Biochem. 269, 255–272 (1999).
Article CAS Google Scholar - Chen, P. et al. Raman signature from brain hippocampus could aid Alzheimer’s disease diagnosis. Appl. Optics 48, 4743–4748 (2009).
Article CAS ADS Google Scholar - Krafft, C., Neudert, L., Simat, T. & Salzer, R. Near infrared Raman spectra of human brain lipids. Spectrochim. Acta Mol. Biomol. Spectros. 61, 1529–1535 (2005).
Article ADS Google Scholar - Milanesi, L. et al. Direct three-dimensional visualization of membrane disruption by amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 109, 20455–20460 (2012).
Article CAS ADS Google Scholar - Coutinho, A., Loura, L. M. S. & Prieto, M. FRET studies of lipid-protein aggregates related to amyloid-like fibers. J. Neurochem. 116, 696–701 (2011).
Article CAS Google Scholar - Hellstrand, E., Nowacka, A., Topgaard, D., Linse, S. & Sparr, E. Membrane Lipid Co-Aggregation with alpha-Synuclein Fibrils. Plos One 8, e77235 (2013). 10.1371/journal.pone.0077235
Article CAS ADS PubMed PubMed Central Google Scholar - Joshi, P. et al. Microglia convert aggregated amyloid-beta into neurotoxic forms through the shedding of microvesicles. Cell Death Differ. 21, 582–593 (2014).
Article CAS Google Scholar - Verghese, P. B. et al. ApoE influences amyloid-beta (A beta) clearance despite minimal apoE/A beta association in physiological conditions. Proc. Natl. Acad. Sci. U.S.A. 110, E1807–E1816 (2013).
Article CAS Google Scholar - Liu, C. C., Kanekiyo, T., Xu, H. X. & Bu, G. J. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat. Rev. Neurol. 9, 106–118 (2013).
Article CAS Google Scholar - Galli, R. et al. Effects of tissue fixation on coherent anti-Stokes Raman scattering images of brain. J. Biomed. Opt. 19, 071402–071402 (2014).
Article ADS Google Scholar - Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Article CAS Google Scholar - Zhang, Z. M., Chen, S. & Liang, Y. Z. Baseline correction using adaptive iteratively reweighted penalized least squares. Analyst 135, 1138–1146 (2010).
Article CAS ADS Google Scholar
Acknowledgements
The research leading to these results has received funding from the Swedish Research Council and the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme FP7/2007-2013/ under REA grant agreement № 607842.
Author information
Authors and Affiliations
- Department of Biology and Biological Engineering, Chalmers University of Technology, Göteborg, Sweden
Juris Kiskis, Helen Fink, Lena Nyberg & Annika Enejder - K-Analys, Salagatan 16 A, 753 30, Uppsala, Sweden
Jacob Thyr - Department of Experimental Medical Science, Lund University, Lund, Sweden
Jia-Yi Li
Authors
- Juris Kiskis
You can also search for this author inPubMed Google Scholar - Helen Fink
You can also search for this author inPubMed Google Scholar - Lena Nyberg
You can also search for this author inPubMed Google Scholar - Jacob Thyr
You can also search for this author inPubMed Google Scholar - Jia-Yi Li
You can also search for this author inPubMed Google Scholar - Annika Enejder
You can also search for this author inPubMed Google Scholar
Contributions
J.K. analyzed data, prepared figures and wrote the manuscript. H.F. analyzed data and contributed to the manuscript, L.N. performed CARS and fluorescence microscopy measurements. J.T. performed Raman spectroscopy measurements. J.Y.L. prepared and analyzed samples and contributed to the manuscript. A.E. designed the study, analyzed data and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Kiskis, J., Fink, H., Nyberg, L. et al. Plaque-associated lipids in Alzheimer’s diseased brain tissue visualized by nonlinear microscopy.Sci Rep 5, 13489 (2015). https://doi.org/10.1038/srep13489
- Received: 03 March 2015
- Accepted: 27 July 2015
- Published: 27 August 2015
- DOI: https://doi.org/10.1038/srep13489