Structural basis of potent Zika–dengue virus antibody cross-neutralization (original) (raw)
Main
Zika virus (ZIKV) is an arthropod-borne enveloped virus belonging to the Flavivirus genus in the family Flaviviridae, which also includes the human pathogenic yellow fever, dengue, West Nile and tick-borne encephalitis viruses1. Flaviviruses have two structural glycoproteins, prM and E (for precursor membrane and envelope proteins, respectively), which form a heterodimer in the endoplasmic reticulum (ER) of the infected cell and drive the budding of spiky immature virions into the ER lumen. These particles transit through the cellular secretory pathway, during which the _trans_-Golgi-resident protease furin cleaves prM. This processing is required for infectivity, and results in the loss of a large fragment of prM and reorganization of E on the virion surface. The mature particles have a smooth aspect, with 90 E dimers organized with icosahedral symmetry in a ‘herringbone’ pattern2,3.
Three-dimensional cryo-electron microscopy (cryo-EM) structures of the mature ZIKV particles have recently been reported to near atomic resolution (3.8 Å)4,5, showing that the virus has essentially the same organization as the other flaviviruses of known structure, such as dengue virus (DENV)3 and West Nile virus6,7. The E protein is about 500 amino acids long, with the 400 N-terminal residues forming the ectodomain essentially folded as β-sheets with three domains, named I, II and III, aligned in a row with domain I at the centre. The conserved fusion loop is at the distal end of the rod in domain II, buried at the E dimer interface. At the C terminus, the E ectodomain is followed by the ‘stem’, featuring two α-helices lying flat on the viral membrane (the stem helices), which link to two C-terminal transmembrane α-helices. The main distinguishing feature of the ZIKV virion is an insertion within a glycosylated loop of E (the ‘150’ loop), which protrudes from the mature virion surface4,5.
Flaviviruses have been grouped into serocomplexes based on cross-neutralization studies with polyclonal immune sera8. The E protein is the main target of neutralizing antibodies, and is also the viral fusogen; cleavage of prM allows E to respond to the endosomal pH by undergoing a large-scale conformational change that catalyses membrane fusion and releases the viral genome into the cyotosol. Loss of the precursor fragment of prM lets the E protein fluctuate from its tight packing at the surface of the virion, transiently exposing otherwise buried surfaces. One surface exposed by this ‘breathing’ is the fusion-loop epitope (FLE), which is a dominant cross-reactive antigenic site9. Although antibodies to this site can protect by complement-mediated mechanisms, as shown in a mouse model for West Nile virus10, they are poorly neutralizing and lead to antibody-dependent enhancement11,[12](/articles/nature18938#ref-CR12 "Dejnirattisai, W. et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol. http://dx.doi.org/10.1038/ni.3515
(2016)"),[13](/articles/nature18938#ref-CR13 "Dejnirattisai, W. et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 16, 170–177 (2015)"),[14](/articles/nature18938#ref-CR14 "Goncalvez, A. P., Engle, R. E., St Claire, M., Purcell, R. H. & Lai, C. J. Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention. Proc. Natl Acad. Sci. USA 104, 9422–9427 (2007)"),[15](/articles/nature18938#ref-CR15 "Halstead, S. B. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J. Infect. Dis. 140, 527–533 (1979)"), thereby aggravating Flavivirus pathogenesis and complicating the development of safe and effective vaccines.We recently reported the functional and structural characterization of a panel of antibodies isolated from patients with dengue disease13,16. Most of these antibodies target the FLE, but others target a quaternary site readily accessible at the exposed surface of the E protein on the virion, at the interface between the two E subunits in the dimer. These broadly neutralizing antibodies (bnAbs), termed EDE for E-dimer epitope, potently neutralize all four DENV serotypes. Their binding site is conserved across serotypes because it is also the interaction site of prM with E dimers during transport of the immature virus particles through the Golgi apparatus of the cell. There were two subsets of EDE antibodies, characterized by a differential requirement for glycosylation on the 150 loop for binding. The EDE1 bnAbs bind better in the absence of glycan, whereas EDE2 bnAbs bind better when the glycan is present.
In this study, we show that the EDE bnAbs neutralize ZIKV as potently as they neutralize DENV. We also find that the FLE antibodies, which neutralize DENV although not as potently as the EDE bnAbs, do not neutralize ZIKV at concentrations up to 1 μM in spite of a high affinity for the recombinant ZIKV E protein. We further describe the crystal structures of the ZIKV E protein dimer alone and in complex with EDE1 C8 and EDE2 A11, identifying their binding determinants.
A ZIKV–DENV super serogroup
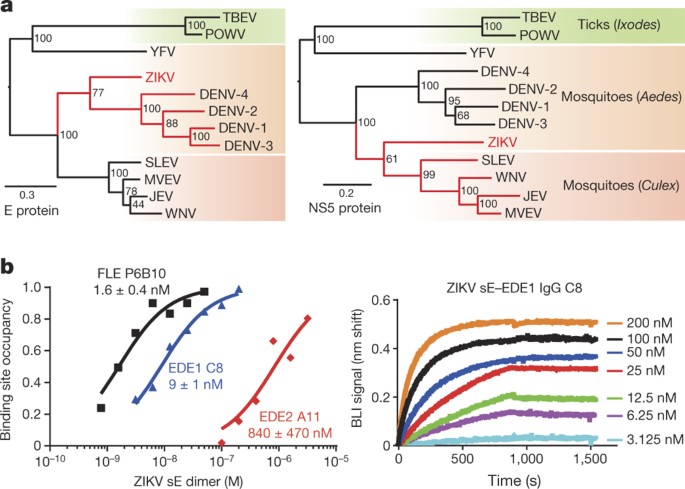
Phylogenetic analyses of the main human pathogenic flaviviruses using the amino acid sequences of the viral RNA polymerase NS5 indicate a clustering of ZIKV with the group of mosquito-borne encephalitic viruses. The clustering is different when the amino acid sequences of the E protein are considered, with ZIKV branching with the DENV group (Fig. 1a). If the sequence clustering extends to the antigenic surface of E, antibodies that cross-react with several DENV serotypes should also bind ZIKV E. To test this hypothesis, we used biolayer interferometry to study the binding properties of a poorly neutralizing, cross-reactive FLE antibody and the potently neutralizing EDE antibodies for recombinant, soluble ZIKV E ectodomain (ZIKV sE) produced in insect cells (see Methods). The FLE antibody (P6B10) bound nearly 10-fold more tightly than did EDE1 C8 (apparent dissociation constant (_K_d) values of 1.5 nM versus 9 nM), and nearly 1,000 times more tightly than EDE2 A11 (Fig. 1b and Extended Data Fig. 1a). Consistent with their affinities, we could isolate a complex of ZIKV sE with a C8 Fab by size-exclusion chromatography, but not with an A11 Fab (Extended Data Fig. 1b).
Figure 1: ZIKV and DENV E protein phylogeny and reactivity with DENV-elicited antibodies.
a, Phylogenetic trees of the main human pathogenic flaviviruses based on the amino acid sequences of the E protein (left) and of the polymerase NS5 protein (right). The arthropod vectors are differentiated by the background colour. JEV, Japanese encephalitis virus; MVEV, Murray Valley encephalitis virus; POWV, Powassan virus; SLEV, Saint Louis encephalitis virus; TBEV, tick-borne encephalitis virus; YFV, yellow fever virus; WNV, West Nile virus. b, ZIKV sE reactivity with human recombinant full-length antibodies FLE P6B10, EDE1 C8 and EDE2 A11. Left, binding properties were monitored by biolayer interferometry (BLI) on Octet-Red (ForteBio). The normalized response values expressed as fraction of binding site occupancy are plotted against concentrations of ZIKV sE dimer shown at logarithmic scale. Lines denote global curve fits used for _K_d evaluation (see Extended Data Fig. 1a for linear concentration range showing concentration-dependent saturation fits). Normalized response values were deduced from individual sensorgrams showing the binding properties for EDE1 C8 measured at different ZIKV sE concentrations.
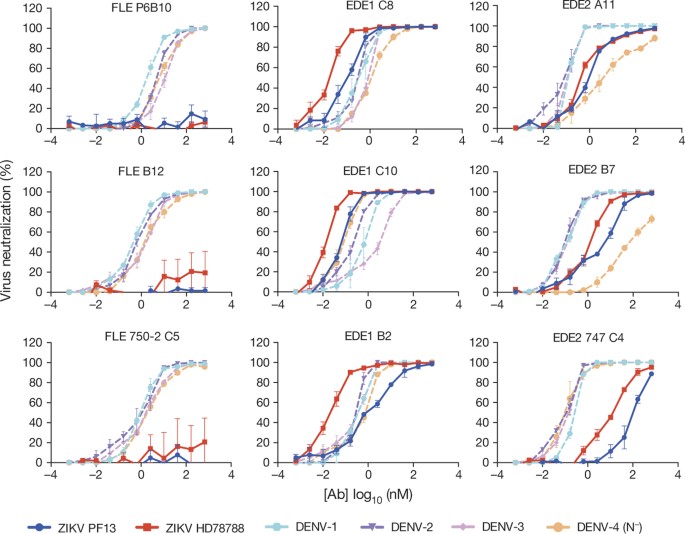
Neutralization assays in African green monkey kidney (Vero) cells using these and other members of the three antibody subsets, showed that the EDE1 antibodies strongly neutralize ZIKV, whereas the EDE2 antibodies were at least one log order less potent. Despite its strong affinity, P6B10 did not neutralize ZIKV in the concentration range used, nor did either of the two other FLE antibodies tested (Fig. 2). The EDE1 bnAbs neutralized best the ZIKV African strain HD78788, with a half-maximum inhibitory concentration (IC50) of about 0.1 nM. This strain has over the years been cell-culture-adapted and passaged in suckling mice brain and lacks E glycosylation. The IC50 against the PF13 strain, isolated in French Polynesia in 2013 and in which the E protein is glycosylated at position 154, was in the nanomolar range and comparable to, or lower than, that against the four DENV serotypes (Table 1). The EDE2 bnAbs showed no difference in neutralization of the two strains, suggesting that the presence of the N154 glycan in the ZIKV E protein did not enhance the interaction.
Figure 2: Neutralization curves using three antibodies each from the three subsets FLE, EDE1 and EDE2.
Results represent the mean ± s.e.m. of four independent experiments performed in triplicate for PF13 and in duplicate for HD78788 strains. The two ZIKV strains are in bright colours, blue and red, respectively. The neutralization data for the four DENV serotypes (dotted lines in pale colours) were taken from ref. 13, and are given here for comparison. The corresponding IC50 values are provided in Table 1. Note that the DENV-4 strain used was a natural isolate lacking the N153 glycosylation site (N−).
Table 1 50% FRNT values of EDE1, EDE2 and FLE antibodies tested against ZIKV and DENV-1–4
The ZIKV–EDE bnAbs immune complexes
We crystallized unliganded ZIKV sE and complexes of ZIKV sE with EDE1 C8 and EDE2 A11 with single-chain Fv (scFv) and Fab fragments, respectively (Extended Data Table 1). In the structure of unliganded ZIKV sE, the 150 loop is ordered, unlike the unglycosylated 150 loop in the recently determined structure of the protein produced in bacteria and refolded in vitro17. In contrast to our insect-cell-secreted protein, which is a dimer (Extended Data Fig. 1b), the refolded protein was reported to be monomeric in solution, suggesting that the glycan may help to structure the loop and promote sE dimerization.
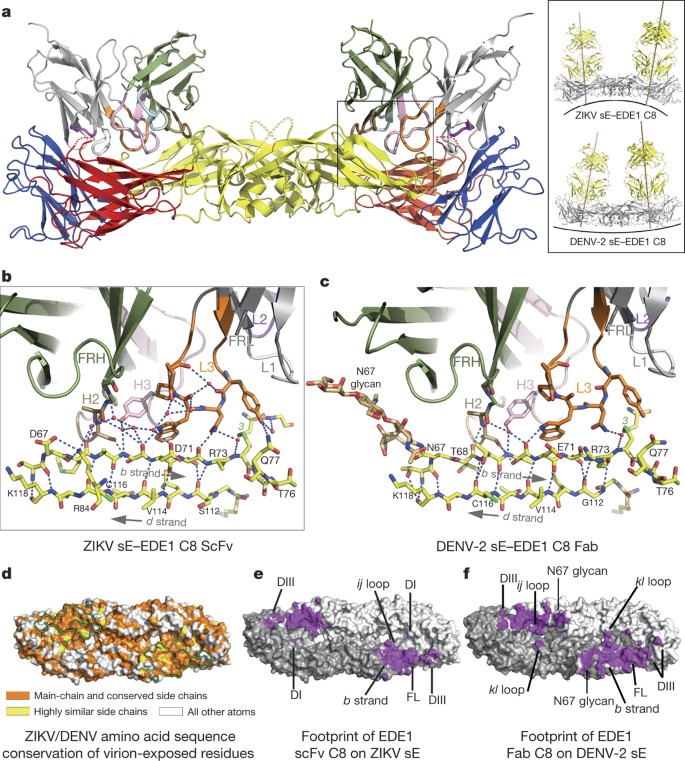
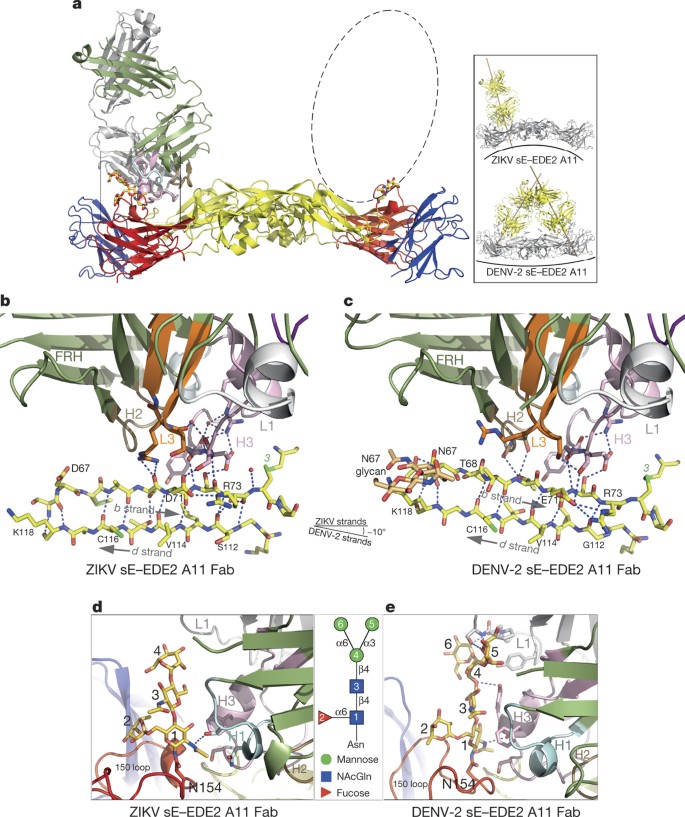
The antibodies recognize a quaternary epitope in the ZIKV sE dimer in the same way that they recognize the DENV serotype 2 (DENV-2) sE dimer described earlier16. The amino acid residues participating in the contacts, for both the ZIKV and DENV-2 structures, are shown in Extended Data Fig. 2. As expected, the pattern is very similar, with the few differences highlighted in red frames in Extended Data Fig. 2b. Both epitopes in the sE dimer are occupied in the case of the complex with C8 (Fig. 3a), whereas only one is occupied in the case of A11 (Fig. 4a). Inspection of the crystal environment showed that a second Fab could not be docked at this position without clashing with neighbouring complexes in the crystal. This observation indicates that crystal growth selected for incorporation of sE dimers with a single Fab bound, which is facilitated by the low affinity of A11.
Figure 3: EDE1 C8–ZIKV sE complex.
a, Overall view of the complex, with the sE moiety coloured by domains (I, II and III in red, yellow and blue, respectively); the antibodies in grey and dark green for light and heavy chains, respectively. The CDRs are coloured (H1, light blue; H2, sand; H3, pink; L1, light grey; L2, magenta; L3, orange). The inset shows a comparison with the corresponding DENV-2 complex. For clarity, the variable region of the C8 Fab fragment of the DENV-2 sE–C8 complex was superposed on the C8 scFv in complex with ZIKV sE to draw the Fab axes and show the docking angles better. b, Zoom of the ZIKV sE–C8 interaction to show the recognition of the b strand. Hydrogen bonds are shown as dotted lines and immobilized water molecules as red spheres. c, Same region on the DENV-2 sE–C8 Fab complex. Note that the N67 glycan on DENV also interacts with the antibody. d, The footprint of EDE1 C8 is outlined on the ZIKV sE dimer shown in surface representation (looking from outside the virion) coloured according to conservation of surface-exposed amino acids. Atoms from the main-chain and conserved side chains are orange, highly similar side chains are yellow, and all the others are white. e, f, Footprints of EDE1 C8 on a surface representation of ZIKV sE (e) and DENV-2 sE (f) shown in purple. FL, fusion loop. The two protomers of sE in the dimer are in light and dark grey. Relevant antigenic sE regions are labelled. Note the more confined interacting surface in the ZIKV sE dimer than in DENV-2, for example, N67 glycan is absent in ZIKV sE.
Figure 4: EDE2 A11–ZIKV sE complex.
Colour coding is as in Fig. 3. a, Overall view of the complex, with only one Fab bound per sE dimer, owing to crystal packing. The dashed ellipse represents the position of the missing A11 Fab. The inset compares the angle of binding to the sE dimer in ZIKV and in DENV-2. b, c, Interactions at the b strand in ZIKV (left) and in DENV-2 (right). Note the different angle of the b strand with respect to the antibody (the antibody is in exactly the same orientation in both panels). d, e, Zoom of the glycan on the 150 loop for ZIKV sE (d) and for DENV-2 sE (e), with sugar residue numbers described in the key. The CDR H3 helix is too far to make interactions with the glycan, as is the case in the DENV-2 structure (see Extended Data Figs 2b and 5b).
The bnAbs dock on ZIKV sE at different angles than they do on DENV-2 sE (see insets in Figs 3a and 4a). In the case of the C8 complex, the difference in docking results mainly from an altered curvature of the sE dimer. We note that the conformation of ZIKV sE in complex with the antibodies is very similar to the one it adopts on the virus particle, with roughly 1.5 Å root mean square deviation (r.m.s.d.) for 790 Cα atoms (see Extended Data Table 2). The unbound ZIKV sE crystallized here displays a more distant conformation (2.5 Å r.m.s.d. when comparing to both virion ZIKV E and either of the sE antibody complexes), suggesting that the antibodies stabilize a conformation close to that on the viral particle. By contrast, the same comparisons done for DENV-2 sE, alone or in complex with the bnAbs, result in r.m.s.d. values of 5–7 Å with respect to the E conformation on the DENV virion observed by cryo-EM3. For comparison, superposition of the ectodomain of virion E from ZIKV5 and DENV-2 (ref. 3) results in a similar 1.5 Å r.m.s.d. value, indicating that they are presented roughly in the same way, but that DENV sE is more deformable in solution. This malleability may reflect the conformational breathing reported for DENV E18. Instead, ZIKV sE remains in a similar conformation in the absence of the interactions with the underlying stem α-helices and with the M protein (the membrane-anchored remnant of prM after furin cleavage) on the virion, in line with the higher stability of the ZIKV particles described recently4.
EDE1 C8 complex
The total buried surface area of EDE1 C8 in the complex with ZIKV sE is about 900 Å2, compared to about 1,300 Å2 in the DENV-2 sE complex (Extended Data Table 3). Figure 3d shows the conservation of the epitope, and Fig. 3e and f compare the C8 footprint on ZIKV and DENV-2 sE. The DENV-specific glycan at position N67, which is ordered in the DENV-2 sE structure (Fig. 3c), accounts for around two-thirds of the overall difference in footprint area. The N67 glycan interacts with the framework region 2 of the heavy chain (FRH2), and its absence in ZIKV sE shows that these contacts are not essential for binding. The key cluster of interactions that is maintained is centred on β-strand b of domain II, with side chains from complementarity determining regions (CDRs) H2, H3 and L3 recognizing all the available hydrogen bond donors (main-chain carbonyls) and acceptors (main-chain carbonyls) of the bdc β-sheet edge (Fig. 3b, c). In addition, the fusion loop main chain (which contains several glycine residues) and the disulfide bond between Cys74 and Cys105 are framed by aromatic side chains of the CDRs L1 and L3 (see also Extended Data Fig. 3). Residues from these two CDRs also recognize strictly conserved side chains of the fusion loop (Arg99) or nearby residues (Gln77).
Across the dimer interface, and as in the complex with DENV-2, the 150 loop is partially disordered, with no detectable density for the N154 glycan (Fig. 3a and Extended Data Fig. 3d). As shown in Extended Data Fig. 3, the interacting residues across the dimer interface are different, reflecting the more limited sequence conservation in these regions of the E protein: in the DENV-2 sE complex, these contacts are with residues from β-strands A and B of domain III, but in ZIKV they mainly involve Lys373 from β-strand E interacting with CDRs L1 and L2, via a network of direct or water-mediated hydrogen bonds (Extended Data Fig. 3b, c). Similarly, several charged residues in domain I and from the nearby kl loop of domain II across the interface, contribute to the binding and interact with the heavy chain CDRs H2 and H3 (Extended Data Fig. 3e, f). All of the polar interactions between C8 and ZIKV sE are listed in Extended Data Tables 4 and 5, and the electrostatic surface of the epitope is shown in Extended Data Fig. 4, left panel. In summary, these observations identify the conserved cluster of contacts with the b strand and the fusion loop in domain II as the main binding determinants of C8, with additional contacts from across the dimer interface—or from the N67 glycan in DENV—further stabilizing but not determining the interaction.
EDE2 A11 complex
Extended Data Fig. 4 compares the footprint of C8 and A11 on ZIKV sE, together with the surface electrostatic potential of the complexes, which shows a strong basic patch on sE in the C8 complex due to the disorder of the 150 loop. As shown in Extended Data Fig. 5, C8 would clash with the glycan had the loop remained in place, as was the case in the complex with DENV-2 sE16. In the A11 complex, the 150 loop remains in the same conformation as in the cryo-EM structures of the virion (Extended Data Fig. 5a) and in the X-ray structure of glycosylated unliganded sE reported here. In the DENV-2 sE–A11 complex, the glycan is recognized by an α-helix in the long CDR H3 loop (Fig. 4e). The difference in length in the 150 loop of E in ZIKV compared to DENV shifts the glycan position by about 6–7 Å, such that it cannot make the same interactions with the CDR H3 α-helix (Fig. 4d, e and Extended Data Fig. 5b). As a consequence, the A11 antibody docks at a different angle on ZIKV sE than it does on DENV-2 sE, even accounting for the difference in sE dimer curvature (Fig. 4a, inset). The contacts along the b strand are preserved (Fig. 4b, c). Compared to C8, the b strand is recognized only along half its length (residues 71 and 73), whereas C8 recognizes it all along, from residue 68 (or from 67 in DENV).
Discussion
Our results identify the structural details of a quaternary epitope that provides a previously unrecognized link of potent cross-neutralization between Zika and dengue viruses, and thus identifies an antigenic Flavivirus cluster beyond the traditional serocomplexes. This relationship defines a super serogroup on the basis of strong cross-neutralization through a conserved epitope that had not been recognized using polyclonal sera8. This finding thus introduces the possibility of developing a universal vaccine protecting against all the viruses from this group.
Vaccine design against dengue virus has been hampered by the heterogeneity of DENV particles and the need to use polyvalent formulas to immunize against all four serotypes19,20. One feature of DENV is that it undergoes incomplete furin maturation cleavage of prM in many cell types, giving rise to heterogeneous mosaic particles with an immature-like spiky patch on one side and a smooth mature-like region on the opposite side21. These particles are infectious, as they can fuse with the cellular membrane through the smooth, mature side. Because the FLE is exposed in immature regions22, most of the antibody response in DENV-infected patients is directed against it23. These cross-reactive antibodies coat the particles on the immature side22 but neutralize only weakly, because they can bind the mature side only when the E protein ‘breathes’24,25,26. A recently published structure of monomeric ZIKV sE in complex with an FLE-specific monoclonal mouse antibody of low neutralizing activity indeed shows that its binding site would be occluded in the dimeric E protein on mature infectious virions17. The observation that P6B10 and other FLE antibodies still neutralize DENV13 suggests that E in the mature patches on DENV spends more time in conformations that expose the FLE than does E in those patches on ZIKV. This inference is consistent with the higher thermal stability of ZIKV reported recently4.
Our results suggest that the epitope targeted by the EDE1 bnAbs is better suited for developing an epitope-focused vaccine for viruses in the ZIKV/DENV super serogroup than is the FLE, which induces poorly neutralizing and strongly infection-enhancing antibodies[12](/articles/nature18938#ref-CR12 "Dejnirattisai, W. et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol. http://dx.doi.org/10.1038/ni.3515
(2016)"),[13](/articles/nature18938#ref-CR13 "Dejnirattisai, W. et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 16, 170–177 (2015)"),[14](/articles/nature18938#ref-CR14 "Goncalvez, A. P., Engle, R. E., St Claire, M., Purcell, R. H. & Lai, C. J. Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention. Proc. Natl Acad. Sci. USA 104, 9422–9427 (2007)"). The EDE1 is also better suited than the related EDE2 epitope: although the EDE1 bnAbs require an E dimer to bind, the actual binding determinants are centred on the _b_ strand and on the highly conserved, E-dimer-exposed elements of the fusion loop, as shown by the comparison between their binding to DENV-2 and ZIKV sE. The fact that EDE2 bnAbs rely heavily on their contact points on the adjacent subunit—on the variable 150 loop in which glycosylation is not always present—is a drawback, as demonstrated by their poor affinity ([Fig. 1](/articles/nature18938#Fig1)) and their strong induction of antibody-dependent enhancement[12](/articles/nature18938#ref-CR12 "Dejnirattisai, W. et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol.
http://dx.doi.org/10.1038/ni.3515
(2016)").Targeting the b strand and the E-dimer-exposed elements of the fusion loop appears as a powerful alternative to the multi-immunogen approaches against the DENV cluster that have had limited success in clinical trials27. As the E protein polypeptide chain displays neither insertions nor deletions in the region of the b strand for any medically relevant Flavivirus, this region presents a low risk of inducing escape mutations, most likely because it is also the interacting site with prM during virus maturation. Finally, in a more immediate application, our study also suggests that the EDE1 antibodies, perhaps carrying the ‘LALA’ mutation28 if effector functions are to be avoided, could be useful for immune prophylaxis for pregnant women at risk of contracting ZIKV infection.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment.
Recombinant production of ZIKV sE protein
Recombinant Zika virus sE protein (strain H/PF/2013, GenBank accession number KJ776791) was produced with a tandem strep-tag in the Drosophila Expression System (Invitrogen) as described previously29,30. A chemically synthesized DNA fragment (GeneArt) containing the Zika sE sequence (amino acids 1–408) was cloned into the expression vector pT389 (ref. 31) that encodes the export signal sequence BIP, an enterokinase cleavage site and the strep-tag. Drosophila Schneider 2 (S2) cells were stably transfected using blasticidin for selection. Protein expression was induced by the addition of CuSO4 and supernatants were collected 7–10 days after induction. Antigens were purified via affinity chromatography with Streptactin columns (IBA) according to the manufacturer’s instructions. A final purification gel filtration step used a Superdex increase 200 10/300 GL column equilibrated in 50 mM Tris, pH 8, 500 mM NaCl.
Production of antigen-binding (Fab) and scFv fragments of the bnAbs
The bnAb fragments were cloned into plasmids for expression as Fab32 and scFv33 in Drosophila S2 cells. The constructs contain a tandem strep-tag fused at the C terminus (only of the heavy chain in the case of the Fab) for affinity purification. The purification protocol included a Streptactin affinity column followed by gel filtration as described above.
Expression of human monoclonal anti-DENV E antibodies
Full IgG antibodies were produced in 293T cells (a gift from C. Lee), which were free from mycoplasma contamination tested by Lookout Mycoplasma PCR detection kit (Sigma MP0035). These cells were co-transfected with plasmids containing heavy and light chains of immunoglobulin G1 as described previously13.
Immune complex formation and isolation
The purified ZIKV sE protein was mixed with Fab A11 or scFv C8 (in approximately twofold molar excess) in standard buffer (500 mM NaCl, Tris 50 mM, pH 8.0). The volume was brought to 0.5 ml by centrifugation in a Vivaspin 10 kDa cut-off; after a 30-min incubation at 4 °C, the complex was separated from excess Fab or scFv by size-exclusion chromatography for ZIKV sE and scFv C8. For ZIKV sE and Fab A11, no apparent complex formation could be seen in size-exclusion chromatography; therefore a solution containing sE at a concentration of 1.5 mg ml−1 and Fab A11 at a concentration of 3 mg ml−1 (corresponding to a molar ratio ~1:2 antigen:antibody) was directly used for crystallization. In all cases, the buffer was exchanged to 150 mM NaCl, 15 mM Tris, pH 8, for crystallization trials. The protein concentrations used for crystallization, determined by measuring the absorbance at 280 nm and using an extinction coefficient estimated from the amino acid sequences, are listed in Extended Data Table 1.
Real-time biolayer interferometry binding assays
The interactions of purified ZIKV E protein with IgG FLE P6B10, IgG EDE1 C8, IgG EDE2 A11, and control IgG 28C (an anti-influenza virus) were monitored in real-time using a bio-layer interferometry Octet-Red384 device (Pall ForteBio). Anti-human IgG Fc capture biosensors (Pall ForteBio) were loaded for 10 min at 1,000 r.p.m. shaking speed using antibodies at 5 μg ml−1 in assay buffer (PBS plus 0.2 mg ml−1 BSA and tween 0.01%). Unbound antibodies were washed away for 1 min in assay buffer. IgG-loaded sensors were then incubated for 15 min at 1,200 r.p.m. in the absence and presence of twofold serially diluted ZIKV sE protein concentrations in assay buffer. Molar concentrations were calculated for the sE protein in a dimeric form. For the antibodies FLE P6B10, EDE1 C8 and EDE2 A11, the following ZIKV sE concentration ranges: 0.78–50 nM, 3.125–200 nM and 50–3,200 nM, were respectively used. Reference binding experiments were carried out in parallel on sensors loaded with control IgG 28C. Dissociation of the complexes formed was then monitored for 10 min by dipping sensors in assay buffer alone. Operating temperature was maintained at 25 °C. The real-time data were analysed using Scrubber 2.0 (Biologic Software) and Biaevaluation 4.1 (GE Healthcare). Specific signals were obtained by double-referencing, that is, subtracting non-specific signals measured on non-specific IgG-loaded sensors and buffer signals on specific IgG-loaded sensors. Association and dissociation profiles, as well as steady-state signal versus concentration curves, were fitted assuming a 1:1 binding model.
Crystallization and X-ray structure determinations
The crystallization and cryo-cooling conditions for diffraction data collection are listed in Extended Data Table 1. Crystallization trials were performed in sitting drops of 400 nl. Drops were formed by mixing equal volumes of the protein and reservoir solution in 96-well Greiner plates, using a Mosquito robot and monitored by a Rock-Imager. Crystals were optimized using a robotized Matrix Maker and Mosquito setups on 400 nl sitting or hanging drops, or manually in 24-well plates using 2–3 μl hanging drops.
Because of the strong anisotropy of the crystals (see results for anisotropy in Extended Data Table 1), an important number of crystals was tested at several beam lines at different synchrotrons (SOLEIL, St Aubin, France; ESRF, Grenoble, France; SLS, Villigen, Switzerland). The crystals having the less anisotropic diffraction data were used to determine the structures. The data sets were indexed, integrated, scaled and merged using XDS34 and AIMLESS35. A preliminary model of ZIKV sE protein was built from the DENV-2 sE (4UTA) structure using the structure homology-modelling server SWISS-MODEL36. The structures of the complexes were then determined by molecular replacement with PHASER37 using the search models listed in Extended Data Table 1. AIMLESS and PHASER programs were used within the CCP4 suite38.
The DEBYE and STARANISO programs developed by Global Phasing Ltd were applied to the data scaled with AIMLESS without applying a resolution limit, using the STARANISO server (http://staraniso.globalphasing.org/). These programs perform an anisotropic cut-off of merged intensity data on the basis of an analysis of local I/σ(I), compute Bayesian estimates of structure amplitudes, taking into account their anisotropic fall-off, and apply an anisotropic correction to the data. These corrected anisotropic amplitudes were then used for further refinement of the structures with BUSTER/TNT39. Please note that the Extended Data Table 1 shows the refinement statistics against the full sets of reflections truncated at the best high-resolution along the h, k or l axis.
The models were then alternatively manually corrected and completed using COOT40 and refined using BUSTER/TNT against the amplitudes corrected for anisotropy. Refinements were constrained using non-crystallographic symmetry. The refined structures have the following final _R_work/_R_free (in %) values: ZIKV sE–EDE1 C8 scFv (19.5/22.1), ZIKV sE–EDE2 A11 Fab (22.3/23.7) and unliganded ZIKV sE (20.8/23.6) (see Extended Data Table 1).
Analysis of the atomic models and illustrations
Each complex was analysed with the CCP4 suite of programs and the polar contacts were computed with the PISA website41. For the intermolecular interactions shown in Extended Data Figs 2 and 3 and Extended Data Tables 4 and 5, the maximal cut-off distances used were 4 Å and 4.75 Å for polar and van der Waals contacts, respectively. Multiple sequence alignments were calculated using Clustal W and Clustal X version 2 (ref. 42) on the EBI server43. The figures illustrating the structural models were prepared using ESPript44 and the PyMOL Molecular Graphics System, version 1.5.0.4 (Schrödinger) (http://pymol.sourceforge.net).
Phylogenic trees
The maximum likelihood phylogenetic trees were inferred using 12 representative amino acid sequences of Flavivirus envelope protein E or RNA polymerase NS5 proteins, using the LG model available in PhyML45 and a combination of SPR+NNI branch-swapping. Bootstrap values were calculated from 100 bootstrap replicates. The trees were visualized using Figtree (http://tree.bio.ed.ac.uk/software/figtree/). The accession codes of sequences used in the tree: Zika virus (ZIKV, KJ776791, strain H-PF-2013_French_Polynesia); dengue virus serotype 1 (DENV-1, NC_001477); dengue virus serotype 2 (DENV-2, NC_001474); dengue virus serotype 3 (DENV-3, NC_001475); dengue virus serotype 4 (DENV-4, NC_002640); Saint Louis encephalitis virus (SLEV, NC_007580); Japanese encephalitis virus (JEV, NC_001437; Murray Valley encephalitis virus (MVEV, NC_000943); West Nile virus (WNV, NC_001563); yellow fever virus (YFV, NC_002031); tick-borne encephalitis virus (TBEV, NC_001672); and Powassan virus (POWV, NC_003687).
Viral stocks
The African strain Zika HD78788 was obtained from the Institut Pasteur collection and the Asian strain Zika PF13, isolated from a patient during ZIKV outbreak in French Polynesia in 2013, was obtained through the DENFREE (FP7/2007-2013) consortium. Viral stocks were prepared from supernatant of infected C6/36 cells (ATCC CRL-1660) clarified by centrifugation at 3,000_g_ at 4 °C and titrated on Vero cells (ATCC CRL-1586) by a focus-forming assay. Stocks were kept at −80 °C until use. All cell lines were free from mycoplasma contamination.
Neutralization assays
Virus neutralization by the tested human antibodies was evaluated using a focus reduction neutralization test (FRNT). About 100 focus-forming units from virus stocks were incubated with a serial dilution of antibody for 1 h at 37 °C. The mixture was then added to Vero cells and foci were left to develop in presence of 1.5% methylcellulose for 2 days. Foci were then stained after fixation with 4% formaldehyde using anti-E 4G2 antibody (ATCC HB-112) and anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (ThermoFisher 31430). The foci were visualized by diaminobenzidine (DAB) (Sigma D5905) staining and plates were counted using the ImmunoSpot S6 Analyser (Cellular Technology Limited, CTL). Neutralization curves and 50% FRNT values were calculated by nonlinear regression analysis using Prism 6, GraphPad software.
Accession codes
Primary accessions
Protein Data Bank
Referenced accessions
GenBank/EMBL/DDBJ
Data deposits
Atomic coordinates and structure factors amplitudes have been deposited in the Protein Data Bank (PDB) under accession numbers 5LBS (for complex ZIKV sE–EDE1 C8 scFv), 5LCV (for complex ZIKV sE–EDE2) and 5LBV (for A11 Fab and unliganded ZIKV sE).
References
- Lindenbach, B., Murray, C., Thiel, H. & Rice, C. Flaviviridae: The Viruses and Their Replication 6th edn, Vol. 1 1101–1152 (Lippincott Williams & Wilkins, 2013)
Google Scholar - Kuhn, R. J. et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108, 717–725 (2002)
Article CAS PubMed PubMed Central Google Scholar - Zhang, X. et al. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 20, 105–110 (2013)
Article PubMed CAS Google Scholar - Kostyuchenko, V. A. et al. Structure of the thermally stable Zika virus. Nature 533, 425–428 (2016)
Article ADS CAS PubMed Google Scholar - Sirohi, D. et al. The 3.8 Å resolution cryo-EM structure of Zika virus. Science 352, 467–470 (2016)
Article ADS CAS PubMed PubMed Central Google Scholar - Mukhopadhyay, S., Kim, B. S., Chipman, P. R., Rossmann, M. G. & Kuhn, R. J. Structure of West Nile virus. Science 302, 248 (2003)
Article CAS PubMed Google Scholar - Zhang, W., Kaufmann, B., Chipman, P. R., Kuhn, R. J. & Rossmann, M. G. Membrane curvature in flaviviruses. J. Struct. Biol. 183, 86–94 (2013)
Article CAS PubMed PubMed Central Google Scholar - Calisher, C. H. et al. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J. Gen. Virol. 70, 37–43 (1989)
Article PubMed Google Scholar - Stiasny, K., Kiermayr, S., Holzmann, H. & Heinz, F. X. Cryptic properties of a cluster of dominant flavivirus cross-reactive antigenic sites. J. Virol. 80, 9557–9568 (2006)
Article CAS PubMed PubMed Central Google Scholar - Vogt, M. R. et al. Poorly neutralizing cross-reactive antibodies against the fusion loop of West Nile virus envelope protein protect in vivo via Fcγ receptor and complement-dependent effector mechanisms. J. Virol. 85, 11567–11580 (2011)
Article CAS PubMed PubMed Central Google Scholar - Balsitis, S. J. et al. Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLoS Pathog. 6, e1000790 (2010)
Article PubMed PubMed Central CAS Google Scholar - Dejnirattisai, W. et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol. http://dx.doi.org/10.1038/ni.3515 (2016)
- Dejnirattisai, W. et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 16, 170–177 (2015)
Article CAS PubMed Google Scholar - Goncalvez, A. P., Engle, R. E., St Claire, M., Purcell, R. H. & Lai, C. J. Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention. Proc. Natl Acad. Sci. USA 104, 9422–9427 (2007)
Article ADS CAS PubMed PubMed Central Google Scholar - Halstead, S. B. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J. Infect. Dis. 140, 527–533 (1979)
CAS PubMed Google Scholar - Rouvinski, A. et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 520, 109–113 (2015)
Article ADS CAS PubMed Google Scholar - Dai, L. et al. Structures of the Zika virus envelope protein and its complex with a flavivirus broadly protective antibody. Cell Host Microbe 19, 696–704 (2016)
Article CAS PubMed Google Scholar - Kuhn, R. J., Dowd, K. A., Beth Post, C. & Pierson, T. C. Shake, rattle, and roll: Impact of the dynamics of flavivirus particles on their interactions with the host. Virology 479-480, 508–517 (2015)
Article CAS PubMed Google Scholar - Sabchareon, A. et al. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380, 1559–1567 (2013)
Article CAS Google Scholar - Vannice, K. S., Durbin, A. & Hombach, J. Status of vaccine research and development of vaccines for dengue. Vaccine 34, 2934–2938 (2016)
Article PubMed Google Scholar - Plevka, P. et al. Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO Rep. 12, 602–606 (2011)
Article CAS PubMed PubMed Central Google Scholar - Cherrier, M. V. et al. Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody. EMBO J. 28, 3269–3276 (2009)
Article CAS PubMed PubMed Central Google Scholar - Beltramello, M. et al. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8, 271–283 (2010)
Article CAS PubMed Google Scholar - Dowd, K. A., Mukherjee, S., Kuhn, R. J. & Pierson, T. C. Combined effects of the structural heterogeneity and dynamics of flaviviruses on antibody recognition. J. Virol. 88, 11726–11737 (2014)
Article PubMed PubMed Central CAS Google Scholar - Lee, P. D. et al. The Fc region of an antibody impacts the neutralization of West Nile viruses in different maturation states. J. Virol. 87, 13729–13740 (2013)
Article CAS PubMed PubMed Central Google Scholar - Mukherjee, S. et al. Mechanism and significance of cell type-dependent neutralization of flaviviruses. J. Virol. 88, 7210–7220 (2014)
Article PubMed PubMed Central CAS Google Scholar - Capeding, M. R. et al. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet 384, 1358–1365 (2014)
Article CAS PubMed Google Scholar - Hessell, A. J. et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature 449, 101–104 (2007)
Article ADS CAS PubMed Google Scholar - Vratskikh, O. et al. Dissection of antibody specificities induced by yellow fever vaccination. PLoS Pathog. 9, e1003458 (2013)
Article CAS PubMed PubMed Central Google Scholar - Jarmer, J. et al. Variation of the specificity of the human antibody responses after tick-borne encephalitis virus infection and vaccination. J. Virol. 88, 13845–13857 (2014)
Article PubMed PubMed Central CAS Google Scholar - DuBois, R. M. et al. Functional and evolutionary insight from the crystal structure of rubella virus protein E1. Nature 493, 552–556 (2013)
Article ADS CAS PubMed Google Scholar - Backovic, M. et al. Efficient method for production of high yields of Fab fragments in Drosophila S2 cells. Protein Eng. Des. Sel. 23, 169–174 (2010)
Article CAS PubMed Google Scholar - Gilmartin, A. A. et al. High-level secretion of recombinant monomeric murine and human single-chain Fv antibodies from Drosophila S2 cells. Protein Eng. Des. Sel. 25, 59–66 (2012)
Article CAS PubMed Google Scholar - Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
Article CAS PubMed PubMed Central Google Scholar - Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013)
Article CAS PubMed PubMed Central Google Scholar - Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014)
Article CAS PubMed PubMed Central Google Scholar - McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Article CAS PubMed PubMed Central Google Scholar - Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011)
Article CAS PubMed PubMed Central Google Scholar - Bricogne, G. et al. BUSTER version 2.10.2 (Global Phasing Ltd, 2016)
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Article CAS PubMed PubMed Central Google Scholar - Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007)
Article CAS PubMed Google Scholar - Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007)
Article CAS PubMed Google Scholar - Goujon, M. et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 (2010)
Article ADS CAS PubMed PubMed Central Google Scholar - Gouet, P., Courcelle, E., Stuart, D. I. & Métoz, F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 (1999)
CAS PubMed Google Scholar - Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010)
Article CAS PubMed Google Scholar
Acknowledgements
We thank M. van Kerkhove, the ‘working group ZIKA’ and our colleagues in the Unité de Virologie Structurale at Institut Pasteur for help and discussions; the staff at the crystallogenesis platform and the recombinant proteins platform of the Institut Pasteur; P. Sylvestre and the Risk Prevention Unit of the Institut Pasteur for their biosafety expertise; W.Shepard and P. Legrand for help during data collection and for discussions; the staff at beamlines PX1 and PX2 at SOLEIL synchrotron (Saclay, France), at beamline ID23-2 of the European Synchrotron Radiation Facility (Grenoble, France) and at beamline PX1 of the Swiss Light Source synchrotron (Villigen, Switzerland); G. Bricogne for suggestions on processing anisotropic diffraction data; A. Reiter and H. Prechler for help in recombinant sE production. G.R.S. is a Wellcome Trust Senior Investigator. We acknowledge support from the European Commission FP7 Programme for the DENFREE project under Grant Agreement number 282 378FP7 (F.A.R., J.M., G.R.S. and A.Sa.); the “Integrative Biology of Emerging Infectious Diseases” Labex (Laboratoire d’Excellence) grant number ANR-10-LABX-62-IBEID (French Government’s “Investissements d’Avenir” program) (F.A.R.); the transnational ANR/FWF grant FlaviStem/I1378 (F.A.R. and K.S.), the Medical Research Council, UK (J.M.); the National Institute for Health Research Biomedical Research Centre, Funding Scheme, UK (G.R.S.); and the NEUTRAVIR grant from Région ile-de-France (DIM-Maladies Infectieuses) (P.E., A.H. and F.A.R.)
Author information
Author notes
- Giovanna Barba-Spaeth, Wanwisa Dejnirattisai, Alexander Rouvinski and Marie-Christine Vaney: These authors contributed equally to this work.
Authors and Affiliations
- Département de Virologie, Institut Pasteur, Unité de Virologie Structurale, Paris Cedex 15, F-75724, France
Giovanna Barba-Spaeth, Alexander Rouvinski, Marie-Christine Vaney, Arvind Sharma & Félix A. Rey - CNRS UMR 3569 Virologie, Paris Cedex 15, F-75724, France
Giovanna Barba-Spaeth, Alexander Rouvinski, Marie-Christine Vaney, Arvind Sharma & Félix A. Rey - Division of Immunology and Inflammation, Department of Medicine, Hammersmith campus, Imperial College London, W12 0NN, UK
Wanwisa Dejnirattisai, Juthathip Mongkolsapaya & Gavin R. Screaton - Department of Virology, Medical University of Vienna, Kinderspitalgasse 15, Vienna, A-1095, Austria
Iris Medits, Karin Stiasny & Franz X. Heinz - Département de Génomes et Génétique, Institut Pasteur, Unité de Génétique fonctionnelle des maladies infectieuses, Paris Cedex 15, F-75724, France
Etienne Simon-Lorière & Anavaj Sakuntabhai - CNRS URA 3012, F-75724, Paris Cedex 15, France
Etienne Simon-Lorière & Anavaj Sakuntabhai - Unit of Emerging Infectious Diseases, Institut Louis Malardé, 98713 Papeete, Tahiti, French, Polynesia
Van-Mai Cao-Lormeau - Département de Biologie Structurale et Chimie, Institut Pasteur, Plateforme de Cristallographie, CiTech, Paris Cedex 15, F-75724, France
Ahmed Haouz - CNRS UMR 3528, Paris Cedex 15, F-75724, France
Ahmed Haouz & Patrick England - Département de Biologie Structurale et Chimie, Institut Pasteur, Plateforme de Biophysique des Macromolécules et de leurs Interactions, CiTech, Paris Cedex 15, F-75724, France
Patrick England - Dengue Hemorrhagic Fever Research Unit, Office for Research and Development, Siriraj Hospital, Faculty of Medicine, Mahidol University, Bangkok, 10700, Thailand
Juthathip Mongkolsapaya
Authors
- Giovanna Barba-Spaeth
You can also search for this author inPubMed Google Scholar - Wanwisa Dejnirattisai
You can also search for this author inPubMed Google Scholar - Alexander Rouvinski
You can also search for this author inPubMed Google Scholar - Marie-Christine Vaney
You can also search for this author inPubMed Google Scholar - Iris Medits
You can also search for this author inPubMed Google Scholar - Arvind Sharma
You can also search for this author inPubMed Google Scholar - Etienne Simon-Lorière
You can also search for this author inPubMed Google Scholar - Anavaj Sakuntabhai
You can also search for this author inPubMed Google Scholar - Van-Mai Cao-Lormeau
You can also search for this author inPubMed Google Scholar - Ahmed Haouz
You can also search for this author inPubMed Google Scholar - Patrick England
You can also search for this author inPubMed Google Scholar - Karin Stiasny
You can also search for this author inPubMed Google Scholar - Juthathip Mongkolsapaya
You can also search for this author inPubMed Google Scholar - Franz X. Heinz
You can also search for this author inPubMed Google Scholar - Gavin R. Screaton
You can also search for this author inPubMed Google Scholar - Félix A. Rey
You can also search for this author inPubMed Google Scholar
Contributions
The first four authors (G.B.-S., W.D., A.R. and M.-C.V.) contributed equally to this work and are listed in alphabetical order. G.B.-S. did the first neutralization experiments and contributed throughout the project. V.-M.C.-L., A.Sa. and E.S.L. provided ZIKV from both PF13 and HD78788 strains. I.M., K.S. and F.X.H. provided the recombinant sE protein. A.R. and A.Sh. prepared recombinant antibody fragments; A.R., G.B.-S. and W.D. did neutralization and binding experiments; A.R. grew the crystals and collected synchrotron data together with A.H.; M.-C.V. collected synchrotron data, processed the data, built, refined and analysed the atomic models. E.S.-L. did the phylogenetic analyses; A.R., A.Sh. and P.E. did the biolayer interferometry experiments with recombinant sE. A.R., G.B.-S., K.S., J.M., F.X.H., G.R.S. and F.A.R. planned the experiments. F.A.R. wrote the paper.
Corresponding authors
Correspondence toFranz X. Heinz, Gavin R. Screaton or Félix A. Rey.
Ethics declarations
Competing interests
The EDE antibodies and epitope are the subject of a patent application by Imperial College and Institut Pasteur on which G.S., J.M., F.R., A.R., M.C.V. and G.B.S. are named as inventors.
Extended data figures and tables
Extended Data Figure 1 Antibody binding to recombinant ZIKV protein.
a, Biolayer interferometry experiments plotted on a linear scale. The antibodies were immobilized on the biosensor tip, and the ZIKV sE protein was in solution at the indicated concentrations. The antibody used is indicated in each plot. Note that the horizontal scale is different for the three antibodies. The estimated dissociation constant (_K_d) and estimated dissociation rate (_K_off) values are indicated. b, Size-exclusion chromatography results for isolated sE, isolated Fab fragments, and ZIKV sE plus Fab fragments, as indicated.
Extended Data Figure 2 Residues involved in bnAb–antigen interactions.
a, Antibody contacts on the amino acid sequence alignment of ZIKV and DENV-2 sE. A red background highlights identical residues. Secondary structure elements are indicated together with their labels above (ZIKV) and below (DENV-2) the sequences. The domain organization of ZIKV and DENV-2 sE is symbolized by a coloured bar above the sequences (domain I red, domain II yellow, domain III blue and the fusion loop orange). Residues involved in polar and van der Waals protein–protein contacts are marked using blue and green symbols, respectively, as indicated in the inset key, displayed above and below the alignment for ZIKV and DENV-2 sE, respectively. Full and empty symbols correspond to antibody contacts on the reference subunit of sE (defined as the one contributing the fusion loop to the epitope) and the opposite subunit of sE, respectively. Residues contacted only by the heavy or light chain are marked with squares or triangles, respectively, and those contacted by both antibody chains with circles. Dots above the sequences mark every 10 residues on the ZIKV sE sequence. Disulfide bridges are numbered in green above the sequences. b, Amino acid sequence of the heavy and light chains variable domains (vH and vL) of bnAbs EDE1 C8 (top) and EDE2 A11 (bottom) with the framework (FRW) indicated by black bars and IMGT CDR regions by thin dashed lines. The secondary structure elements of the Ig vH and vL β-barrels are indicated above the sequences. Somatic mutations are in red and residues arising from recombination at the V-D-J junction are in green. Symbols above and below the sequences mark residues involved in contacts with ZIKV and DENV-2 sE, respectively, coded for the contacted site in sE as indicated in the key (inset at the bottom). Polar and van der Waals contacts are shown in blue and green, respectively. The antibody residues contacting the reference sE subunit (defined as the one contributing the fusion loop to the epitope) are marked by plain colour symbols while those making contact across the dimer interface by empty coloured symbols. Red boxes highlight the contacts found in the DENV-2 sE complex and absent in the ZIKV sE complex, involving N67 glycan, kl and 150 loops. The details of the polar contacts are listed in the Extended Data Tables 4 and 5 (see also Fig. 3e, f). The predicted vH and vL germline alleles are indicated with the corresponding CDR lengths (see Table 1 in ref. 16).
Extended Data Figure 3 Details of EDE1 C8 bnAb contact across the dimer interface.
a, Overall view of the ZIKV sE–EDE1 C8 scFv complex. The box indicates the region zoomed in b. b, Details of the interactions of the C8 light chain with domain III across the dimer interface. c, Same region for the DENV-2 sE–EDE1 C8 Fab complex. Note that the sE residues involved are different. d, The complex rotated by 120° (as indicated by the arrow) to show the interaction in the ij loop, enlarged in e. e, The ij loop is displayed in sticks, to show the interaction of its main chain with the antibody. Domain II from the subunit across is coloured green to distinguish from domain II of the reference subunit; the dashed sticks for the arginine residue indicate that it has poor electron density in the crystal. f, Same view of the complex with DENV-2. Note that the residues from across the dimer interface that contact the antibody are different. The residues in the various CDRs are coloured coded, matching their label colour (as in Figs 3 and 4).
Extended Data Figure 4 Surface electrostatic potential on an open-book representation of the immunocomplexes.
The electrostatic potential is coloured according to the bar underneath. The antibody footprints are outlined in green. The disordered 150 loop in the complex with C8 (left) results in a positive surface patch at one edge of the epitope, which is counteracted by the residues in the 150 loop, as shown on the right in the complex with A11 where this loop is ordered.
Extended Data Figure 5 Details of the A11 interaction with the glycan on the 150 loop.
a, Superposition of the ZIKV sE–A11 complex (in colours) on the E protein from the cryo-EM structure of the mature virion (ref. 5) (PDB code 5IRE) in white. The E protein was superimposed on the tip of domain II of the reference subunit together with domain III from the opposite subunit. It shows that the 150 loop adopts essentially the same conformation, although fewer sugar residues are visible in the absence of the antibody. b, Superposition of the ZIKV sE–A11 complex (in colours) on the DENV-2 sE–A11 complex (in white). The variable domains of the antibody from the two structures were superimposed on each other. Note that in DENV-2 the glycan packs against the α-helix of the CDR H3, whereas in ZIKV sE the glycan is too far to make the same interaction. c, The ZIKV sE–C8 complex (in pink) was superimposed on the ZIKV sE–A11 complex (in colours), to show the clash of the C8 light chain with the glycan, forcing it to move out of the way and be disordered. The superposition also shows that EDE1 C8 reaches further in to contact the ij loop and the kl loop of the adjacent subunit, as well as domain III. As in a, the superposition was done using the tip of domain II of the reference subunit and domain III of the adjacent subunit in the dimer as anchors. The two black asterisks mark the places where the electron density of the 150 loop is lost, resulting in no density in the ZIKV sE–C8 crystal for the short helix, nor for the glycan.
Extended Data Table 1 Crystallization conditions, data collection and refinement statistics
Extended Data Table 2 The r.m.s.d. values between sE dimers in the various structures of ZIKV and DENV-2
Extended Data Table 3 Buried surface areas and surface complementarity in the ZIKV sE dimer–EDE complexes and in DENV-2 sE dimer–EDE complexes
Extended Data Table 4 Polar and salt-bridge interactions for ZIKV sE–EDE2 A11 Fab, DENV-2 sE–EDE2 A11 Fab, ZIKV sE–EDE1 C8 scFv and DENV-2 sE–EDE1 C8 Fab
Extended Data Table 5 Polar and salt-bridge interactions for ZIKV sE–EDE2 A11 Fab, DENV-2 sE–EDE2 A11 Fab, ZIKV sE–EDE1 C8 scFv and DENV-2 sE–EDE1 C8 Fab
PowerPoint slides
Rights and permissions
About this article
Cite this article
Barba-Spaeth, G., Dejnirattisai, W., Rouvinski, A. et al. Structural basis of potent Zika–dengue virus antibody cross-neutralization.Nature 536, 48–53 (2016). https://doi.org/10.1038/nature18938
- Received: 19 May 2016
- Accepted: 17 June 2016
- Published: 23 June 2016
- Issue Date: 04 August 2016
- DOI: https://doi.org/10.1038/nature18938