Common variants in HNF-1 α and risk of type 2 diabetes (original) (raw)
Abstract
Aims/hypothesis
Mutations in the hepatocyte nuclear factor 1-α gene (HNF-1α, now known as the transcription factor 1 gene [_TCF1_]) cause the most common monogenic form of diabetes, MODY3, but it is not known if common variants in HNF-1a are associated with decreased transcriptional activity or phenotypes related to type 2 diabetes, or whether they predict future type 2 diabetes.
Subjects and methods
We studied the effect of four common polymorphisms (rs1920792, I27L, A98V and S487N) in and upstream of the HNF-1α gene on transcriptional activity in vitro, and their possible association with type 2 diabetes and insulin secretion in vivo.
Results
Certain combinations of the I27L and A98V polymorphisms in the HNF-1α gene showed decreased transcriptional activity on the target promoters glucose transporter 2 (now known as solute carrier family 2 [facilitated glucose transporter], member 2) and albumin in both HeLa and INS-1 cells. In vivo, these polymorphisms were associated with a modest but significant impairment in insulin secretion in response to oral glucose. Insulin secretion deteriorated over time in individuals carrying the V allele of the A98V polymorphism (n = 2,293; p = 0.003). In a new case–control (n = 1,511 and n = 2,225 respectively) data set, the I27L polymorphism was associated with increased risk of type 2 diabetes, odds ratio (OR) = 1.5 (p = 0.002; multiple logistic regression), particularly in elderly (age > 60 years) and overweight (BMI > 25 kg/m2) patients (OR = 2.3, p = 0.002).
Conclusions/interpretation
This study provides in vitro and in vivo evidence that common variants in the MODY3 gene, HNF-1α, influence transcriptional activity and insulin secretion in vivo. These variants are associated with a modestly increased risk of late-onset type 2 diabetes in subsets of elderly overweight individuals.
Similar content being viewed by others
Introduction
Type 2 diabetes is a late-onset polygenic disease, in which an affluent westernised environment interacts with genetic factors to bring about disease manifestation. The prevalence of the disease increases with age and bodyweight [1]. It is likely that genetic factors can identify individuals susceptible to changes in the environment. The greatest success in the genetics of type 2 diabetes has been the identification of genes causing MODY, whereas dissection of the genetic causes of late-onset type 2 diabetes has been less rewarding.
It has been speculated that milder variants in the same genes that cause early-onset monogenic forms can increase risk of late-onset type 2 diabetes. In support of this view, rare mutations in the genes encoding peroxisome proliferator-activated receptor-γ and potassium inwardly rectifying channel, subfamily J, member 11 (previously known as Kir6.2) have been shown to cause rare early-onset forms, whereas common variants in these genes increase the risk of late-onset type 2 diabetes [2, 3].
Hepatocyte nuclear factor-1 α (HNF-1_α_, now known as transcription factor-1 [TCF1]), which is responsible for MODY3, is a transcription factor regulating a number of liver- and beta-cell-specific genes. The gene is located on chromosome 12q24, which has shown linkage to late-onset type 2 diabetes [4–10], and a private G319S missense variant has been associated with type 2 diabetes in Canadian Oji-Cree individuals [11]. Common variations in the HNF-1α gene have been associated with impaired insulin secretion [12, 13]. We recently studied genetic variation in and across the gene by genotyping 21 common single nucleotide polymorphisms (SNPs) in 4,100 individuals. Several SNPs showed nominal association with type 2 diabetes, including rs1920792 and two missense mutations, rs1169288 (I27L) and rs1800574 (A98V) in the Scandinavian samples. However, these SNPs were not associated with type 2 diabetes, either in another sample of 4,400 individuals from North America and Poland, or in the combined sample of ∼ 9,000 persons [14]. A concurrent large association study testing 29 SNPs in HNF-1α in 5,307 individuals from the UK also concluded that common variation in HNF-1α is not associated with type 2 diabetes [15], with the exception of the rare A98V polymorphism (3%) [14, 15].
It has generally been thought that variants influencing beta cell function would result in earlier onset, while variants influencing insulin sensitivity would result in later onset of diabetes. However, this does not take into account the physiological interplay between insulin secretion and action, i.e. subtle defects in beta cell function should become more easily manifested during conditions of enhanced demand, e.g. insulin resistance as in elderly overweight individuals, as is the case for MODY mutations in the same gene [16].
To test our hypotheses we studied: (1) the role played by three common missense variants, I27L, A98V and S487N in transcriptional activity in vitro in terms of expression in cell systems; (2) their in vivo effect on insulin secretion; (3) their risk potential, in a new large case–control study, for type 2 diabetes in elderly overweight individuals; and (4) whether these variants affect changes in diabetes-related traits or increase risk of subsequent type 2 diabetes, in individuals followed prospectively for 7 to 20 years. In total 11,108 individuals were included in the study.
Subjects and methods
Plasmid constructs
In vitro mutagenesis was performed on full-length human HNF-1α cDNA using a site-directed mutagenesis kit (QuickChange [Multi]; Stratagene, La Jolla, CA, USA) (Electronic supplementary material [ESM] Table 1). Wild-type and mutant HNF-1α, L at position 27 (denoted L27), V at position 98 (V98), N at position 487 (N487), were subcloned into a pcDNA3.1 expression vector (Invitrogen, NV Leek, the Netherlands). DNA sequencing before expression studies verified sequences of the created constructs.
Transactivation assay
We cultured 1.5 × 105 HeLa cells in DMEM and 2.5 × 105 INS-1 cells in RPMI 1640 medium. Cultured cells were transfected using a reagent (LipofectAMINE PLUS; Life Technologies, Rockville, MD, USA) with 50 ng _HNF-1α-_pcDNA3.1, 0.5 μg hGLUT2-promoter/pGL3-basic luciferase vector (Promega, Madison, WI, USA) and 25 ng pRL-TK internal control vector (Promega). The transcriptional activity was measured after 24 h using the Dual Luciferase Assay System (Promega) on a multilabel counter (Victor2 Wallac 1420; PerkinElmer, Stockholm, Sweden). Each experiment was performed in triplicate, i.e. independently repeated three times.
Western blot analysis
HeLa cells and INS-1 cells were transfected with 5 μg wild-type or mutant _HNF-1α-_pcDNA3.1. Blotting was performed as described [17] using anti-HNF-1α (N-19) antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and horseradish peroxidase conjugate anti-goat IgG (Santa Cruz Biotechnology).
Electrophoretic mobility shift assay
HeLa cells were transfected with 5 μg of wild-type or mutant _HNF-1α -pcDNA3.1. After 24 h, nuclear protein extracts were prepared as previously described [18] and the electrophoretic mobility shift assay (EMSA) was performed [17] using 4 μg of nuclear extract with 32P-labelled glucose transporter type 2 (GLUT2, now known as solute carrier family 2 [facilitated glucose transporter], member 2 [SLC2A2]) promoter HNF-1_α binding site sequence (CTCAGTAAAGATTAACCAT) as the probe. A rabbit anti-serum was used for supershift, raised against the DNA-binding domain of HNF-1α.
Subjects
The case–control study
This included 1,511 Scandinavian cases with type 2 diabetes (from a local Diabetes Registry) and 2,225 unrelated ethnically matched control persons (from the Malmö Diet and Cancer Study [19]. Type 2 diabetes was diagnosed according to WHO criteria [20], with C-peptide concentrations ≥ 0.3 nmol/l, no GAD antibodies and age-at-onset > 35 years (Table 1). The control persons had fasting blood glucose 5.6 mmol/l, and no known family history of type 2 diabetes (Table 1).
Table 1 Clinical characteristics of participating subjects
Prospective studies
We monitored 2,293 (1,051 male/1,242 female) non-diabetic subjects participating in the Botnia study with repeated OGTTs every 2 to 3 years for a median period of 6 years (Table 1) [21]. Of these subject, 132 developed type 2 diabetes. To avoid the confounding effect of overt hyperglycaemia on beta cell function, only subjects who did not convert to type 2 diabetes were included in the study of change in insulin secretion over time.
A total of 4,873 subjects (2,836 males, 2,043 females) from the Malmö Prevention Project (MPP), followed for 15 to 25 years [22] (Table 1), were also included in a prospective analysis; of these subjects, 491 (372 males, 119 females) developed type 2 diabetes. Diabetes diagnosis was either based upon a clinical diagnosis or measurement of fasting plasma glucose (FPG) concentration during re-examination.
All subjects gave their informed consent to the study, which was approved by local ethics committees. Subjects with genetically verified MODY or type 1 diabetes were excluded.
Measurements
In the Botnia prospective study a 75-g OGTT with measurements of plasma glucose and insulin was performed as previously described [23]. Insulin resistance was estimated by the homeostasis model assessment (HOMA) (fasting serum insulin × FPG/22.5), while beta cell function was estimated as insulinogenic index (I/G30) (insulin 30 min-fasting insulin/glucose 30 min) and disposition index (DI) (I/G30/HOMA), which is a measure of beta cell function corrected for insulin resistance.
Arginine-glucose-stimulated insulin secretion
A total of 200 Swedish men (66 ± 2 years) underwent a stepwise assessment of acute insulin response to arginine at three different glucose concentrations (fasting, 14 and 28 mmol/l) [24]. The insulin response to arginine at a glucose concentration of 28 mmol/l was used as a surrogate measure of total insulin secretory capacity or beta cell mass.
Hyperinsulinaemic–euglycaemic clamp
The same 200 Swedish men underwent a hyperinsulinaemic–euglycaemic clamp, to obtain measures of insulin-stimulated glucose uptake (M value) [24].
Genotyping
Genotyping of rs1920792, rs1169288 (I27L), rs1800574 (A98V) and rs2464196 (S487N) was performed using the allelic discrimination technique (Applied Biosystems, Foster City, CA, USA) on an Applied Biosystems 7900HT instrument using the standard protocol (PCR primers and discrimination primers, see ESM Table 2). Genotyping error was determined to 0.1%, using duplicate genotypes with 5% re-genotyping.
Statistical analysis
The data from the dual luciferase assay were analysed by Mann–Whitney and expressed as mean ± SD. A p value of < 0.05 was considered statistically significant if the normalised luciferase activity was at least 1.5 times that of its comparator [25].
Normally distributed continuous variables of in vivo measurements are presented as mean ± SD, whereas non-normally distributed data were logarithmically transformed before analysis and presented as median with interquartile range. Nominal p values for regression analysis, general estimation equation (GEE) and Cox survival analysis are presented. A p value of < 0.05 was considered statistically significant.
Power for the association study was calculated using the genetic Power Calculator (http://statgen.iop.kcl.ac.uk/gpc/) [26]. Type 2 diabetes disease prevalence was assumed to be 6% both under a dominant model (relative risk [RR] = 1.25) and a recessive model (RR = 1.45) (α = 0.05). Multivariate logistic regression analyses were performed adjusting for age (controls), age-at-onset (cases), BMI and sex.
Prospective studies
Baseline levels and rate change of phenotype residuals between different genotype carriers in the Botnia prospective study were calculated using linear regression analysis adjusted for age, BMI, sex and family history of type 2 diabetes. Standard errors were adjusted for repeated measurements from the same individual by the GEE method [27, 28], since the repeated measurements of I/G30 were obtained at different time points in different subjects. Phenotypic data from each visit, except the visit of diagnosis for converters and the last visit for the non-converters, were used in the analysis.
Cox proportional hazards model was used to estimate relative genotype and phenotype effect on the risk of developing type 2 diabetes. Survival analyses were stratified for sex and adjusted for family history of diabetes and BMI [29]. A robust variance estimate was used to adjust for within-pedigree dependence, treating each pedigree as an independent entity when calculating the variance of the estimates. Power calculations were performed using the asymptotic normality of the estimates.
Interaction analysis between genotype and phenotype was performed in the Botnia prospective cohort and analysed by multivariate Cox analysis with the model for the hazard h(t) = h 0(t)*_e_^(β1*genotype + β2*phenotype + β3*genotype*phenotype). In this formula, β1 is the single effect of the genotype, β2 the single effect of the phenotype and β3 measures the interaction between genotype and phenotype [29].
All statistical analyses were performed using STATA (StataCorp, College Station, TX, USA) and/or Number Cruncher Statistical Systems, version 2000 (NCSS, Kaysville, UT, USA).
Results
Common variants in the HNF-1α gene influence transcription
Functional consequences of three non-synonymous polymorphisms (I27L, A98V and S487N) were studied comparing the ability of in vitro mutated HNF-1α and the wild-type protein to activate the human GLUT2 promoter. Two cell lines were used: HeLa cells which lack endogenous HNF-1α, and INS-1 cells expressing the corresponding rat Hnf-1α.
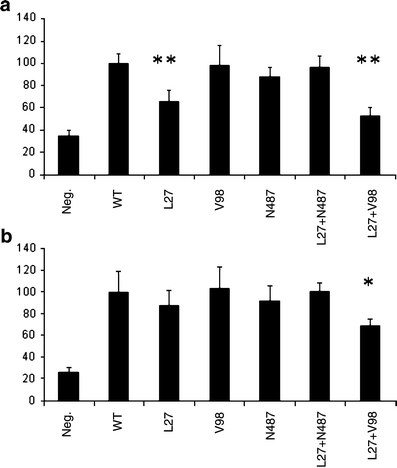
Using the GLUT2 promoter in the reporter construct, we observed significantly decreased transcriptional activity in HeLa cells for the L27 (−30%, p < 0.0005) and for the combination of L27 and V98 (−50%, p < 0.0005). In INS-1 cells only the combination of L27 and V98 (−30%, p < 0.05) showed significantly decreased activity (Fig. 1). Western blot analysis confirmed equal expression of the mutated and wild-type protein (data not shown).
Fig. 1
Results from in vitro experiments using HeLa (a) and INS-1 (b) cells with genetically different HNF-1α using the GLUT2 promoter as the reporter gene. Transcriptional activity was measured in both cell types. Neg, empty vector; WT, wild-type. *p < 0.05, **p < 0.005
The different variants of HNF-1α were analysed by EMSA to exclude the possibility that this effect was due to impaired DNA-binding capacity. None of the mutated HNF-1α proteins showed differences in binding compared with wild-type protein (ESM Fig. 1).
Genotype frequencies
Both the genotyping of the four SNPs rs1920792 (C/T), I27L (T/G), A98V (C/T) and S487N (C/T) in 6,270 individuals from Botnia, Finland and Malmö, Sweden, and the genotyping, in an additional cohort of 4,873 individuals (MPP), of I27L and A98V gave similar genotype frequencies (Table 2) and D′ values (measure of linkage disequilibrium [LD]), as previously published [14], with results in Hardy–Weinberg equilibrium.
Table 2 Genotype frequencies
Effects of common variants in the HNF-1α gene on measures of insulin secretion and action in vivo
The effect of the polymorphisms on insulin secretion in vivo was assessed (1) as insulin response to oral glucose, and (2) as insulin response to arginine at 28 mmol/l of glucose, which is a measure of maximum insulin secretory capacity and was used as a surrogate measure of beta cell mass in a subgroup of individuals. The effect of the polymorphisms on insulin sensitivity was assessed by a hyperinsulinaemic–euglycaemic clamp in a subgroup of individuals and expressed as insulin-stimulated glucose uptake (M value).
Multiple regression analyses were performed including 2,293 individuals from the Botnia prospective cohort at baseline and last visits (Table 1). At baseline visit, individuals carrying the LL genotype of the I27L polymorphism showed reduced I/G30 (p = 0.03) and DI (p = 0.01) compared with the II-genotype carriers. The IL-genotype carriers also had reduced I/G30 (p < 0.05) and DI (p = 0.03). Individuals with the V allele of the A98V polymorphism showed reduced I/G30 (p = 0.004) compared with individuals with the AA genotype. At the last visit, carriers of the LL genotype had lower I/G30 (p = 0.01) and DI (p = 0.01) than individuals with the II genotype. The DI was also lower for the heterozygous IL-genotype carriers (p = 0.02). Carriers of the V allele of the A98V polymorphism showed both significantly reduced I/G30 (p = 0.0004) and DI (p = 0.0003), compared with carriers of the AA genotype (Table 3).
Table 3 Results from prospective and case–control cohorts
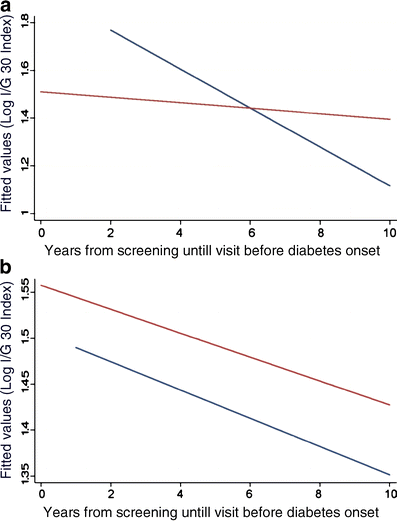
Although these data suggested impaired beta cell function both at entry and at the end of follow-up, they did not show whether there was a true deterioration of beta cell function over time. To address this question, we applied a linear regression analysis with GEE adjusted for age, BMI, sex and family clustering in the 2,293 subjects followed for 6 years. In fact, individuals with the V allele of the A98V polymorphism showed significantly reduced I/G30, (p = 0.003) compared with AA-genotype carriers (Fig. 2a), whereas no significant changes either in I/G30 or DI over time were observed for the I27L and S487N polymorphisms (Fig. 2b).
Fig. 2
Change in insulin secretion (insulinogenic index) over time in the Botnia prospective cohort using a linear regression analysis in (a) carriers of the AV or VV genotypes compared with carriers of the AA genotype (p = 0.003) of the A98V polymorphism. Blue line, AA genotype; red line, AV and VV genotypes. (b) Change as above in carriers of the IL and LL genotypes compared with carriers of the II genotype of the I27L polymorphism (p = 0.4), in whom there was a parallel decline in insulin secretion over time. Blue line, II genotype; red line, IL and LL genotypes
Glucose-potentiated arginine-stimulated maximal insulin secretion among 200 men from the Malmö Preventive Trial study [24, 30, 31] showed no significant differences in insulin secretion between the different genotype carriers, suggesting that the polymorphisms had no large effect on this surrogate measure of beta cell mass (data not shown). Also, carriers of the V allele of the A98V polymorphism showed higher M values corrected for lean body mass than carriers of the AA genotype (7.1 ± 2.9 vs 5.5 ± 2.7 mg kg fat-free mass−1 min−1, p = 0.03), suggesting enhanced insulin sensitivity. In keeping with these findings, control individuals carrying the V allele in the case–control study had lower HOMA-IR values, indicating better insulin sensitivity than the AA-genotype carriers (1.5 ± 0.8 vs 1.7 ± 1.5, p = 0.02).
Association between SNPs in the HNF-1α gene and type 2 diabetes in elderly overweight individuals
The rs1920792, I27L, A98V and S487N were genotyped in 1,511 type 2 diabetic patients and 2,225 controls of Scandinavian origin (Table 2). Two of the investigated SNPs (I27L and S487N) showed nominally significant allelic association to type 2 diabetes (odds ratio [OR] = 1.1, p = 0.02 and OR = 1.1, p = 0.03, Table 3); however, when permutated 10,000 times, this was no longer significant (p = 0.06 and p = 0.08), although power calculations showed > 75% power to detect an association under a dominant (RR = 1.25) and recessive model (RR = 1.45). We also evaluated haplotype block structure between the four SNPs using the Haploview 3.2 program [32], in which D′ values were calculated with 95% CIs [33]. The four SNPs were not part of a common haplotype block, although three of the four variants (rs1920792, I27L and S487N) showed strong LD with D′ between 0.93 and 1 (ESM Table 3). The I27L and S487N were not in LD with A98V (D′ < 0.3), which is in line with other findings [14].
In multivariate logistic regression analysis without adjustments, the LL genotype of I27L was significantly associated with type 2 diabetes compared with the II genotype with an OR = 1.4 (1.2–1.8) (p = 0.0009) and when adjusting for sex, BMI and age, OR = 1.5 (1.2–1.9) (p = 0.002). Meta analyses of published data support the association with type 2 diabetes (ESM Figs. 2 and 3). Initially we hypothesised that the HNF-1α variants might play a role in deterioration of insulin secretion, especially in elderly and overweight individuals, since age and BMI are strong predictors of type 2 diabetes. This was in fact the case in individuals above the median of 60 years of age (410 cases, age at onset 66 ± 6 years; 1,036 controls, age at visit 64 ± 2 years) where the OR increased to 2.1 (1.3–3.2) (p = 0.002) for the LL-genotype carriers. The risk was also more pronounced in overweight individuals, BMI > 25, (1,129 cases, BMI 31 ± 5; 1,076 controls, BMI 28 ± 3), OR = 1.5 (1.1–2.0) (p = 0.009). The combined analysis of individuals above 60 years with BMI >25 (336 cases, BMI 31 ± 4, age at onset 67 ± 5; 520 controls, BMI 28 ± 2, age at visit 64 ± 2) yielded an OR = 2.2 (1.3–3.7) (p = 0.003) (Table 3). Of note, re-analysis of previously published case–control data from Botnia [14] revealed OR = 1.4, p = 0.2 (n = 866) in all individuals, whereas in overweight individuals OR = 1.7, p = 0.06 (n = 596). Combining these published case–controls (n = 4,433) and our data further strengthened the results to OR = 1.5, p = 0.0008 for all individuals and OR = 1.5, p = 0.001 (n = 2,801) for the overweight individuals.
The I27L polymorphism in the HNF-1α predicts future risk of type 2 diabetes
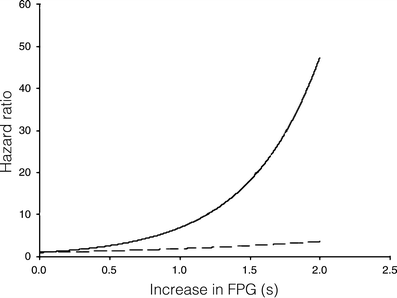
In the Botnia prospective study, 132 of the 2,293 individuals converted to type 2 diabetes during the median follow-up of 6 years [29]. The I27L polymorphism did not confer an increased risk of future type 2 diabetes (hazard ratio [HR] = 0.8, p = 0.2) in this population. Interestingly, given the low number of converters, the power was only 30%. However, the LL genotype of the I27L polymorphism showed a significant interaction with FPG concentration (β1 [I27L] = −8.3, p = 0.03; β2 [FPG] = 0.6, p < 0.0001; β3 [I27L*FPG] = 1.3, p = 0.03), i.e. the risk of type 2 diabetes increased more rapidly for an individual with the LL genotype and increasing fasting glucose concentrations than for carriers of the II genotype with the same increase in fasting glucose (Fig. 3). Similarly the NN genotype of the S487N polymorphism showed a significant interaction with FPG (β1 [S487N] = −7.5 p = 0.05; β2 [FPG] = 0.7 p < 0.001; β3 [S487N*FPG] = 1.2, p = 0.05).
Fig. 3
Interaction analysis in the Botnia prospective cohort showing a more rapid increase in type 2 diabetes risk with increasing fasting plasma glucose (FPG) levels (s, units of FPG increase) for individuals with the LL genotype (risk genotype, solid line) of HNF-1α compared with individuals with the IL or II genotypes (non-risk genotypes, dashed line) of the I27L polymorphism; β1(I27L) = −8.3, p = 0.03; β2(FPG) = 0.6, p < 0.0001; β3(I27L*FPG) = 1.3, p = 0.03, where β1 is the single effect of the genotype, β2 is the single effect of the phenotype and β3 gives the interaction between genotype and phenotype
In the MPP cohort the L allele was associated with a modestly increased risk of type 2 diabetes as compared with the II genotype in overweight individuals (BMI > 25), HR = 1.3 (1.0–1.6), p = 0.04 (Table 3). The power to find an association to diabetes was 33% for the L allele of the I27L polymorphism and 21% for the V allele of the A98V polymorphism. No interaction between the polymorphisms was observed in any of the two cohorts (data not shown).
Discussion
In this study we found that certain polymorphisms in the HNF-1α gene, i.e. I27L, A98V and S487N were associated with: (1) reduced transcriptional activity in vitro; (2) lower glucose-stimulated insulin secretion in vivo; (3) a further deterioration of insulin secretion over a 6-year period; (4) enhanced insulin sensitivity; and (5) increased risk of type 2 diabetes in elderly overweight individuals.
When expressing combinations of HNF-1α variants in HeLa cells, we observed a reduction in transcriptional activity for the L27 variant irrespective of the variant at codon 98 (Fig. 1a). Similar results were observed using a rat albumin promoter (data not shown). The reduction in transcriptional activity in carriers of L27 was about 40%, which should be compared with an approximately 90% reduction in carriers of the most common MODY 3 mutations, the P291fsinsC mutation (data not shown). Although the finding was, as expected, less pronounced in INS-1 cells (Fig. 1b) expressing endogenous HNF-1α, this clearly suggests that both L27 and V98 influence transcription. Some caution is, however, warranted in generalising data from transient expression studies to the in vivo situation. The I27L polymorphism resides in the dimerisation domain and the A98V is located two amino acids upstream of the DNA-binding domain, which are well-conserved regions between human, rat and mouse. Replacing the amino acids could result in a small conformational change and thereby alter the function or the expression of the protein. Theoretically, impairment in dimerisation could influence DNA binding. Although we did not directly measure dimerisation, no change in DNA binding was observed using EMSA. The nearest mutation previously studied is the L107I mutation, which has an effect on transcriptional activity and causes MODY [34]. No functional studies have previously been performed on mutations between amino acids 12 and 107.
The in vitro observation of decreased transcriptional activity was reflected in vivo by a reduced insulin response to oral glucose. We observed a consistent decline in the 30-min insulin response to the OGTT (I/G30) over time in carriers of the A98V polymorphism. This is in line with earlier cross-sectional findings of a 20% decrease in C-peptide concentrations at 30 min in carriers of the A98V polymorphism [35] and reduced first- and second-phase insulin secretion during a hyperglycaemic clamp in carriers of the I27L polymorphism [13, 36]. The 30-min insulin response reflects an early dynamic insulin response most probably dependent upon glucose being metabolised in the beta cells. This could be compatible with the decreased transcription of GLUT2 in the cell lines. It has also been suggested that HNF-1α might influence islet size and, indeed, the Hnf-1α −/− mouse is characterised by decreased islet mass [37]. We used the insulin response to arginine, measured at high glucose concentrations of 28 mmol/l, as an in vivo surrogate measure of islet mass, as this maximum response should be dependent upon total islet mass. We could not, however, demonstrate any effect of the variants on the maximum insulin response, suggesting that these variants may not have a great effect on islet mass. Of note, this was a post hoc analysis in a limited number of subjects.
Importantly, the V-allele carriers also showed increased insulin sensitivity, as estimated from increased M value during the clamp and reduced HOMA-IR values. This is in line with earlier findings that carriers of mutations that cause MODY3 show increased insulin sensitivity [16, 38]. Nothing, however, is known of the molecular mechanisms by which variations in HNF-1α could influence insulin sensitivity. The enhanced insulin sensitivity might be an important means of counteracting the untoward effects of impaired beta cell function on glucose tolerance [16].
In our previous large study [14], we found a nominal association between SNPs in HNF-1α and type 2 diabetes in the Scandinavian cohort, but there was no significant effect in the entire sample of ∼ 9,000 individuals, which also included patients from Poland and USA, and this despite the fact that a nominal test of heterogeneity was negative. To further address this issue in Scandinavians, a new case–control study with cases from the local Diabetes Registry and ethnically matched controls from the Malmö European Prospective Investigation into Cancer and Nutrition cohort [22] were collected. Although we found no allelic association to type 2 diabetes, individuals carrying the LL genotype had an increased risk of developing the disease, particularly if they were overweight and > 60 years of age.
These findings are compatible with the hypothesis that the risk of developing type 2 diabetes for any variant influencing insulin secretion should be increased in obese elderly individuals, whose beta cells have been exposed to the stress of insulin resistance for a long period of time. In support of this, in the prospective Malmö cohort, the I27L polymorphism predicted subsequent type 2 diabetes in overweight individuals followed for 25 years. These findings suggest that the metabolic consequences of impaired beta cell function induced by the polymorphisms in HNF-1α are greatest in insulin-resistant individuals, but could be alleviated by the simultaneous enhancement of insulin sensitivity.
Clearly, we cannot deny the possibility of false positive results, given the number of tests performed for association of insulin secretion with four genetic variants in HNF-1α, and the level of significance used. Since we restricted the analysis to two measures of insulin secretion (I/G30, DI; r 2 = 0.7), most of the results would stand a Bonferroni correction. However, it was reassuring to see that the associations found at nominal p values were observed with consistency in the different subsets, supporting the notion that common polymorphisms in HNF-1α could be associated with impaired transcriptional activity and consequently with impaired insulin secretion.
In conclusion, our data show that certain combinations of common variants in HNF-1α are associated with decreased transcriptional activity in vitro and a decreased insulin response to oral glucose in vivo, which further decreases with time. The I27L polymorphism seems to be associated with an increased risk of type 2 diabetes, particularly in elderly overweight individuals. In future studies of polymorphisms influencing insulin secretion, the central role of insulin resistance in triggering the disease should be considered.
Abbreviations
DI:
disposition index
EMSA:
electrophoretic mobility shift assay
FPG:
fasting plasma glucose
GEE:
general estimation equation
GLUT2:
glucose transporter type 2 (now known as solute carrier family 2 [facilitated glucose transporter], member 2 [SLC2A2])
HNF-1α :
hepatocyte nuclear factor 1-α (now known as transcription factor 1 [_TCF1_]
HOMA:
homeostasis model assessment
HR:
hazard ratio
I/G30:
insulinogenic index
LD:
linkage disequilibrium
MPP:
Malmö Preventive Project
OR:
odds ratio
RR:
relative risk
SNP:
single nucleotide polymorphism
References
- Zimmet P, Alberti KGMM, Shaw J (2001) Global and societal implications of the diabetes epidemic. Nature 414:782–787
Article PubMed CAS Google Scholar - Gloyn AL, Weedon MN, Owen KR et al (2003) Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes 52:568–572
PubMed CAS Google Scholar - Altshuler D, Hirschhorn JN, Klannemark M et al (2000) The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26:76–80
Article PubMed CAS Google Scholar - Mahtani MM, Widen E, Lehto M et al (1996) Mapping of a gene for type 2 diabetes associated with an insulin secretion defect by a genome scan in Finnish families. Nat Genet 14:90–94
Article PubMed CAS Google Scholar - Lindgren CM, Mahtani MM, Widen E et al (2002) Genomewide search for type 2 diabetes mellitus susceptibility loci in Finnish families: the Botnia study. Am J Hum Genet 70:509–516
Article PubMed CAS Google Scholar - Ehm GM, Karnoub MC, Sakul H et al, the American Diabetes Association GENNID Study Group (2000) Genomewide search for type 2 diabetes susceptibility genes in four American populations. Am J Hum Genet 66:1871–1881
Article PubMed CAS Google Scholar - Bowden DW, Sale M, Howard TD et al (1997) Linkage of genetic markers on human chromosomes 20 and 12 to NIDDM in Caucasian sib pairs with a history of diabetic nephropathy. Diabetes 46:882–886
PubMed CAS Google Scholar - Shaw JT, Lovelock PK, Kesting JB et al (1998) Novel susceptibility gene for late-onset NIDDM is localized to human chromosome 12q. Diabetes 47:1793–1796
PubMed CAS Google Scholar - Wiltshire S, Frayling TM, Groves CJ et al (2004) Evidence from a large UK family collection that genes influencing age of onset of type 2 diabetes map to chromosome 12p and to the MODY3/NIDDM2 locus on 12q24. Diabetes 53:855–860
PubMed CAS Google Scholar - Reynisdottir I, Thorleifsson G, Benediktsson R et al (2003) Localization of a susceptibility gene for type 2 diabetes to chromosome 5q34–q35.2. Am J Hum Genet 73:323–335
Article PubMed CAS Google Scholar - Hegele RA, Cao H, Harris SB, Hanley AJG, Zinman B (1999) The hepatic nuclear factor-1{alpha} G319S variant is associated with early-onset type 2 diabetes in Canadian Oji-Cree. J Clin Endocrinol Metab 84:1077–1082
Article PubMed CAS Google Scholar - Urhammer S, Fridberg M, Hansen T et al (1997) A prevalent amino acid polymorphism at codon 98 in the hepatocyte nuclear factor-1alpha gene is associated with reduced serum C-peptide and insulin responses to an oral glucose challenge. Diabetes 46:912–916
PubMed CAS Google Scholar - Chiu KC, Chuang L-M, Chu A, Wang M (2003) Transcription factor 1 and beta-cell function in glucose-tolerant subjects. Diabet Med 20:225–230
Article PubMed CAS Google Scholar - Winckler W, Burtt NP, Holmkvist J et al (2005) Association of common variation in the HNF1{alpha} gene region with risk of type 2 diabetes. Diabetes 54:2336–2342
PubMed CAS Google Scholar - Weedon MN, Owen KR, Shields B et al (2005) A large-scale association analysis of common variation of the HNF1{alpha} gene with type 2 diabetes in the UK Caucasian population. Diabetes 54:2487–2491
PubMed CAS Google Scholar - Tripathy D, Carlsson Ã-L, Lehto M, Isomaa B, Tuomi T, Groop L (2000) Insulin secretion and insulin sensitivity in diabetic subgroups: studies in the prediabetic and diabetic state. Diabetologia 43:1476–1483
Article PubMed CAS Google Scholar - Bjorkhaug L, Ye H, Horikawa Y, Sovik O, Molven A, Njolstad PR (2000) MODY associated with two novel hepatocyte nuclear factor-1[alpha] loss-of-function mutations (P112L and Q466X). Biochem Biophys Res Commun 279:792–798
Article PubMed CAS Google Scholar - Vaxillaire M, Abderrahmani A, Boutin P et al (1999) Anatomy of a homeoprotein revealed by the analysis of human MODY3 mutations. J Biol Chem 274:35639–35646
Article PubMed CAS Google Scholar - Lindholm E, Agardh E, Tuomi T, Groop L, Agardh CD (2001) Classifying diabetes according to the new WHO clinical stages. Eur J Epidemiol 17:983–989
Article PubMed CAS Google Scholar - Alberti KG, Zimmet PZ (1998) Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med 15:539–553
Article PubMed CAS Google Scholar - Groop L, Forsblom C, Lehtovirta M et al (1996) Metabolic consequences of a family history of NIDDM (the Botnia study): evidence for sex-specific parental effects. Diabetes 45:1585–1593
PubMed CAS Google Scholar - Berglund G, Nilsson P, Eriksson K-F et al (2000) Long-term outcome of the Malmö Preventive Project: mortality and cardiovascular morbidity. J Intern Med 247:19–29
Article PubMed CAS Google Scholar - Lyssenko V, Almgren P, Anevski D et al (2005) Predictors of and longitudinal changes in insulin sensitivity and secretion preceding onset of type 2 diabetes. Diabetes 54:166–174
PubMed CAS Google Scholar - Tripathy D, Eriksson KF, Orho-Melander M, Fredriksson J, Ahlqvist G, Groop L (2004) Parallel manifestation of insulin resistance and beta cell decompensation is compatible with a common defect in Type 2 diabetes. Diabetologia 47:782–793
Article PubMed CAS Google Scholar - Hoogendoorn B, Coleman SL, Guy CA et al (2003) Functional analysis of human promoter polymorphisms. Hum Mol Genet 12:2249–2254
Article PubMed CAS Google Scholar - Purcell S, Cherny SS, Sham PC (2003) Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19:149–150
Article PubMed CAS Google Scholar - Liang K, Zeger S (1986) Longitudinal data analysis using generalized linear models. Biometrika 73:13–22
Article Google Scholar - Liang K, Zeger S (1993) Regression analysis for correlated data. Annu Rev Public Health 4:43–68
Article Google Scholar - Lyssenko V, Almgren P, Anevski D et al (2005) Genetic prediction of future type 2 diabetes. PLoS Medicine 12:1299–1308
Google Scholar - Eriksson K, Lindgarde F (1990) Impaired glucose tolerance in a middle-aged male urban population: a new approach for identifying high-risk cases. Diabetologia 33:526–531
Article PubMed CAS Google Scholar - Eriksson K, Lindegarde F (1991) Prevention of type 2 (non-insulin-dependent) diabetes mellitus by diet and physical exercise. The 6-year Malmö feasibility study. Diabetiologia 34:891–898
Article CAS Google Scholar - Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265
Article PubMed CAS Google Scholar - Gabriel SB, Schaffner SF, Nguyen H et al (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229
Article PubMed CAS Google Scholar - Cervin C, Orho-Melander M, Ridderstrale M et al (2002) Characterization of a naturally occurring mutation (L107I) in the HNF1 alpha (MODY3) gene. Diabetologia 45:1703–1708
Article PubMed CAS Google Scholar - Urhammer SA, Moller AM, Nyholm B et al (1998) The effect of two frequent amino acid variants of the hepatocyte nuclear factor-1{alpha} gene on estimates of the pancreatic {beta}-cell function in Caucasian glucose-tolerant first-degree relatives of type 2 diabetic patients. J Clin Endocrinol Metab 83:3992–3995
Article PubMed CAS Google Scholar - Chiu KC, Chuang L-M, Ryu JM, Tsai GP, Saad MF (2000) The I27L amino acid polymorphism of hepatic nuclear factor-1{alpha} is associated with insulin resistance. J Clin Endocrinol Metab 85:2178–2183
Article PubMed CAS Google Scholar - Pontoglio M, Sreenan S, Roe M et al (1998) Defective insulin secretion in hepatocyte nuclear factor 1alpha-deficient mice. J Clin Invest 101:2215–2222
Article PubMed CAS Google Scholar - Stride A, Ellard S, Clark P et al (2005) {beta}-Cell dysfunction, insulin sensitivity, and glycosuria precede diabetes in hepatocyte nuclear factor-1{alpha} mutation carriers. Diabetes Care 28:1751–1756
PubMed CAS Google Scholar
Acknowledgements
This work was supported by grants from the Swedish Research Council, the Novo Nordisk Foundation, the Söderberg Foundation, the Sigrid Juselius Foundation, The Folkhälsan Research Foundation, the Lundberg Foundation, af Ugglas Stiftelse, Alex and Eva Wallströms Stiftelse, the Heart- and Lung Foundation Sweden and the Diabetes Program Lund University. The supershift antibody was kindly provided R. Cortese (Department of Cellular and Molecular Biology, IRMB, Rome, Italy), the GLUT2/pGL3 was provided by J. Takeda (Department of Social Environmental Medicine H3, Osaka University, Osaka, Japan). DNA extraction of the MPP samples was performed at Swegene PPD, Lund University. We thank K. F. Eriksson (Department of Clinical Sciences — Diabetes & Endocrinology, Malmö University Hospital, Lund University, Malmö, Sweden ) for the data from the Malmö Preventive Trial study, and naturally all participants for making this project possible.
Duality of interest
The authors of this manuscript have no dualities of interest. T. Tuomi is a member of the Nordic Cardiovascular Advisory Board, Novartis. L. Groop has been a consultant for and served on advisory boards for Aventis-Sanofi, Bristol Myers Squibb, GSK, Kowa and Roche. These consultancies are not directly related to this manuscript.
Author information
Authors and Affiliations
- Department of Clinical Sciences, Diabetes and Endocrinology, Clinical Research Center, Malmö University Hospital, Lund University, S-205 02, Malmö, Sweden
J. Holmkvist, C. Cervin, V. Lyssenko, D. Anevski, C. Cilio, P. Almgren & L. Groop - Program in Medical and Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MA, USA
W. Winckler & D. Altshuler - Department of Molecular Biology, Center for Human Genetic Research and Diabetes Unit, Massachusetts General Hospital, Boston, MA, USA
W. Winckler & D. Altshuler - School of Mathematical Sciences, Chalmers University of Technology, Gothenburg, Sweden
D. Anevski - Department of Medicine, Malmö University Hospital, Lund University, Malmö, Sweden
G. Berglund & P. Nilsson - Department of Medicine, Helsinki University Central Hospital, and Research Program of Molecular Medicine, University of Helsinki, Helsinki, Finland
T. Tuomi & L. Groop - Folkhalsan Research Centre, Helsinki, Finland
T. Tuomi - Department of Biosciences at Novum, Karolinska Institute, Huddinge, Sweden
C. M. Lindgren - Clinical Research Centre, Karolinska University Hospital, Huddinge, Sweden
C. M. Lindgren
Authors
- J. Holmkvist
You can also search for this author inPubMed Google Scholar - C. Cervin
You can also search for this author inPubMed Google Scholar - V. Lyssenko
You can also search for this author inPubMed Google Scholar - W. Winckler
You can also search for this author inPubMed Google Scholar - D. Anevski
You can also search for this author inPubMed Google Scholar - C. Cilio
You can also search for this author inPubMed Google Scholar - P. Almgren
You can also search for this author inPubMed Google Scholar - G. Berglund
You can also search for this author inPubMed Google Scholar - P. Nilsson
You can also search for this author inPubMed Google Scholar - T. Tuomi
You can also search for this author inPubMed Google Scholar - C. M. Lindgren
You can also search for this author inPubMed Google Scholar - D. Altshuler
You can also search for this author inPubMed Google Scholar - L. Groop
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toL. Groop.
Additional information
Authors J. Holmkvist and C. Cervin contributed equally to this work.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Holmkvist, J., Cervin, C., Lyssenko, V. et al. Common variants in HNF-1 α and risk of type 2 diabetes.Diabetologia 49, 2882–2891 (2006). https://doi.org/10.1007/s00125-006-0450-x
- Received: 30 March 2006
- Accepted: 25 July 2006
- Published: 11 October 2006
- Issue Date: December 2006
- DOI: https://doi.org/10.1007/s00125-006-0450-x