Noninvasive Visualization of microRNA by Bioluminescence Imaging (original) (raw)
MicroRNAs (miRNAs) are non-coding endogenous RNA regulators of gene expression that are approximately 21-nucleotide long [1]. Since its discovery in 1993 [2], the regulatory function of miRNA has been extensively studied. MiRNAs silence gene expression mainly by suppressing mRNA translation and enhancing mRNA degradation [3]. MiRNA dysregulation is common in cancer cells and has been implicated in carcinogenesis via its role in regulating the expression of a slew of well-known oncogenes and tumor suppressor genes [4]. Specific subsets of overexpressed or downregulated miRNAs have been identified in various cancer types, including those originating from the lung, breast and thyroid [5–7]. For instance, Nikiforova et al. [7] investigated the expression patterns of miRNA in all major types of thyroid tumors and found that various histopathological types of thyroid tumors, including those derived from the same cell type, showed significantly different profiles of miRNA expression. A set of seven miRNAs, including miR-221, were most differentially overexpressed in thyroid tumors versus hyperplastic nodules in the surgical samples. Another study found that miR-146b was the most consistently overexpressed miRNA in both classical papillary carcinoma and follicular variants of thyroid origin [8].
Owing to their important regulatory function in both malignant diseases and other pathological processes, miRNAs have been investigated as therapeutic targets. A class of chemically engineered oligonucleotides, termed “antagomirs”, has been utilized as specific silencers of endogenous miRNAs in mice [9]. Intravenous administration of antagomirs against miR-16, miR-122, miR-192 and miR-194 resulted in a marked reduction of corresponding miRNA levels in various organs. Gene expression and bioinformatic analysis of mRNA from antagomir-treated animals revealed that the 3′ untranslated regions of upregulated genes were strongly enriched in miR-122 recognition motifs, whereas downregulated genes were depleted in these motifs. The miR-122 was also inhibited in mice with a 2′-_O_-methoxyethyl phosphorothioate ASO (antisense oligonucleotide) [10]. Recently, the experiments performed in non-human primates showed that the systemic delivery of an unconjugated, PBS-formulated locked nucleic-acid-modified oligonucleotide (LNA-antimiR) effectively antagonized the liver-expressed miR-122 [11].
Endogenous miRNA levels can be evaluated by techniques such as northern blotting, RT-PCR, and microarrays after the extraction of total RNAs from biopsy samples [5–7]. Taylor and Gercel-Taylor [12] have analyzed miRNA profile in isolated circulating tumor-derived exosomes. They found that the levels of eight specific microRNAs for human ovarian cancer were similar between cellular and exosomal microRNAs. This technique can be used as potential surrogate diagnostic markers for biopsy profiling. However, the aforementioned methods are time-consuming and cannot provide real-time information of miRNA changes.
In this issue of Molecular Imaging & Biology, Kim et al. [[13](/article/10.1007/s11307-008-0190-z#ref-CR13 "Kim HJ, Chung JK, Lee DS, Kim SH (2008) In vivo imaging of miR-221 biogenesis in papillary thyroid carcinoma. Mol Imaging Biol doi: 10.1007/s11307-008-0188-6
")\] reported a novel bioluminescence imaging (BLI) strategy to evaluate and visualize the expression level of miR-221\. MiR-221 is upregulated in human hepatocellular carcinoma (HCC) as well as in other malignancies \[[7](/article/10.1007/s11307-008-0190-z#ref-CR7 "Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE (2008) MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab 93:1600–1608")\]. The direct targets of miR-221 include CDKN1B/p27, a key negative regulator of the cell cycle, and CDKN1C/p57, a cyclin-dependent kinase inhibitor (CDKI) \[[14](/article/10.1007/s11307-008-0190-z#ref-CR14 "Gillies JK, Lorimer IA (2007) Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle 6:2005–2009"), [15](/article/10.1007/s11307-008-0190-z#ref-CR15 "Fornari F, Gramantieri L, Ferracin M et al (2008) MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 27:5651–5661")\]. Kim et al. constructed a miR-221-specific vector in which Gaussian luciferase (Gluc) is driven by a CMV promoter. After the Gluc stop codon, three repeat perfect complementary target sequences (3×PT) of mature miR-221 were inserted. Endogenous miR-221 is expected to bind with the transcript of this vector, thus suppressing the translation of Gluc or degrading the transcript, resulting in a decrease of BLI signal. In this way, miR-221 expression level can be indirectly visualized and assessed. The results demonstrated that Gluc activities were repressed in a dose-dependent manner after co-transfected CMV/Gluc-3×PT\_miR221with cognate pre-miRNA. The specificity of this method was confirmed by the fact that the Gluc activities of CMV/Gluc-3×PT\_miR221 in HT-ori3, NPA and TPC-1 cells were dramatically retrieved when they were co-transfected with anti-miR-221\. _In vivo_ real-time bioluminescence imaging of miR-221 production and specificity related to papillary thyroid carcinoma (PTC) was also successfully achieved \[[13](/article/10.1007/s11307-008-0190-z#ref-CR13 "Kim HJ, Chung JK, Lee DS, Kim SH (2008) In vivo imaging of miR-221 biogenesis in papillary thyroid carcinoma. Mol Imaging Biol doi:
10.1007/s11307-008-0188-6
")\].This bioluminescence imaging strategy can be expanded to image other miRNAs. Indeed, the same group has applied the same technique to monitor the expression and regulation of another miRNA, miR-23a [16]. They developed a dual-luciferase reporter system in which the firefly luciferase (Fluc) was under the control of miR-23a promoter to monitor the transcriptional level of miR-23a, and a cytomegalovirus (CMV) promoter driven Gluc gene was fused with three copies of the target sequence of miR-23a before the poly(A) tail to monitor the targeting activity of mature miR-23a. With the seminal dual-luciferase reporter system, the endogenous expression of pre-miRNA and the activity of mature miRNA within the same cell population can be visualized simultaneously [16]. In another study, they successfully monitored the changes of both miR-9 and miR-9* during the neuronal differentiation of P19 cells by bioluminescent imaging [17].
The paper from Kim et al. in this issue of MIB provides a novel imaging strategy to non-invasively monitor miR-221 expression level in real-time manner. The high sensitivity of luciferase reporter gene in combination with miRNA-specific vectors facilitates the quantitative visualization of miRNA expression. Although luciferase reporter system has limited clinical application [18] at the present, this cutting-edge technique will greatly assist our understanding of the biological function of miRNAs in a living organism. In addition, the BLI imaging system can work as a platform for high-throughput screening of drugs related to miRNAs. Finally, the non-invasive imaging strategy provides a powerful tool to monitor the efficacy of anti-miRNA therapy because the miRNA levels can be monitored longitudinally along with the therapeutic process.
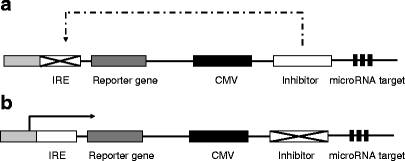
However, there are several concerns about this particular reporter gene imaging strategy. For example, it is a “negative” response between luciferase signals and miRNA expression level because the target miRNA suppresses luciferase translation and promotes its degradation under the aforementioned miRNA-specific reporter system. A suggested construct containing an inhibitory element, shown in Fig. 1, may be considered in its place. In the presence of specific miRNAs, the inhibitor expression will be suppressed and the reporter gene expression will be allowed. Thus, the correlation between reporter gene and miRNA will be “positive.” A second problem is that the vector itself serves as an antagonist to the target miRNA and therefore could deplete the endogenous miRNA reservoir as both the transcripts of the vector and target miRNA may be degraded simultaneously when they form a complex. Finally, there is also a possibility that miRNA will bind to the vector itself and result in accelerated degradation of the vector, which may further complicate the quantification of miRNAs based on BLI imaging.
Fig. 1
A possible reporter gene construct to image miRNA. a Without the presence of complementary endogenous miRNA, the expressed inhibitor will bind with IRE (inhibitor response element) and prohibit reporter gene expression. b With the presence of miRNA, mRNA encoding the inhibitor will be degraded and the reporter gene will be allowed.
Despite these potential limitations, the results from this paper shed new light on non-invasive imaging and quantification of endogenous miRNAs. The luciferase reporter can be replaced by the more clinically relevant positron emission tomography (PET) or magnetic resonance imaging (MRI) reporters such as thymidine kinase, sodium iodide symporter or ferritin [19–21]. Fluorescent dyes or radioisotopes labeled oligos for targeting miRNAs may also be used to image endogenous miRNA level after systemic administration. In addition to investigating the roles of miRNAs in the initiation and progression of diseases, we expect that molecular imaging of miRNAs will also be critical to advance our understanding of the interactions of miRNAs with their target genes and signaling pathways, and eventually to evaluate the use of miRNAs as a novel class of therapeutic targets.
References
- Rana TM (2007) Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol 8:23–36
Article PubMed CAS Google Scholar - Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Article PubMed CAS Google Scholar - Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114
Article PubMed CAS Google Scholar - Esquela-Kerscher A, Slack FJ (2006) Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6:259–269
Article PubMed CAS Google Scholar - Yanaihara N, Caplen N, Bowman E et al (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9:189–198
Article PubMed CAS Google Scholar - Iorio MV, Ferracin M, Liu CG et al (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65:7065–7070
Article PubMed CAS Google Scholar - Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE (2008) MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab 93:1600–1608
Article PubMed CAS Google Scholar - Chen YT, Kitabayashi N, Zhou XK, Fahey TJ 3rd, Scognamiglio T (2008) MicroRNA analysis as a potential diagnostic tool for papillary thyroid carcinoma. Mod Pathol 21:1139–1146
Article PubMed CAS Google Scholar - Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M (2005) Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438:685–689
Article PubMed Google Scholar - Esau C, Davis S, Murray SF et al (2006) miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3:87–98
Article PubMed CAS Google Scholar - Elmen J, Lindow M, Schutz S et al (2008) LNA-mediated microRNA silencing in non-human primates. Nature 452:896–899
Article PubMed CAS Google Scholar - Taylor DD, Gercel-Taylor C (2008) MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 110:13–21
Article PubMed CAS Google Scholar - Kim HJ, Chung JK, Lee DS, Kim SH (2008) In vivo imaging of miR-221 biogenesis in papillary thyroid carcinoma. Mol Imaging Biol doi:10.1007/s11307-008-0188-6
- Gillies JK, Lorimer IA (2007) Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle 6:2005–2009
PubMed CAS Google Scholar - Fornari F, Gramantieri L, Ferracin M et al (2008) MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 27:5651–5661
Article PubMed CAS Google Scholar - Lee JY, Kim S, Hwang do W, Jeong JM, Chung JK, Lee MC, Lee DS (2008) Development of a dual-luciferase reporter system for in vivo visualization of MicroRNA biogenesis and posttranscriptional regulation. J Nucl Med 49:285–294
Article PubMed CAS Google Scholar - Ko MH, Kim S, Hwang do W, Ko HY, Kim YH, Lee DS (2008) Bioimaging of the unbalanced expression of microRNA9 and microRNA9* during the neuronal differentiation of P19 cells. Febs J 275:2605–2616
Article PubMed CAS Google Scholar - Contag CH, Bachmann MH (2002) Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng 4:235–260
Article PubMed CAS Google Scholar - Iyer M, Barrio JR, Namavari M et al (2001) 8-[18F]Fluoropenciclovir: an improved reporter probe for imaging HSV1-tk reporter gene expression n vivo using PET. J Nucl Med 42:96–105
PubMed CAS Google Scholar - Niu G, Gaut AW, Ponto LL, Hichwa RD, Madsen MT, Graham MM, Domann FE (2004) Multimodality noninvasive imaging of gene transfer using the human sodium iodide symporter. J Nucl Med 45:445–449
PubMed CAS Google Scholar - Cohen B, Dafni H, Meir G, Harmelin A, Neeman M (2005) Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia 7:109–117
Article PubMed CAS Google Scholar
Author information
Authors and Affiliations
- The Molecular Imaging Program at Stanford (MIPS), Department of Radiology and Bio-X Program, Stanford University School of Medicine, 1201 Welch Rd, P095, Stanford, CA, 94305-5484, USA
Gang Niu & Xiaoyuan Chen
Authors
- Gang Niu
You can also search for this author inPubMed Google Scholar - Xiaoyuan Chen
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toXiaoyuan Chen.
Rights and permissions
About this article
Cite this article
Niu, G., Chen, X. Noninvasive Visualization of microRNA by Bioluminescence Imaging.Mol Imaging Biol 11, 61–63 (2009). https://doi.org/10.1007/s11307-008-0190-z
- Received: 21 October 2008
- Accepted: 21 October 2008
- Published: 27 November 2008
- Issue Date: March 2009
- DOI: https://doi.org/10.1007/s11307-008-0190-z