Implication of a novel multiprotein Dam1p complex in outer kinetochore function (original) (raw)
Abstract
Dam1p, Duo1p, and Dad1p can associate with each other physically and are required for both spindle integrity and kinetochore function in budding yeast. Here, we present our purification from yeast extracts of an ∼245 kD complex containing Dam1p, Duo1p, and Dad1p and Spc19p, Spc34p, and the previously uncharacterized proteins Dad2p and Ask1p. This Dam1p complex appears to be regulated through the phosphorylation of multiple subunits with at least one phosphorylation event changing during the cell cycle. We also find that purified Dam1p complex binds directly to microtubules in vitro with an affinity of ∼0.5 μM. To demonstrate that subunits of the Dam1p complex are functionally important for mitosis in vivo, we localized Spc19–green fluorescent protein (GFP), Spc34-GFP, Dad2-GFP, and Ask1-GFP to the mitotic spindle and to kinetochores and generated temperature-sensitive mutants of DAD2 and ASK1. These and other analyses implicate the four newly identified subunits and the Dam1p complex as a whole in outer kinetochore function where they are well positioned to facilitate the association of chromosomes with spindle microtubules.
Keywords: microtubule; spindle; mitosis; kinetochore; Saccharomyces cerevisiae
Introduction
The mitotic spindle facilitates chromosome segregation by capturing sister chromatids and segregating them to opposite poles during anaphase. This process requires that spindle microtubules form a physical connection with the chromosomes at kinetochores, proteinaceous structures assembled upon centromeric DNA. Analysis of the 125 bp budding yeast centromere has revealed three conserved sequence elements: CDEI, CDEII, and CDEIII (Fitzgerald-Hayes et al., 1982). Kinetochore function both in vivo and in vitro requires the CBF3 complex (Ndc10p, Cep3p, Ctf13p, and Skp1p [Goh and Kilmartin, 1993; Sorger et al., 1994]). This complex binds to CDEIII and provides a scaffold that allows the association of other kinetochore proteins including the CenpC-like protein Mif2p (Meluh and Koshland, 1995), the Ctf19 complex (Ctf19p, Mcm21p, and Okp1p [Ortiz et al., 1999]), and the Ndc80 complex (Ndc80p, Spc24p, Spc25p, and Nuf2p [Janke et al., 2001; Wigge and Kilmartin, 2001]). Current models of the kinetochore suggest that CDEII wraps around a core histone containing the H3-related protein, Cse4p (Espelin et al., 1997; Meluh and Koshland, 1997), allowing proteins bound to CDEI (Cbf1p) to interact with those bound to CDEIII (CBF3).
Despite the large number of kinetochore proteins identified in budding yeast, some of the most important questions about the kinetochore remain unanswered. Although the organization of the inner kinetochore proteins, which associate directly with DNA, is well established (Espelin et al., 1997; Meluh and Koshland, 1997), the organization and identity of the outer kinetochore proteins, which associate with spindle microtubules, remains to be determined. In addition, the precise nature of the connection between the kinetochore and microtubules and how this attachment is formed and regulated are still unclear. Although the proteins mentioned above are required for correct kinetochore function in vivo, none of these proteins has been shown to bind to microtubules directly.
One protein that might function to generate kinetochore–microtubule connections is Dam1p. Dam1p binds to microtubules directly in vitro (Hofmann et al., 1998) and plays an important role in multiple aspects of spindle structure (Hofmann et al., 1998; Jones et al., 1999; Cheeseman et al., 2001). Analysis of dam1 mutants has also demonstrated that Dam1p plays an essential role in chromosome segregation with phenotypes that are similar to mutants defective for kinetochore function (Cheeseman et al., 2001). In addition, Dam1p associates with kinetochores (Cheeseman et al., 2001; Enquist-Newman et al., 2001), indicating that it is well positioned to facilitate microtubule–kinetochore interactions.
Our previous work demonstrated that Dam1p associates with at least two other proteins, Duo1p and Dad1p (Cheeseman et al., 2001; Enquist-Newman et al., 2001). To better understand how Duo1p, Dam1p, and Dad1p function together to contribute to spindle and kinetochore function, we purified a native complex containing these proteins and at least four others. The identification of these additional subunits and the purification of this complex as a functional unit allows for a more complete understanding of the organization, activities, and regulation of the outer kinetochore in budding yeast.
Results
Duo1p, Dam1p, and Dad1p are components of a discrete multiprotein complex containing at least seven subunits
Using coimmunoprecipitation, we have demonstrated previously that Duo1p, Dam1p, and Dad1p are able to associate with each other in budding yeast (Cheeseman et al., 2001; Enquist-Newman et al., 2001). To purify these proteins from yeast extracts, we used a modified version of the tandem affinity purification tag (Rigaut et al., 1999) integrated at the DAD1 locus (Dad1–S tag–TEV-ZZ; see Materials and methods). A total of nine polypeptides ranging in size from 4 to 50 kD were observed to specifically copurify with Dad1p (Fig. 1 A). Mass spectrometric analysis of this purified complex confirmed the presence of Duo1p, Dam1p, and Dad1p and additionally identified Spc19p, Spc34p, Ask1p (YKL052c), and Dad2p (for Duo1 and Dam1 interacting; YKR083c). Spc19p and Spc34p were identified previously by mass spectrometry of purified spindle poles (Wigge et al., 1998). Despite repeated attempts, we were not able to establish the identity of the bands marked with an asterisk in Fig. 1 A. Although these polypeptides were reproducibly isolated in our purification, it is possible that they are cleavage products of a larger subunit. We conclude that Duo1p, Dam1p, and Dad1p are components of a larger multiprotein complex present in yeast protein extracts. Because Dam1p is the most well characterized of these proteins, we refer to this set of proteins as the Dam1p complex.
Figure 1.
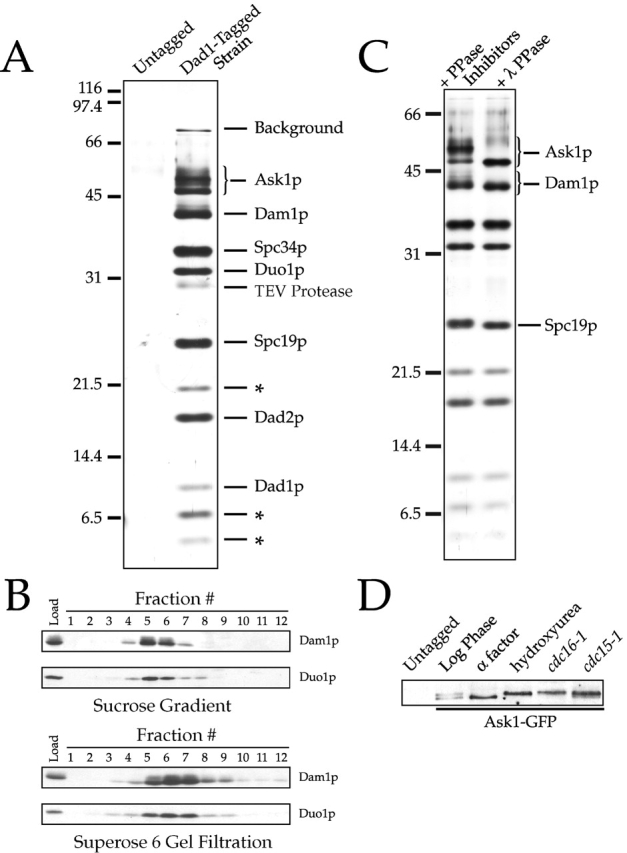
Purification of a 245 kD Dam1p complex. (A) Purification of the Dam1p complex using a tagged Dad1p reveals 10 polypeptides. Purified Dam1p complex (as described in Materials and methods) and protein from an identical purification using an untagged control strain were separated on a 13.5% SDS-PAGE gel and silver stained. Polypeptides identified by mass spectrometric analysis of the complex are indicated. Those not yet identified are denoted by an asterisk. Bands labeled “background” are the highly homologous heat shock proteins Ssb1 and Ssb2 (66 kD) and Ssa1, Ssa2, Ssa3, and Ssa4 (70 kD). (B) Determination of S value and Stoke's radius of the Dam1p complex in yeast extracts. Sucrose gradient and Superose 6 gel filtration fractions (as described in Materials and methods) were probed with antibodies against Duo1p and Dam1p. The S value for Duo1p/Dam1p-containing complex was estimated as 6.5 from a linear fit of the S values versus peak fraction number of standards, and the Stoke's radius was estimated as 90.1 Å from a Porath correlation, relating to the elution volumes of the standard proteins to their known Stoke's radii (Siegel and Monty, 1966). (C) Multiple subunits of the Dam1p complex are phosphorylated in vivo. Dam1p complex was purified in the presence of phosphatase inhibitors and then split into two aliquots, one of which was treated with lambda protein phosphatase. (D) Ask1p phosphorylation changes over the cell cycle. Ask1-GFP–tagged strains were grown to log phase and arrested in either α-factor (G1), 0.2 M hydroxyurea (S phase), or by using temperature-sensitive cdc16–1 (metaphase) and cdc15–1 (telophase) mutants. Protein samples were run on an 8% SDS-PAGE gel and immunoblotted with antibodies against GFP.
We next wanted to test whether the components of the Dam1p complex exist solely in this large multiprotein complex or if subcomplexes and monomeric forms were also present. Sucrose gradient sedimentation and gel filtration chromatography revealed that Duo1p and Dam1p comigrate as a single peak in yeast protein extracts (Fig. 1 B). By comparison to standards, we estimated that the Duo1p/Dam1p-containing complex has an S value of 6.5 and a Stoke's radius of 90 Å, corresponding to a native molecular weight of ∼245 kD. Assuming equal dye binding, densitometry of Coomassie-stained gels further indicated an approximately equimolar stoichiometry of Ask1p, Dam1p, Spc34p, Duo1p, Spc19p, Dad2p, and Dad1p within this complex (Table I). We were not able to estimate the stoichiometry of the two smallest proteins due to insufficient sensitivity, and the unidentified 22-kD band is present at a stoichiometry of 0.6:1, possibly reflecting degradation or partial disassociation of this protein from the complex. Together, these results indicate that Duo1p and Dam1p are present exclusively in an ∼245 kD complex, which based on the stoichiometries determined above contains a single copy of each known subunit.
Table I. Subunit stoichiometry for the Dam1p complex.
| Protein | Predicted molecularweight (SGD)a | Intensity/predictedmolecular weightb |
|---|---|---|
| kD | ||
| Ask1p | 32 | 1.0 |
| Dam1p | 38.4 | 0.9 |
| Spc34p | 34 | 1.2 |
| Duo1p | 27.4 | 1.2 |
| Spc19p | 19 | 1.3 |
| 22-kD band | 22 | 0.6 |
| Dad2p | 15 | 0.8 |
| Dad1-Stag | 12 | 1.0 |
| 8-kD band | 8 | |
| 4-kD band | 4 | |
| Total mass | 212 |
The Dam1p complex undergoes cell cycle–specific phosphorylation
Although multiple bands at ∼50 kD were observed on SDS-PAGE gels (Fig. 1 A), Ask1p was the only protein identified by mass spectrometry in that size range. A possible explanation for this discrepancy is that some subunits of the Dam1p complex are modified by phosphorylation. In fact, when we compared the electrophoretic mobilities of these proteins isolated in the presence of phosphatase inhibitors with those for the same proteins treated with lambda protein phosphatase, we saw a change in mobility for Ask1p, Dam1p, and Spc19p on SDS-PAGE gels (Fig. 1 C), indicating that these proteins are phosphorylated in vivo. Although Dam1p appears to be phosphorylated on multiple sites, the degree of phosphorylation did not appear to change during the cell cycle (unpublished data). In contrast, Ask1–green fluorescent protein (GFP)* (Fig. 1 D) did show evidence of cell cycle–specific phosphorylation. During G1 (α-factor arrest), Ask1p appeared as a fast migrating (dephosphorylated) form. However, during arrest in S phase or mitosis (using hydroxyurea and temperature-sensitive cdc16–1 and cdc15–1 mutants, respectively) Ask1p appeared as a slowly migrating (phosphorylated) form. Similar results were also observed when cells were released from α-factor arrest and allowed to progress synchronously through the cell cycle with the phosphorylated form appearing coincident with the initiation of budding (unpublished data).
The Dam1p complex binds to microtubules in vitro
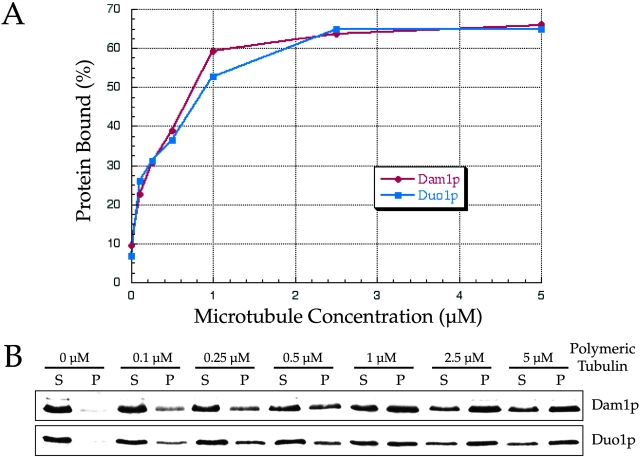
Since a Dam1p in vitro translation product was shown previously to bind to microtubules directly (Hofmann et al., 1998), this suggested that the entire Dam1p complex may also bind to microtubules. To test this possibility, we incubated limiting amounts of purified Dam1p complex (∼5–10 nM) in the presence of varying concentrations of taxol-stabilized bovine brain microtubules (0–5 μM). We then subjected these reactions to ultracentrifugation to determine the percentage of Dam1p complex bound to each concentration of microtubules (determined by examining Duo1p and Dam1p). We determined that the Dam1p complex binds directly to microtubules in vitro with an affinity of ∼0.5 μM (Fig. 2). This value is consistent with the affinity that we determined previously for the binding of in vitro–translated Dam1p (1 μM [Hofmann et al., 1998]) or _Escherichia coli_–purified Dam1p (∼0.5 μM; unpublished data) to microtubules. Interestingly, we found that both phosphorylated and unphosphorylated forms of Dam1p complex bind to microtubules with similar affinities (unpublished data), suggesting that protein phosphorylation of the Dam1p complex regulates an activity other than microtubule binding.
Figure 2.
Purified Dam1p complex binds to microtubules directly with ∼0.5 μM affinity. Purified Dam1p complex (∼5–10 nM) was incubated with varying concentrations of microtubules, which were then pelleted by centrifugation. The amount of complex bound to microtubules was determined by quantifying Duo1p and Dam1p in the pellet and supernatant fractions. (A) Percentage of protein bound to microtubules plotted with respect to the concentration of microtubules in the reaction. (B) Western blots showing the amount of Dam1p or Duo1p that is unbound (S, supernatant) or bound (P, pellet) at each concentration of microtubules.
Spc34p, Spc19p, Dad2p, and Ask1p localize to spindles and kinetochores
The purification described above isolated a tight complex of proteins that behaved as a functional unit in vitro with respect to binding to microtubules. We next wanted to establish whether these proteins formed a functionally relevant complex in vivo. Based on our previous analyses of Duo1p, Dam1p, and Dad1p (Hofmann et al., 1998; Cheeseman et al., 2001; Enquist-Newman et al., 2001), we predicted that the newly identified subunits would localize to the mitotic spindle. In fact, COOH-terminal fusions between Spc34p, Spc19p, Dad2p, or Ask1p, and GFP all localized to spindle poles and along the length of both short and long mitotic spindles (Fig. 3 A). Interestingly, Dad2-GFP showed strong punctate localization along longer spindles in addition to a weaker uniform straining. Colocalization with tubulin confirmed the localization of these proteins to the spindle but not to cytoplasmic microtubules (unpublished data). GFP fusions of Spc19p and Spc34p have also been localized along the mitotic spindle by immunofluorescence and immunoelectron microscopy (Wigge et al., 1998).
Figure 3.
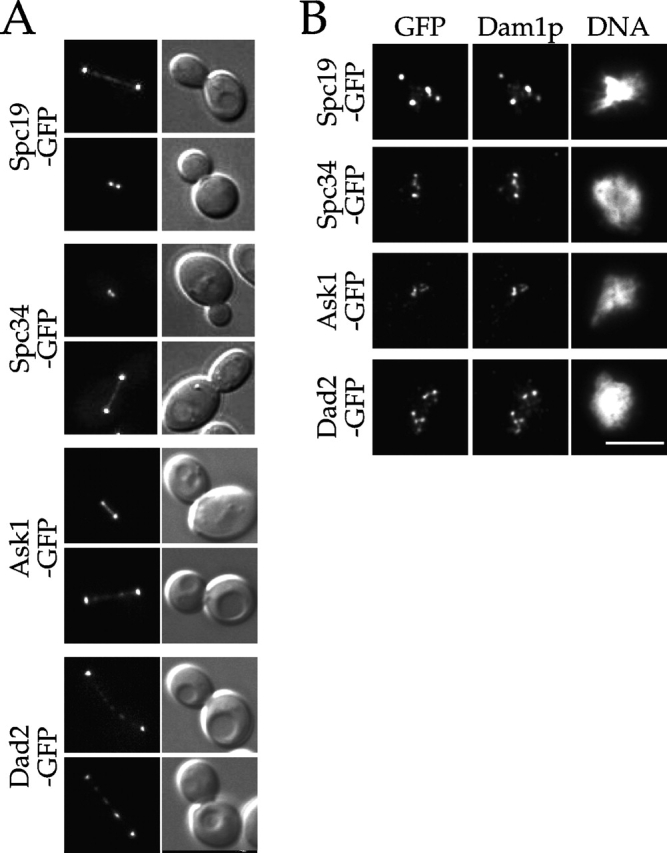
Spc19p, Spc34p, Dad2p, and Ask1p localize to spindles and kinetochores. (A) GFP fluorescence and corresponding DIC images showing the localization of the indicated fusion protein to the mitotic spindle. (B) Cells expressing the indicated GFP fusion proteins were prepared for chromosome spreads as described (Loidl et al., 1998). They were then processed for immunofluorescence and stained with anti-GFP and anti-Dam1p antibodies. Bar, 5 μm.
In addition to localizing to the mitotic spindle, immunofluorescence microscopy of chromosome spreads (Cheeseman et al., 2001) and chromatin immunoprecipitation analysis (Enquist-Newman et al., 2001) demonstrated previously the association of Duo1p, Dam1p, and Dad1p with kinetochores. To determine whether the newly identified subunits of this complex also localize to kinetochores, we performed chromosome spreads on strains expressing Spc19-GFP, Spc34-GFP, Dad2-GFP, and Ask1-GFP. All of these proteins colocalized precisely with Dam1p to punctate foci (Fig. 3 B) but not to bulk DNA, consistent with localization to kinetochores, which are known to be clustered in these preparations (Jin et al., 1998). Since we showed previously that Dam1p colocalizes with the kinetochore components Ndc10p and Mtw1p in these spreads (Cheeseman et al., 2001), these results indicate that the additional subunits localize with Dam1p to kinetochores. Recently, Spc19p and Spc34p were shown by chromatin immunoprecipitation to localize to centromeric DNA (He et al., 2001). In total, the above results provide strong in vivo support for the inclusion of these additional subunits in the Dam1p complex.
ask1 and dad2 mutants show spindle defects
Previous analyses of temperature-sensitive duo1, dam1, and dad1 mutants revealed a range of defects in spindle integrity (Hofmann et al., 1998; Cheeseman et al., 2001; Enquist-Newman et al., 2001). If the newly identified subunits are functionally relevant, we predicted that they would show similar mutant phenotypes. SPC19, SPC34, DAD2, and ASK1 are all essential genes (Wigge et al., 1998; Winzeler et al., 1999). To determine the loss of function phenotypes of some of these additional subunits, we generated degron-tagged alleles of ask1 and dad2 (termed td, for temperature-degron [Dohmen et al., 1994]). When ask1 td and dad2 td mutants were shifted to the restrictive temperature, the Ask1p and Dad2p fusion proteins were targeted for degradation (Fig. 4 A), and the mutants arrested with a high proportion of large-budded cells. When the mutant spindles were examined, we found that the majority of ask1 td and dad2 td cells arrested with a short mitotic spindle and a single mass of DNA (Fig. 4 B) similar to what we have described previously for duo1-2, dam1-9, and dad1-1 mutants. A smaller percentage of ask1 td and dad2 td mutant cells showed spindles that had broken down partially or completely in the middle and elongated beyond the short spindle stage (Fig. 4 B, insets). In total, these results provide strong phenotypic evidence that Ask1p and Dad2p function as components of the Dam1p complex to maintain spindle integrity.
Figure 4.
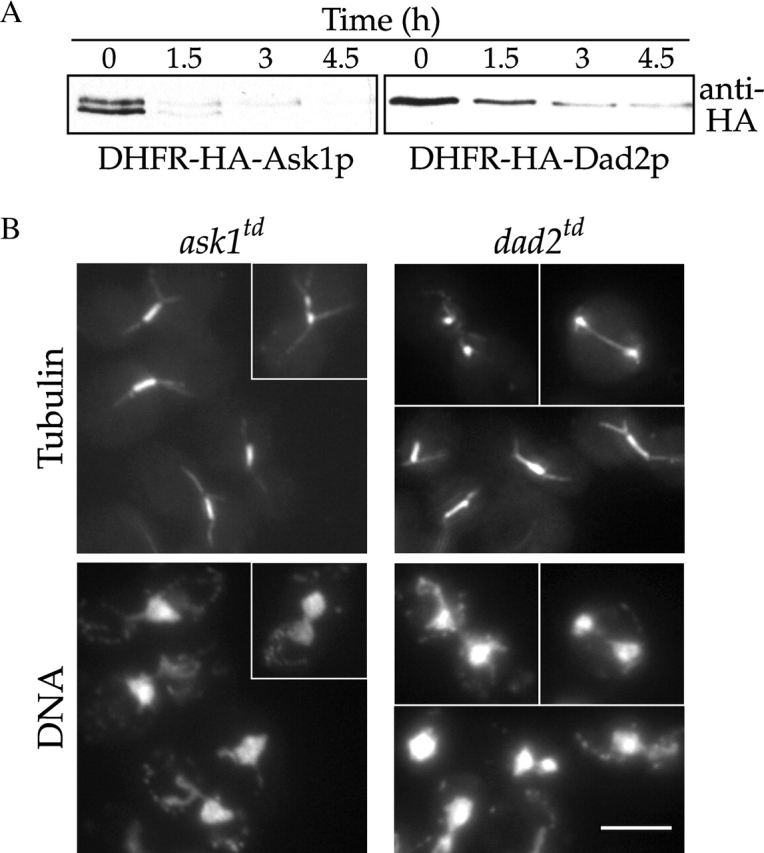
dad2 and ask1 mutants show spindle defects. (A) Immunoblots of ask1 td and dad2 td strains showing degradation of the fusion protein. Degron-tagged alleles of ask1 and dad2 were grown at 25°C and shifted to 37°C at t = 0 h Protein samples were immunoblotted with anti-HA antibodies to detect the DHFRts–HA fusion protein. (B) ask1 td and dad2 td mutant phenotypes. Degron-tagged alleles of ask1 and dad2 were grown at 25°C and shifted to 37°C for 3 h. They were then processed for tubulin immunofluorescence and DNA staining (DAPI). At this time point, 90% of large budded ask1 td and 82% of large budded dad2 td cells showed short spindle structures and a single mass of DNA, whereas 10% of ask1 td and 15% of dad2 td cells showed broken down spindles (n = 100 cells/sample). Bar, 5 μm.
A genetic context for the Dam1p complex at the outer kinetochore
The identification of the additional subunits of the Dam1p complex in combination with data from large scale two-hybrid experiments (Ito et al., 2000, 2001; Uetz et al., 2000) suggested that the Dam1p complex might interact physically with multiple components of the kinetochore (see Discussion). Examining genetic interactions in budding yeast provides a functional context for such physical interactions. We showed previously that dam1-1 interacts genetically with the kinetochore components _ctf19_Δ, _bir1_Δ, ipl1-2, and sli15-3 but not with mutants in the CBF3 complex or mif2-3 (Cheeseman et al., 2001; Kang et al., 2001). To extend this genetic analysis, we crossed dam1-1 to a wide range of mutants that affect kinetochore function. dam1-1 showed synthetic lethality in combination with _mcm16_Δ (Sanyal et al., 1998), _chl4_Δ (also known as _ctf17_Δ or _mcm17_Δ [Roy et al., 1997]), _mcm21_Δ, and _mcm22_Δ (Poddar et al., 1999) but not with _cbf1_Δ, cse4-1 (Stoler et al. 1995), _mcm19_Δ (also known as _iml3_Δ [Ghosh et al., 2001]), or mtw1-1 (Goshima and Yanagida, 2000). Strikingly, although several genetic interactions were observed with established kinetochore components (see indicated references, and for Mcm16 and Mcm22 [V. Measday and P. Hieter, personal communication]) no interactions were detected with mutants of the inner kinetochore proteins that bind directly to centromeric DNA (such as CBF3 mutants, _cbf1_Δ, cse4-1, or mif2-3). Therefore, these genetic results considered in combination with our biochemical demonstration of microtubule binding described above strongly implicate the Dam1p complex in outer kinetochore function.
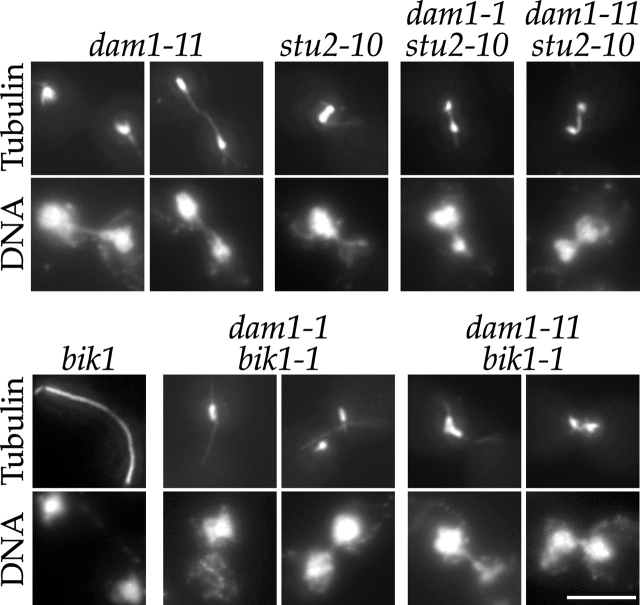
Our previous work indicated that Dam1p is a microtubule-associated protein involved in attaching the kinetochore to spindle microtubules (Cheeseman et al., 2001). Recent work has suggested a similar role for the microtubule-associated proteins Stu2p and Bik1p (He et al., 2001). Therefore, we also conducted crosses between dam1-1 and stu2-10 (Kosco et al., 2001; Severin et al., 2001) or bik1-1 (Pellman et al., 1995). dam1-1 was synthetically sick in combination with both stu2-10 and bik1-1; however, bik1-1 dam1-1 double mutants were much sicker, growing poorly even at 25°C. To determine whether these genetic interactions indicated a shared function in kinetochore–microtubule attachments or in spindle structure, we performed tubulin immunofluorescence on bik1-1 dam1-1, bik1-1 dam1-11, stu2-10 dam1-1, and stu2-10 dam1-11 double mutants grown at 37°C (Fig. 5). In contrast to dam1-1 and dam1-11 mutants that arrest in metaphase and show premature spindle elongation (Cheeseman et al., 2001; Fig. 5), dam1-1 stu2-10 and dam1-11 stu2-10 double mutants showed shorter broken down spindles. This lack of elongated spindles is consistent with the role that has been ascribed to Stu2p in mediating microtubule dynamics and spindle elongation (Kosco et al., 2001; Severin et al., 2001). dam1-11 bik1-1 and dam1-1 bik1-1 double mutants arrested primarily with short highly abnormal spindles, although some DNA segregation was observed. This severe spindle phenotype is not observed in either dam1 or bik1 single mutants, suggesting an overlapping role for Bik1p and Dam1p in spindle structure. Therefore, although Stu2p and Bik1p may have roles in kinetochore function the results described here indicate that both proteins play important roles in spindle structure.
Figure 5.
Analysis of stu2 dam1 and dam1 bik1 double mutants reveals shared roles in spindle structure. dam1-11, stu2-10, and bik1-1 single mutants and dam1-11 stu2-10, dam1-1 stu2-10, dam1-11 bik1-1, and dam1-1 bik1-1 double mutants were grown at 25°C and shifted to 37°C for 3 h. They were then processed for tubulin immunofluorescence and DNA staining (DAPI). Bar, 5 μm.
Discussion
Purification of the Dam1p complex
Here, we presented evidence that Duo1p, Dam1p, and Dad1p exist as components of an ∼245 kD complex that additionally contains Spc19p, Spc34p, Dad2p, and Ask1p. Spc19p and Spc34p were identified previously (Wigge et al., 1998), but their specific functions during yeast mitosis were not determined. We have also isolated two novel and previously uncharacterized proteins, Dad2p and Ask1p. Based on the localization of these additional subunits to the spindle and kinetochores and on the phenotypic analysis of ask1 td and dad2 td, we believe that this set of proteins forms a functionally important mitotic spindle complex in vivo. Significantly, the identification of homologs for all subunits of the Dam1p complex in diverse fungal species, including Candida albicans, Aspergillus nidulans, and Schizosaccharomyces pombe (Cheeseman et al., 2001; Enquist-Newman et al., 2001; unpublished data), suggests that this complex has conserved spindle and kinetochore functions.
Kinetochore functions of the Dam1p complex
We showed previously that the Dam1p complex contributes to both spindle and kinetochore function in vivo (Cheeseman et al., 2001). Here, we demonstrated that intact Dam1p complex is able to bind to microtubules directly in vitro. Since the Dam1p complex associates with both kinetochores and microtubules, it may play a direct role in mediating kinetochore–microtubule attachments. In fact, some duo1 and dam1 mutants show a premature spindle elongation/chromosome segregation phenotype during a metaphase arrest (Cheeseman et al., 2001) and altered chromosome dynamics (He et al., 2001), both of which appear to reflect a monopolar spindle attachment of paired sister chromatids. Our previous analyses suggested that these monopolar attachments result from a failure to form new kinetochore–microtubule attachments after DNA replication (Cheeseman et al., 2001), supporting a role for the Dam1p complex in facilitating the establishment but not necessarily the maintenance of microtubule–kinetochore interactions. Once such an attachment is formed through the activity of the Dam1p complex, other microtubule-associated proteins (such as those described by He et al., 2001) may play roles in maintaining this attachment. This hypothesis is supported by the genetic interactions that we observed between dam1-1 and stu2-10 or bik1-1. However, based on the spindle phenotypes presented here for these double mutants it is also likely that these genetic interactions reflect a shared role in spindle structure.
Regulation of the Dam1p complex
The activities of the Dam1p complex are specifically required for a period of the cell cycle between S phase and the end of mitosis (Cheeseman et al., 2001). Given the key roles that this complex plays in spindle and kinetochore function, it is likely that the cell would need to regulate these activities. We found previously that Dam1p associates with Ipl1p, an aurora protein kinase, and that Dam1p is a target of Ipl1p both in vivo and in vitro (Kang et al., 2001). dam1-1 also shows genetic interactions with Mps1p (Jones et al., 1999), a protein kinase involved in spindle pole body duplication and the spindle assembly checkpoint, suggesting that this complex may be regulated by Mps1p. Here, we presented evidence that Ask1p and Spc19p are also phosphorylated in vivo. Importantly, the phosphorylation of Ask1p changes in a cell cycle stage–specific manner with the highest levels of phosphorylation observed during S phase and mitosis.
A physical and genetic context for the Dam1p complex at the outer kinetochore
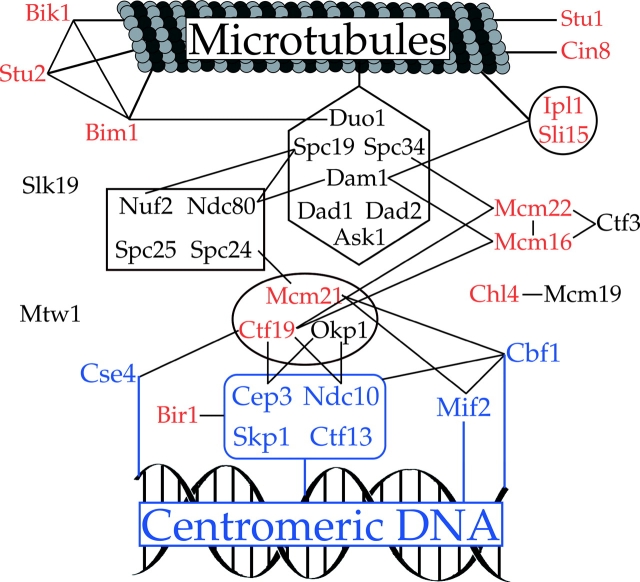
The identification of four additional subunits of the Dam1p complex allows for a more complete understanding of the roles that this complex plays in kinetochore function. Genome-wide two-hybrid screens identified multiple physical interactions between subunits of the Dam1p complex and other components of the kinetochore (Ito et al., 2000, 2001; Uetz et al., 2000). For example, this complex shows multiple interactions with components of the Ndc80 complex (Dam1p–Ndc80p, Spc19p–Ndc80p, and Spc19p–Nuf2p), suggesting that the functions of these two complexes may be closely associated. In addition, Dam1p interacts with Mcm16p (Ito et al., 2001), and Spc34p interacts by two-hybrid with Mcm22p (V. Measday and P. Hieter, personal communication). The interactions revealed by our genetic studies lend support to the biological relevance to the two-hybrid interactions with Mcm16p and Mcm22p and also point to an interaction with the Ctf19 complex. These data combined with recent work on other complexes within the kinetochore and the extensive protein–protein interactions identified by genome-wide two-hybrid studies, have helped to generate an appreciation of the large network of physical interactions that exist within the kinetochore. Fig. 6 represents the first attempt to model the physical interactions that may serve to connect microtubules and centromeric DNA in budding yeast. Although much work remains to be done, the elucidation of this network of interactions is an important first step toward understanding the organization and function of the outer kinetochore.
Figure 6.
Protein interactions establishing the connectivity between spindle microtubules and centromeric DNA. Physical interactions are indicated by lines between proteins. Shapes indicate distinct complexes within the kinetochore. Proteins that interact directly with centromeric DNA are shown in blue. Proteins whose loss of function results in genetic interactions with _dam1_-1 are shown in red. Protein interaction data are from Chen et al. (1998), Ghosh et al. (2001), Ito et al. (2001), Ito et al. (2000), Newman et al. (2000), Ortiz et al. (1999), Uetz et al. (2000), Yoon and Carbon (1999), and Kang et al. (2001). Physical interactions between Mcm16-Ctf19, Mcm22-Ctf19, Mcm16-Ctf3, Mcm22-Ctf3, and Spc34-Mcm22 represent coimmunoprecipitation and two-hybrid data from V. Measday and P. Hieter (personal communication).
Materials and methods
Strains and growth conditions
Yeast strains used in this study are listed in Table II. COOH-terminal GFP tags were generated at the endogenous SPC19, SPC34, DAD2, and ASK1 loci using a method similar to that described by Longtine et al. (1998). Degron-tagged alleles (Dohmen et al., 1994) of ASK1 and DAD2 were constructed by cloning the ASK1 and DAD2 ORFs into the HindIII sites of pPW66R. The resulting plasmids were cut with NcoI and integrated at the URA3 locus of a corresponding heterozygous deletion strain (Table II). These strains were sporulated, and haploid integrants in a null background were recovered. Yeast were grown on either yeast extract/peptone or synthetic medium supplemented with the appropriate nutrients and 2% glucose using standard procedures. All growth experiments were conducted in yeast extract/peptone plus dextrose. Geneticin (GIBCO BRL) was used at an effective concentration of 0.4 mg/ml.
Table II. Yeast strains used in this study.
| Name | Genotype | Source |
|---|---|---|
| DDY1810 | MATa, leu2, ura3-52, trp1, prb1-1122, pep4-3, pre1-451 | Drubin/Barnes lab |
| DDY2369 | MATa, leu2, ura3-52, trp1, prb1-1122, pep4-3, pre1-451, DAD1-STag-TEV-ZZ::KanMX | This study |
| CUY1266 | MATa, his3Δ200, leu2-3,112, ura3-52, SPC34-GFP::HIS3 | This study |
| CUY1268 | MAT_α, his3_Δ_200, leu2-3,112, ura3-52, DAD2-GFP::HIS3_ | This study |
| CUY1291 | MATa, his3Δ200, leu2-3,112, ura3-52, SPC19-GFP::HIS3 | This study |
| DDY2370 | MATa, his3Δ200, leu2-3,112, ura3-52, ade2-1, ASK1-GFP::HIS3 | This study |
| DDY2371 | MATa/MAT_α, his3_Δ_200/his3_Δ_200, leu2-3,112/leu2-3,112, ura3-52/ura3-52, ade2-1/ADE2, lys2-801/LYS2, ask1Δ::HIS3/_+ | This study |
| DDY2372 | MATa/MAT_α, his3_Δ_200/his3_Δ_200, leu2-3,112/leu2-3,112, ura3-52/ura3-52, ade2-1/ADE2, lys2-801/LYS2, dad2_Δ_::HIS3/_+ | This study |
| DDY2373 | MATa, his3Δ200, leu2-3,112, ade2-1, ask1Δ::HIS3, ura3-52::ask1td::URA3 | This study |
| DDY2374 | MATa, his3Δ200, leu2-3,112, ade2-1, dad2Δ::HIS3, ura3-52::dad2td::URA3 | This study |
| DDY1909 | MATa, his3Δ200, leu2-3,112, ura3-52, ade2-1, dam1-11::KanMX | Drubin/Barnes lab |
| DDY1913 | MATa, his3Δ200, leu2-3,112, ura3-52, ade2-1, dam1-1::KanMX | Drubin/Barnes lab |
| PY434 | MAT_α, leu2, ura3, trp1, ade2, ade3, bik1-1::TRP1_ | D. Pellmana |
| CUY1088 | MATa, his3Δ200, leu2-3,112, ura3-52, stu2-10::URA3 | Huffaker lab |
| DDY2375 | MATa, his3Δ200, leu2-3,112, ura3-52, lys2-801, stu2-10::URA3, dam1-11::KanMX | This study |
| DDY2376 | MATa, his3Δ200, leu2-3,112, ura3-52, lys2-801, stu2-10::URA3, dam1-1::KanMX | This study |
| DDY2377 | MATa, his3Δ200, leu2-3,112, ura3-52, ade2-1, bik1-1::TRP1, dam1-11::KanMX | This study |
Purification of the Dam1p complex
A modified version of a tandem affinity purification tag (Rigaut et al., 1999) composed of an S tag (Kim and Raines, 1993) in place of the calmodulin binding peptide, a TEV protease cleavage site, and a ZZ tag (minimal protein A binding domain) was amplified from pKW804 (a gift from K. Weis, University of California) and integrated at the COOH terminus of DAD1 in DDY1810 to generate DDY2369. DDY2369 was grown to OD600 = 1.2, washed with H2O, resuspended in 0.2 vol H2O, and drop frozen in liquid N2. Cells were lysed five times in a Waring blender with liquid N2. An equal volume of 2× lysis buffer (1× lysis buffer: 50 mM bis-Tris propane, pH 7.0, 0.1 M KCl, 5 mM EDTA, 5 mM EGTA, 10% glycerol) plus 2 mM PMSF and 2× protease inhibitors was added to the cell powder. Triton X-100 was added to the thawed extract to a final concentration of 1%. All remaining steps were conducted at 4°C. The extract was centrifuged at 10 k rpm in a SS-34 rotor for 20 min and at 45 k rpm in a Ti70 rotor for 30 min. The supernatant was passed over Q sepharose resin (Amersham Pharmacia Biotech), and the salt concentration of the flow through was adjusted to 0.4 M KCl by the addition of 2.5 M KCl. The flow through was then added to IgG sepharose (Amersham Pharmacia Biotech) and washed with lysis buffer with 0.4 M KCl, 0.6 M KCl, and finally 0.4 M KCl plus 1 mM DTT and 0.1% Tween-20. The IgG sepharose was incubated overnight in a small volume of the final wash buffer plus TEV protease. The supernatant was then removed from the IgG sepharose and added to S protein agarose (Novagen) for 3 h. The S protein agarose was washed with lysis buffer with 0.4 M KCl, and SDS protein sample buffer was added to elute the complex. For biochemical studies, eluate from the IgG sepharose was concentrated in a Cen30 concentration device and separated on a Superdex 200 gel filtration column equilibrated with 1× lysis buffer plus 1 mM DTT. Typically, 100 g of yeast extract (from 16 liter of culture) yielded ∼1 μg of purified Dam1p (or ∼5–6 μg of purified complex).
For studies on the phosphorylated complex, the initial 2× lysis buffer was supplemented with 20 mM Na pyrophosphate, 10 mM NaN3, 20 mM NaF, 0.8 mM Na orthovanadate, and 0.1 M β-glycerophosphate. For studies on the dephosphorylated complex, protein bound to either the IgG sepharose or S protein agarose was washed into lambda phosphatase buffer (50 mM Tris-HCl, pH 7.5, 0.1 mM EDTA, 5 mM DTT, 0.01% Brij 35, 2 mM MnCl2) and incubated with 4 μl of lambda protein phosphatase (New England Biolabs, Inc.) for 1 h at 30°C. It was then washed into 1× lysis buffer, and the purification was completed as described above.
Mass spectrometry analysis
A 50-μl sample of purified Dam1p complex eluted from the S protein agarose in 8 M urea, 50 mM Tris-HCl, pH 8.5, was reduced and alkylated using Tris 2-carboxyethyl phosphine HCL and iodoacetamide. The sample was then sequentially digested with endoproteinase Lys-C (Roche Diagnostics) and Porozyme™ immobilized trypsin (PerSeptive Biosystems) (McCormack et al., 1997). The resulting peptide mixture was then analyzed by MudPIT as described in Link et al. (1999) and Washburn et al. (2001). Tandem mass spectra were searched against a database of predicted ORFs (Saccharomyces Genome Database) to which common contaminants such as keratin and trypsin were added. Search results were filtered and grouped using the DTASelect program (Tabb et al., 2002), and identifications were confirmed through manual evaluation of spectra.
Immunofluorescence microscopy
Chromosome spreads were prepared as described (Loidl et al., 1998). Indirect immunofluorescence microscopy on intact yeast cells was performed as described (Ayscough and Drubin, 1998). The YOL134 antitubulin antibody (Accurate Chemical and Scientific Corporation) was used at a dilution of 1:200, rabbit anti-GFP antibody (a gift from Pam Silver, Dana-Farber Cancer Institute, Boston, MA) at 1:4000, and affinity purified guinea pig anti-Dam1p antibody (Cheeseman et al., 2001) at 1:1,000. Fluorescein or rhodamine-conjugated anti-IgG heavy chain secondary antibodies (Jackson ImmunoResearch Laboratories) were used at 1:500. Light microscopy was performed using a Nikon TE300 microscope equipped with a 100×/1.4 Plan-Apo objective and an Orca-100 cooled CCD camera (Hamamatsu) controlled by Phase-3 Imaging Systems software.
Protein and immunological techniques
For sucrose gradients, 50 μl of yeast extract was loaded onto a 2.1 ml 5–20% sucrose gradient, pelleted at 50,000 rpm for 18 h at 4°C in a TLS55 rotor, and fractions were collected from the top and TCA precipitated. In a parallel gradient, proteins of known S value (ribonuclease A, 1.8S, chymotrypsinogen A, 2.58S, BSA, 4.3S, and aldolase, 7.35S) were fractionated and analyzed by Coomassie staining of SDS-PAGE gels. For the Superose 6 gel filtration column, 0.5 ml of yeast protein extract was fractionated on a 24 ml Superose 6 column, and 1.5-ml fractions were collected. Standards for the gel filtration column include thyroglobulin (85 Å), catalase (52.2 Å), aldolase (48.1 Å), ovalbumin (30.5 Å), and chymotrypsinogen A (20.9 Å).
Immunoblots were performed as described (Cheeseman et al., 2001). Anti-Duo1p antibody (Hofmann et al., 1998) was used at a dilution of 1:2,000, guinea pig or rabbit anti-Dam1p antibody at 1:1,000, mouse anti-HA.11 antibody (Covance) at 1:1,000, and mouse anti-GFP antibody (Roche) at 1:500. HRP-conjugated secondary antibodies against rabbit, mouse (Amersham Life Sciences), and guinea pig (Alpha Diagnostic, Inc.) were used at 1:10,000.
Microtubule binding assays
Purified bovine brain tubulin (60 μM) in BRB80 buffer (80 mM K-Pipes, pH 6.8, 1 mM EGTA, 1 mM MgCl2) was assembled as described previously (Hofmann et al., 1998). For cosedimentation assays on the entire Dam1p complex, peak gel filtration fractions were supplemented with 1 mg/ml BSA and centrifuged at 23°C for 20 min at 60,000 rpm in a TLA100 rotor. 20 μl of soluble complex was added to an equal volume of taxol-stabilized microtubules that had been diluted into 1× lysis buffer with 1 mM DTT. These reactions were incubated for 20 min at 23°C to allow binding to occur and then were centrifuged as described above to pellet the microtubules. Pellets and supernatants were fractionated on SDS-PAGE gels and immunoblotted. At least two independent samples were examined for each concentration of microtubules.
Acknowledgments
The authors thank M. Enquist-Newman, C. Shang, and J. Wong for experimental assistance and extensive discussions, R. Heald and K. Kozminski for critical reading of the article, C. Brune and K. Weis for plasmids and advice on purification, P. Sinha, S. Stoler, M. Fitzgerald-Hayes, and M. Yanagida for strains, P. Silver for the anti-GFP antibody, J. Kilmartin for advice on purification, and V. Measday and P. Hieter for generously allowing us to cite unpublished work.
This work was supported by grants from the National Institute of General Medical Sciences to G. Barnes (GM-47842) and T.C. Huffaker (GM-40479), a grant to S. Fields from the Yeast Research Consortium from the National Center for Research Resources of the National Institutes of Health (Comprehensive Biology: Exploiting the Yeast Genome; PHS no. P41 RR11823), and a National Science Foundation graduate research fellowship to I.M. Cheeseman.
Footnotes
*
Abbreviation used in this paper: GFP, green fluorescent protein.
References
- Ayscough, K.R., and D.G. Drubin. 1998. Immunofluorescence microscopy of yeast cells. Cell Biology: A Laboratory Handbook. Vol. 2. J. Celis, editor. Academic Press, Inc., New York. 477–485.
- Cheeseman, I.M., M. Enquist-Newman, T. Müller-Reichert, D.G. Drubin, and G. Barnes. 2001. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 152:197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X.P., H. Yin, and T.C. Huffaker. 1998. The yeast spindle pole body component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J. Cell Biol. 141:1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen, R.J., P. Wu, and A. Varshavsky. 1994. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 263:1273–1276. [DOI] [PubMed] [Google Scholar]
- Enquist-Newman, M., I.M. Cheeseman, D. Van Goor, D.G. Drubin, P. Meluh, and G. Barnes. 2001. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell. 12:2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin, C.W., K.B. Kaplan, and P.K. Sorger. 1997. Probing the architecture of a simple kinetochore using DNA protein crosslinking. J. Cell Biol. 139:1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes, M., L. Clarke, and J. Carbon. 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 29:235–244. [DOI] [PubMed] [Google Scholar]
- Ghosh, S.K., A. Poddar, S. Hajra, K. Sanyal, and P. Sinha. 2001. The IML3/MCM19 gene of Saccharomyces cerevisiae is required for a kinetochore-related process during chromosome segregation. Mol. Genet. Genomics. 265:249–257. [DOI] [PubMed] [Google Scholar]
- Goh, P.-Y., and J.V. Kilmartin. 1993. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121:503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and M. Yanagida. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 100:619–633. [DOI] [PubMed] [Google Scholar]
- He, X., D.R. Rines, C.W. Espelin, and P.K. Sorger. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 106:195–206. [DOI] [PubMed] [Google Scholar]
- Hofmann, C., I.M. Cheeseman, B.L. Goode, K.L. McDonald, G. Barnes, and D.G. Drubin. 1998. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol. 143:1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., K. Tashiro, S. Muta, R. Ozawa, T. Chiba, M. Nishizawa, K. Yamamoto, S. Kuhara, and Y. Sakaki. 2000. Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. USA. 97:1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 98:4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., J. Ortiz, J. Lechner, A. Shevchenko, M.M. Magiera, C. Schramm, and E. Schiebel. 2001. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20:777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Q., E. Trelles-Sticken, H. Scherthan, and J. Loidl. 1998. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J. Cell Biol. 141:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.H., J.B. Bachant, A.R. Castillo, T.H. Giddings, and M. Winey. 1999. Yeast Dam1p is required to maintain spindle integrity during mitosis and interacts with the Mps1p kinase. Mol. Biol. Cell. 10:2377–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J., I.M. Cheeseman, G. Kallstrom, S. Velmurugan, G. Barnes, and C.S.M. Chan. 2001. Functional cooperation of Dam1, Ipl1, and the INCENP-related Sli15 in chromosome segregation. J. Cell Biol. 155:763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.S., and R.T. Raines. 1993. Ribonuclease S-peptide as a carrier in fusion proteins. Prot. Sci. 2:348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosco, K.A., C.G. Pearson, P.S. Maddox, P.J. Wang, I.R. Adams, E.D. Salmon, K. Bloom, and T.C. Huffaker. 2001. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell. 12:2870–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, A.J., J. Eng, D.M. Schieltz, E. Carmack, G.J. Mize, D.R. Morris, B.M. Garvik, and J.R. Yates, III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17:676–682. [DOI] [PubMed] [Google Scholar]
- Loidl, J., F. Klein, and J. Engebrecht. 1998. Genetic and morphological approaches for the analysis of meiotic chromosomes in yeast. Methods Cell Biol. 53:257–285. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- McCormack, A.L., D.M. Schieltz, B. Goode, S. Yang, G. Barnes, D. Drubin, and J.R. Yates, III. 1997. Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal. Chem. 69:767–776. [DOI] [PubMed] [Google Scholar]
- Meluh, P.B., and D. Koshland. 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 6:793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., and D. Koshland. 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11:3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, J.R.S., E. Wolf, and P.S. Kim. 2000. A computationally directed screen identifying interacting coiled coils from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 97:13203–13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, J., O. Stemmann, S. Rank, and J. Lechner. 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13:1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellman, D., M. Bagget, Y.H. Tu, G.R. Fink, and H. Tu. 1995. Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J. Cell Biol. 130:1373–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poddar, A., N. Roy, and P. Sinha. 1999. MCM21 and MCM22, two novel genes of the yeast Saccharomyces cerevisiae are required for chromosome transmission. Mol. Microbiol. 31:349–360. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Séraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nature Biotechnol. 17:1030–1032. [DOI] [PubMed] [Google Scholar]
- Roy, N., A. Poddar, A. Lohia, and P. Sinha. 1997. The mcm17 mutation of yeast shows a size-dependent segregational defect of a mini-chromosome. Curr. Genet. 32:182–189. [DOI] [PubMed] [Google Scholar]
- Sanyal, K., S.K. Ghosh, and P. Sinha. 1998. The MCM16 gene of the yeast Saccharomyces cerevisiae is required for chromosome segregation. Mol. Gen. Genet. 260:242–250. [DOI] [PubMed] [Google Scholar]
- Severin, F., B. Habermann, T. Huffaker, and T. Hyman. 2001. Stu2 promotes mitotic spindle elongation in anaphase. J. Cell Biol. 153:435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, L.M., and K.J. Monty. 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta. 112:346–362. [DOI] [PubMed] [Google Scholar]
- Sorger, P.K., F.F. Severin, and A.A. Hyman. 1994. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J. Cell Biol. 127:995–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler, S., K.C. Keith, K.E. Curnick, and M. Fitzgerald-Hayes. 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9:573–586. [DOI] [PubMed] [Google Scholar]
- Tabb, D.L., W.H. McDonald, and J.R. Yates. 2002. DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. Journal of Proteone Research. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., L. Giot, G. Cagney, T.A. Mansfield, R.S. Judson, J.R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 403:623–627. [DOI] [PubMed] [Google Scholar]
- Washburn, M.P., D. Wolters, and J.R. Yates, III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242–247. [DOI] [PubMed] [Google Scholar]
- Wigge, P.A., and J.V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge, P.A., O.N. Jensen, S. Holmes, S. Souès, M. Mann, and J.V. Kilmartin. 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141:967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E.A., D.D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J.D. Boeke, H. Bussey, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 285:901–906. [DOI] [PubMed] [Google Scholar]
- Yoon, H.J., and J. Carbon. 1999. Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc. Natl. Acad. Sci. USA. 96:13208–13213. [DOI] [PMC free article] [PubMed] [Google Scholar]