The molecular biology of mixed lineage leukemia (original) (raw)
Mixed-lineage-leukemia is an aggressive leukemia that predominantly occurs in pediatric patients and is characterized by the expression of fusion genes involving the histone methyltransferase MLL and a variety of fusion partners. It is now clear that MLL fusion partners can activate transcription by two different mechanisms, which are discussed in this review article. Insights in these functions may open new avenues for rational drug development. See related papers on page 891 and 918.
Keywords: MLL, proteins, leukemia
Abstract
Mixed lineage leukemia is a very aggressive blood cancer that predominantly occurs in pediatric patients. In contrast to other types of childhood acute leukemias, mixed lineage leukemia presents with a dismal prognosis and despite the availability of advanced treatment methods cure rates have stagnated over the last years. Mixed lineage leukemia is characterized by the presence of MLL fusion proteins that are the result of chromosomal translocations affecting the MLL gene at 11q23. These events juxtapose the amino-terminus of the histone methyltransferase MLL with a variety of different fusion partners that destroy normal histone methyltransferase function of MLL and replace it by heterologous functions contributed by the fusion partner. The resulting chimeras are transcriptional regulators that take control of targets normally controlled by MLL with the clustered HOX homeobox genes as prominent examples. Recent studies suggested that MLL fusion partners activate transcription by two different mechanisms. Some of these proteins are themselves chromatin modifiers that introduce histone acetylation whereas other fusion partners can recruit histone methyltransferases. In particular, histone H3 specific methylation at lysine 79 catalyzed by DOT1L has been recognized as a hallmark of chromatin activated by MLL fusion proteins. Interestingly, several frequent MLL fusion partners seem to coordinate DOT1L activity with a protein complex that stimulates the elongation phase of transcription by phosphorylating the carboxy-terminal repeat domain of RNA polymerase II. The discovery of these novel enzymatic activities that are essentially involved in MLL fusion protein function presents potential new targets for a rational drug development.
Mixed lineage leukemia - a clinical primer
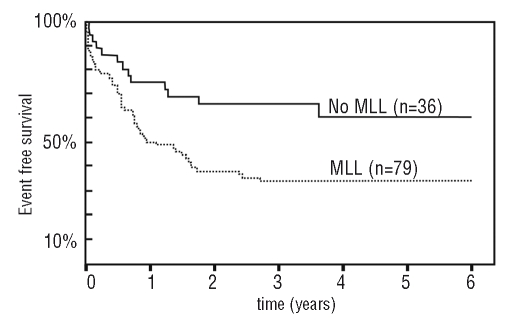
Apart from the fascinating molecular biology underlying the pathogenesis, mixed lineage leukemia mainly presents a clinical challenge. More than 30 years ago, physicians realized that certain subsets of patients initially diagnosed with acute lymphoblastic (ALL) or acute myeloid leukemia (AML) fared far worse than others. In the pediatric field, one of these high-risk leukemias stood out particularly amongst all remaining cases of childhood leukemia. A cohort of ALLs diagnosed in newborns and infants (younger than one year) fell into a group with similar clinical aspects and an extremely dismal prognosis. With the advent of fluorescent activated cell sorting (FACS) it was revealed that the leukemic blasts of these aggressive leukemias frequently expressed surface markers of both the lymphoid and the myeloid lineage. Sometimes even a complete lineage switch was observed during treatment and a leukemia initially diagnosed as ALL could relapse as AML.1 Accordingly the term mixed lineage leukemia was coined.2–4 Even before this, cytogeneticists had noted that translocations affecting the locus 11q23, and in particular the translocation t(4;11), characterize a special subset of ALL that was associated with poor survival.5–7 Soon thereafter it became clear that these translocations of the locus 11q23 are also typical for mixed lineage leukemia. Whereas treatment of non-mixed lineage leukemia in children has become the textbook success story of modern medicine with 5-year survival rates approaching 90%,8 mixed lineage leukemia treatment seems to have hit a roadblock with hardly 40% of all infants surviving five years after diagnosis (Figure 1).8–11
Figure 1.
Event free survival of infants with ALL separated by MLL status. Redrawn after Hilden et al.9 Please note that the data correspond to event free survival (a more stringent criterion) and do not include children older than one year.
Mixed lineage leukemia reaches a second peak of incidence later in life, particularly in patients who have been treated previously for an unrelated neoplastic disease with topoisomerase inhibitors like etoposide (so-called therapy related leukemia; t-AML or t-ALL).12 In total, 11q23 abnormalities occur in up to 70% of infant ALL, and in approximately 10% of all other ALL cases.13 Therapy related leukemia almost always manifests as AML with about 10% of therapy induced AML and 3% of de novo AML carrying an 11q23 translocation.14 Intriguingly, the MLL gene is also the target of a second type of aberration that creates a short repeat within the MLL coding sequence resulting in an internal partial tandem duplication (PTD). As a consequence, an extra amino-terminus is added in frame to full length MLL. MLL-PTD occurs predominantly in AML. Judging from gene expression patterns and clinical parameters, MLL-PTD seems to cause a different disease from that induced by classical MLL fusions. MLL-PTD has been covered by a recent publication15 and therefore this aberration will not be subject of this review.
Normal MLL - a histone methyltransferase necessary for efficient transcription
Guided by the chromosomal aberrations, four groups independently succeeded in cloning the gene spanning the translocation breakpoint at 11q23.16–19 From sequence comparison it became immediately clear that this gene encoded a homolog of a known fly gene named trithorax (Trx). Because of this relationship and the involvement in leukemia, the human gene was initially labeled either HRX (human trithorax), ALL-1 (acute lymphocytic leukemia-1) or MLL (mixed lineage leukemia). Later it was agreed to use MLL as the standard name. Drosophila Trx mutants displayed a very suggestive phenotype with homeotic changes in all three breast segments reminiscent of Hox gene mutations and indeed Hox gene expression was perturbed in Trx negative flies.20–22 This function was conserved in mammals as MLL knockout embryos also showed skeletal transformations and misexpression of Hox genes before they died in utero around day 10.5–16.5 p.c., depending on the particular knockout allele.23–25 Fly Trx was also isolated in a genetic screen that was set up to identify genes that counteracted genetic silencing.26 Since Hox gene expression was correctly initiated in _Trx_−/− flies as well as in _MLL_−/− mice but later deteriorated during embryogenesis, it was thought that Trx/MLL is a specific maintenance factor for Hox genes. However, nowadays it is known that MLL serves a much more general function. The breakthrough came with the identification of the highly conserved SET domain (an acronym for Suppressor of variegation, Enhancer of zeste, Trithorax) at the C-terminus of MLL as the site of a histone methyltransferase activity that specifically methylates histone H3 at lysine 4.27 MLL was found to be incorporated into a large macromolecular complex that was purified from mammalian nuclei.28,29 The complex showed conservation across phyla all the way down to Saccharomyces cerevisiae where SET1, the yeast counterpart of MLL, was also present in a similar complex called COMPASS (complex of proteins associated with Set1).30,31 Interestingly, MLL is post-translationally processed by proteolytic cleavage.
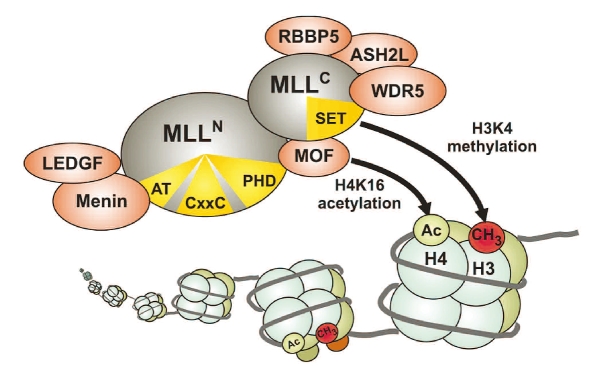
The large MLL protein is cut by an aspartic protease called taspase into an N-terminal 320kDa fragment and a C-terminal 180kDa moiety that are both core components of the MLL complex (Figure 2).32–35 Within this molecular machinery a division of labor exists. The MLLC subunit associates with at least four proteins that help in preparing chromatin for efficient transcription. One of these proteins is the histone H4 lysine 16 specific acetyl-transferase MOF that loosens up chromatin by histone charge neutralization.36 The WDR5 protein in turn recognizes the histone H3 lysine 4 methyl-mark introduced by MLL and it has therefore been suggested that WDR5 ensures the processitivity of histone modification.37–39 And finally the proteins RBBP5 and ASH2L appear to be necessary for efficient methyltransferase activity by stabilizing an active conformation of MLL allowing allosteric control.40 The histone acetyltransferase CBP and the INI1 subunit of the SWI/SNF nucleosome remodeling complex have also been identified as interaction partners of MLLC in interaction screens,41,42 although these proteins did not copurify with MLLC in biochemical experiments.
Figure 2.
The MLL complex. After post-transcriptional proteolytic processing amino-terminal and carboxy-terminal portions of MLL are incorporated in a macromolecular complex with histone methyltransferase and histone acetyltransferase function. Functional domains in MLL are indicated in yellow. AT = AT-hooks, a DNA binding domain, CxxC = motif recognizing unmethylated CpG dinucleotides, PHD = plant homeodomain, SET = histone methyltransferase active site. Proteins associated with MLL are explained in the text.
MLLN on the other side contains features essential for correct targeting of the MLL complex. At the outmost amino-terminal end of MLL a binding site for menin, the product of the tumor suppressor gene multiple endocrine neoplasia is present.43–45 Menin and MLL form an interaction surface for LEDGF (lens epithelium derived growth factor) and LEDGF makes contact to chromatin via a PWWP domain.46 Interestingly, LEDGF is also involved in HIV pathogenesis where it assists integration of HIV proviruses into chromatin.47,48 In addition, MLLN codes for several AT-hooks, a minor groove DNA binding motif that preferentially recognizes DNA with distortions like bends or kinks.49 Further downstream a CxxC domain can be found. CxxC domains occur in proteins that discriminate the methylation status of DNA, and indeed, also the MLL CxxC moiety binds specifically to unmethylated CpG dinucleotides.50 Swap experiments between MLL and the highly homologous MLL2 indicated that the CxxC domain seems to be a major determinant of subnuclear localization and target gene selection.51 In addition, the CxxC region also has been shown to recruit repressive factors like histone deacetylases and polycomb group proteins.52 This interaction appeared to be regulated by conformational changes elicited by the prolylisomerase cyclophilin 33 (Cyp33) that interacts further carboxy-terminal with the plant homeodomain (PHD) of MLLN.
In summary therefore, the MLL complex coordinates three major mechanisms of chromatin modification: methylation, acetylation and nucleosome remodeling. Most likely transcription factors recruit the MLL complex to initiate RNA synthesis. Examples are p53 and β-catenin that have been found to associate with MLL during transcriptional activation.36,53 H3K4 methylation is universally introduced around the transcription start site of all transcribed genes, and next to MLL several other confirmed or putative H3K4 methyltransferases (MLL2, MLL3, MLL5, SET1A, SET1B, and ASH1L) have been identified in mammalian cells. If all of these proteins have a comparable number of cellular targets, and with an estimated 10,000 genes transcribed at any present moment under standard conditions, each H3K4 methyl-transferase should be responsible for more than 1,000 loci. Although this is a greatly oversimplified prediction, nonetheless it seems to be confirmed by emerging experimental data that identified several hundred genomic loci bound by MLL and ASH1L in ChIP experiments.54,55 Obviously certain genes, like the Hox genes, depend more on MLL mediated chromatin modification than others, and therefore they stand out in the MLL loss of function phenotype.
The origin of 11q23 translocations
Much has been speculated about the origins of the chromosomal aberrations that convert an innocuous chromatin modifier into a pernicious oncogene. Several lines of evidence point to a mishap in non-homologous end joining of double strand breaks as the most likely reason for 11q23 translocations.
For one, the characteristic peak of mixed lineage leukemia in patients treated with etoposide is highly suggestive for an involvement of DNA double strand lesions in the etiology of MLL fusions. Etoposide inhibits topoisomerase II and therefore causes breaks in both DNA strands. Indeed, it could be shown that the locus 11q23 is particularly susceptible to this kind of assault in cells treated with topo II inhibitors.56,57 Alternatively, a break might be introduced at early stages of apoptotic DNA fragmentation that was later aborted and repaired. Published data also provide some support for this scenario, as breaks preferentially occur at 11q23 in early apoptotic cells.58
Whatever the reason for the initiating event, an aberrant non-homologous end joining (NHEJ) process most likely causes the cross-wise sealing of the DNA ends. A close examination of the breakpoint junctions revealed that they frequently code for non-templated nucleotides,59 a hallmark of NHEJ repair as known from generation of antibody and T-cell receptor diversity. Despite the attractions of this hypothesis as an explanation for the origin of 11q23 translocations it does not take into consideration that many double strand breaks induced by background radiation are continuously repaired in each cell without dire consequences. In this respect, a publication might be important showing that double strand breaks lead to chromosomal aberrations only in cells with impaired ATM-dependent DNA-damage signaling, whereas normal cells are able to join free ends correctly.60 The potential involvement of DNA repair pathway defects in mixed lineage leukemia is almost completely unexplored and would be a rewarding topic for future research.
MLL fusion proteins; transcriptional elongation and chromatin modification versus dimerization
The first and most striking property of MLL fusion proteins is their incredible diversity.
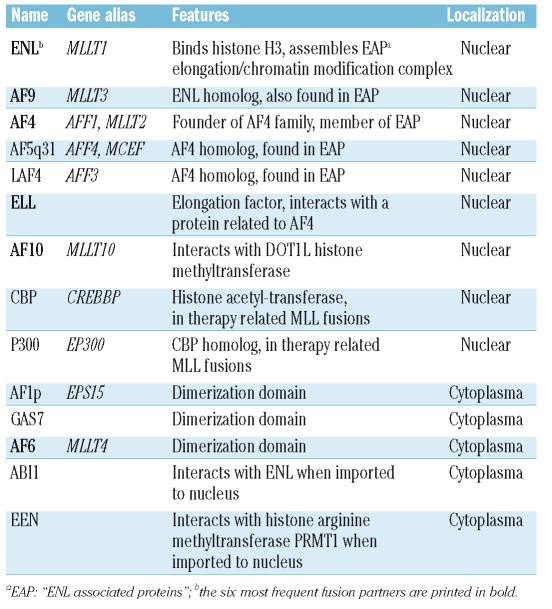
MLL has been found in 73 different translocations and 54 partner genes have been cloned (http://atlasgeneticsoncology.org/Genes/MLL.html; last update 5/08). Despite this variety most cases of mixed lineage leukemia present as a clinical entity and gene expression signatures in leukemic blasts do not separate MLL fusions according to the fusion partner.61–64 Therefore, it was a long standing question how a multitude of different proteins could cause the same disease. Two facts gave early clues to this problem. Firstly, all MLL fusion proteins share a common structure with the respective partners invariably fused in frame to MLLN right after the CxxC domain but excluding the PHD fingers. Secondly, proteins joined to MLL clearly fall into two classes. Only 6 frequent partner proteins (AF4, AF9, ENL, AF10, ELL, AF6) constitute the bulk (> 85%) of all clinical cases of mixed lineage leukemia65,66 (Table 1) whereas the remaining fusions were cloned each from a few isolated, mostly adult patients. This distinction is mirrored by the biology of the respective proteins. With the exception of AF6, all frequent MLL partners are nuclear while cytoplasmatic localization predominates amongst the rarely occurring MLL fusions. Therefore it was expected that at least two different mechanisms should be responsible for MLL fusion function. Further clarification of these pathways was promoted by the development of an in vitro assay that was able to measure the biological readout of MLL fusion activity.67
Table 1.
A selection of MLL fusion partners with known functions or domains.
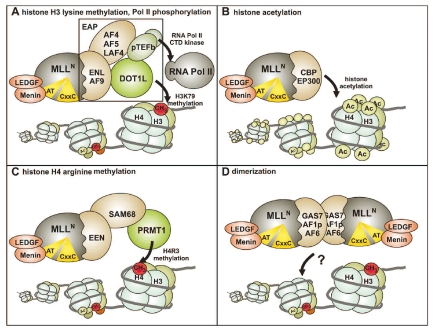
This serial replating assay records an inhibition of hematopoietic differentiation as surrogate parameter for transformation activity. A block in differentiation can be visualized as enhanced clonogenic capacity of hematopoietic precursor cells after repeated replating in semisolid medium. With respect to the MLL portion included in the fusions, deletion studies demonstrated that the LEDGF-menin binding motif and the CxxC domain were absolutely necessary for the overall function of MLL fusions.68,69 In addition, it was mandatory that the breakpoint in MLL was upstream of the PHD fingers because artificial MLL fusions including this domain lost their transforming capacity.70,71 This explains the strict conservation of the fusion breakpoints found in leukemic blasts. Further studies suggested that the leukemogenic potential of truncated MLL could be activated in at least four different ways (Figure 3).
Figure 3.
Molecular pathways leading to oncogenic activity of MLL fusion proteins. MLL fusions are aberrant transcription factors that activate gene expression. Four different mechanisms have been suggested as to how fusion partners might induce transcriptional activation. (A) The most frequent fusion partners of the ENL and AF4 family are members of the EAP complex that combines histone H3K79 methyltransferase activity catalyzed by DOT1L with transcriptional elongation stimulation by pTEFb (positive transcription elongation factor b, a dimer of CDK9 and a cyclinT) that phosphorylates the C-terminal repeat domain of RNA polymerase II. It is speculated that MLL fusion proteins aberrantly recruit this complex to target chromatin. (B) Active histone acetyltransferases are fused to MLL in the MLL-CBP and MLL-p300 proteins. (C) MLL-EEN indirectly recruits the histone H4R3 arginine methyltransferase through binding of the adaptor SAM68. (D) Dimerization of MLL via coiled-coiled or other dimerization domains supplied by the fusion partner activates target genes by unknown mechanisms.
The “common” nuclear fusion partners (ENL, AF9, AF4, ELL, and AF10) - transcriptional elongation meets histone methylation
Early reports showed that MLL fusions function as a novel type of general transcription factor that is able to indiscriminately activate many different promoters.72 The search for a common MLL machinery revealed that the close homologs ENL and AF9 were both able to interact with other MLL fusion partners like AF4, the AF4-homolog AF5 and probably also with AF10. In addition, it was realized that ENL could bind to histone H3, indicating a potential shared link of these proteins with chromatin modification.73 A breakthrough concerning the normal function of these proteins came from the purification of the ENL associated protein complex (EAP).74,75 In this complex, ENL was not only linked with all members of the AF4 protein family that occur as MLL fusion partners (AF4, AF5q31, LAF4) but also with positive transcription elongation factor b (pTEFb) and the histone methyltransferase DOT1L. pTEFb is a dimer of cyclin dependent kinase 9 (CDK9) and a cyclin T that phosphorylates the carboxy-terminal repeat domain of RNA polymerase II. This activity is essential for efficient transcriptional elongation.76 DOT1L methylates histone H3 at lysine 79, a modification that is introduced also during transcriptional elongation.77 This immediately invoked parallels to AF10 as it had been demonstrated that AF10 binds DOT1L and that DOT1L recruitment was essential for the transforming activity of MLL-AF10.78 A dramatically increased H3K79 methylation has been demonstrated for the HOXA9 gene activated by MLL-ENL79 and this result was corroborated on a global scale in two recent studies by genome-wide ChIP where the majority of MLL target genes defined by MLL fusion protein binding also showed increased H3K79 modification.80,81 Another connection to transcriptional elongation is defined by the fusion partner ELL, as this protein had been identified before as elongation factor.82 This idea was later dismissed because domains in ELL necessary for elongation activity were dispensable for transformation by MLL-ELL.83 However, ELL also interacts with proteins that are homologous to AF4, and ELL might therefore make indirect contact to the EAP elongation machine.84,85 In summary, the common MLL fusion partners with clinical importance all seem to participate in the same biological process if not in the same macromolecular complex responsible for control of transcriptional elongation. Although not yet formally proven, it is tempting to speculate that MLL fusions might recruit EAP to genomic loci to achieve ectopic target gene expression. Indeed, there are hints that the association of ENL and AF9 with AF4 family members is essential for survival of mixed lineage leukemia cells as small peptides disrupting this interaction proved to be toxic for MLL transformed cells but not for blasts of a different etiology.86,87 The potential usefulness of these peptides as therapeutic intervention is under active investigation.88
CBP and p300 - MLL fusions and histone acetylation
Interestingly, fusions of MLL with the histone acetyltransferases CREB binding protein (CBP) and the related p300 have been observed. Although these MLL derivatives have only been found in a few cases of therapy induced secondary leukemia, they allowed immediate insight into a possible activation mode for MLL.89,90 Structure function analyses clearly singled out the bromo- and histoneacetyltransferase domains of CBP as necessary and sufficient for the oncogenic function of the respective fusion proteins.91 Unfortunately, the consequences of MLL-CBP/p300 expression for target chromatin have not yet been investigated. However, it seems reasonable to assume that the permanent recruitment of HAT activity will result in a hyperacetylation of chromatin and an increased transcriptional output.
The special case of MLL-EEN arginine specific histone methylation
Although the EEN fusion partner was cloned only from a single case of mixed lineage leukemia and has never been found again, the resulting MLL fusion was studied in great detail. Surprisingly, a biochemical interaction study revealed that EEN bound the arginine methyltransferase PRMT1 through the adaptor protein SAM68.92 PRMT1 is an arginine specific methyltransferase that shuttles between nucleus and cytoplasma. Next to several cytoplasmic substrates, PRMT1 also methylates histone H4 at arginine 3. This modification has in turn been shown to be correlated with an increased histone acetylation.93 Therefore MLL-EEN might feed into the same pathway as suggested for MLL-CBP or MLL-p300.
Cytoplasmatic fusion partners and dimerization
MLL joined to proteins of cytoplasmatic origin seem to be more weakly transforming as fusions with nuclear proteins. Cytoplasmatic fusions are found preferentially in older patients where more time has elapsed from the initiating pre-leukemic event to the outbreak of acute leukemia allowing additional secondary mutations to occur. Consequently several of these MLL fusions do not read out in the standard in vitro assays that measure MLL fusion activity.94,95 Nevertheless MLL fusions with the cytoplasmatic proteins GAS7, AF1p and AF6 could be assayed in this system, and therefore the minimally necessary contributions of the respective fusion partner could be localized to a coiled-coiled dimerization domain.96 In addition, also fusion of MLL to an artificial inducible dimerization domain caused activation of the transforming potential.97 Unfortunately, up to now it is not known how dimerized MLL fusion proteins activate target genes. With respect to cytoplasmatic fusion partners, it must be taken into consideration that all MLL fusions will be imported to the nucleus because of the strong nuclear import signals in MLLN.98 This might cause aberrant protein-protein interaction of the fusion partner, a situation that has been demonstrated for the ABI1 protein. Normally localized in the cytoplasma, ABI1 interacts with ENL after import to the nucleus.99 In this way, also cytoplasmatic fusion partners might feed into pathways used by the nuclear partner proteins.
MLL fusion downstream targets and the problem of pediatric leukemia
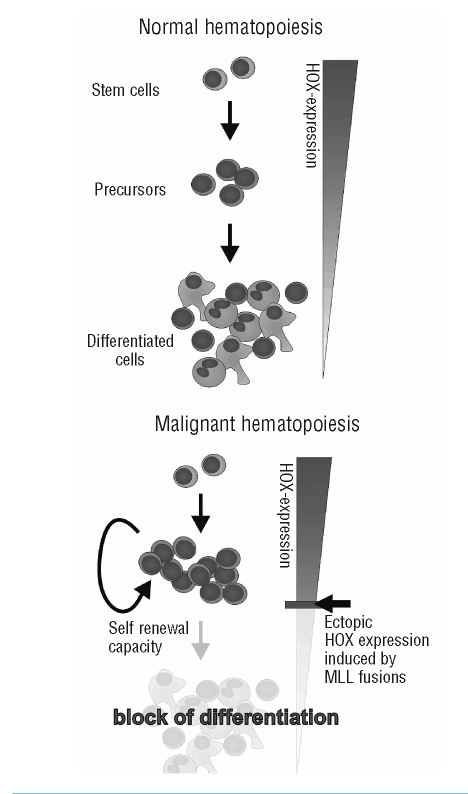
Normal MLL performs an important task necessary for the transcription of many genes. Because all domains within MLLN thought to be involved in target selection are retained in the fusion proteins, it seems likely that MLL fusions will share many target loci with wild type MLL. This assumption has been confirmed for the clustered HOX homeoboxgenes that are under control of MLL as well as of MLL fusion proteins. In addition to the HOX cluster, MLL-AF4 has been found on a genome-wide scale on more than 1,000 promoters that also showed a corresponding H3K79 methylation pattern as an indication for a functional interaction of MLL-AF4 with chromatin.80,81 This number matches approximately the amount of loci occupied by MLL; 55 however, the potential overlap has never been determined. Surprisingly, experiments searching for MLL fusion controlled transcripts on the RNA level uncovered only a relatively small number (<100) of genes with a significant response to MLL fusion presence.64,100,101 Obviously, many genes are largely resistant to MLL fusion induced modifications and only some of them, like the HOX genes, are susceptible to e.g. elongation stimulation. This fits well with the fact that a subset of genes controlling embryonic development and cellular differentiation preferentially are occupied by RNA polymerase II also in the non-transcribed state. These genes are poised for transcription, and release of the stalled polymerase allows a fast response without the need to recruit transcription factors and histone modifiers to free the chromatin and allow assembly of the transcriptional initiation complex.102 Whatever the exact number and identity of all fusion targets will be, undoubtedly, HOX deregulation is the most important factor for MLL fusion induced leukemogenesis.79,100,101,103–109 HOX proteins, especially HOXA9, and its dimerization partner MEIS1, are major hematopoietic oncoproteins that are over-expressed in a wide variety of different leukemias and that act, at least partially, through activation of the proto-oncogene c-Myb.110 In general, HOX transcription factors are not only master controls of embryonic development but they also direct normal hematopoietic differentiation. HOX expression is high in stem cells and early precursors and needs to be down-regulated for maturation. Therefore, a continuous ectopic HOX expression will block differentiation and create a rapidly proliferating pre-leukemic precursor pool (Figure 4). Secondary mutations will have to occur to convert this smoldering state into an acute leukemia. Such mutations have been found in murine experimental models111 and also in patient cells that frequently carry an activating mutation in the receptor tyrosine kinase Flt-3.112,113 In a very surprising development, it has also recently been suggested that increased glycogen-synthase-kinase 3 activity is involved in the etiology of mixed lineage leukemia, an unexpected finding because GSK3 normally is a tumor suppressor gene.114 It is very interesting that mixed lineage leukemia tends to be a pediatric disease in contrast to many other tumors that arise later in life, because several years are needed to accrue the mutations necessary to convert a normal cell into a cancerous state. An attractive hypothesis to answer this question has been brought forward by Greaves and colleagues115. They speculated that a persistent genetic assault during gestation first produces MLL translocations. Once these are transcribed, the presence of the fusion proteins might sensitize cells to further mutations induced by the same mutagens that created the fusion before. In this way, secondary events would accumulate very rapidly and congenital leukemia would ensue. Indeed, there are experimental hints that MLL fusions increase susceptibility to mutagenic influence,115 and a few reports suggest that this might be due to inhibition of the tumor suppressor protein p53.116–118
Figure 4.
The role of HOX proteins in control of hematopoiesis. HOX transcription factors control hematopoietic differentiation. HOX expression must be terminated for maturation to occur, and therefore ectopic presence will block maturation and cause a population of self-renewing precursor cells to expand.
Unsolved questions and future directions
While the molecular pathways triggered by MLL fusion proteins slowly emerge, other major questions still wait for answers.
- Certainly, the most pressing problem with respect to the dismal prognosis of this disease is if new rational treatments can be devised by targeting the enzymatic activities necessary for MLL fusion activity.
One possible approach would be to interfere with the protein interactions necessary for proper MLL fusion protein function. Here, either the interaction with menin or LEDGF could be disrupted or, at least for the fusions employing the EAP complex, the protein-protein interactions stabilizing EAP might be targeted. Proof of principle experiments have been performed with peptides as binding site mimetics86,87 but if this is a feasible approach in patients remains to be seen. Theoretically, also the interaction surface between the MLL CxxC domain and DNA might be a point of attack, since this interaction has been analyzed in detail by X-ray crystallography.119 The largest concern with these strategies, however, is that apart from technical difficulties delivering peptides, likely also the vital natural function of MLL will be abrogated leading to toxicity. A small molecule approach that can be finely tuned to turn down the local hyperactivity of the enzymatic functions of MLL recruited EAP seems more promising. In this regard, specific methyltransferase inhibitors blocking DOT1L might be valuable tools. Also kinases like CDK9 or maybe GSK3 are potential targets for a rational therapeutic approach. Finally, also activities downstream of MLL fusions or cooperative oncogenic pathways might be druggable. It has been shown, for example, that inhibitors of the FLT-3 receptor tyrosine kinase are remarkably efficacious in mixed lineage leukemia animal models,120,121 although the role of FLT-3 in the etiology of MLL fusion induced leukemia is not exactly clear.122 - It is not known how histone modifications introduced by MLL and MLL fusions actually support transcription. In particular, H3K79 methylation still remains enigmatic.
- HOX proteins are transcription factors at the top of a regulatory cascade. We need to know what is downstream of HOX and which other proteins cooperate with these regulators.
- It would be interesting to know if MLL fusions are indeed only hyperactive MLL molecules and how these molecules find their appropriate binding sites.
With advanced molecular biology at hand it is to hope that 18 years of MLL research will finally translate into better survival chances for patients that all but too often face a bleak prospect after their diagnosis.
Acknowledgments:
first I would like to apologize to all scientists whose work could not be recognized here due to space constraints. My special thanks go to all technicians, students, graduates, and post-docs in my laboratory, past and present, and finally generous funding is acknowledged by DFG, Mildred-Scheel Stiftung/Deutsche Krebshilfe, Jose-Carreras Leukemia Fund, Curt-Bohnewald-Fond, and Freifrau v. Fritsch Stiftung.
Footnotes
Authorship and Disclosures
The author reported no potential conflicts of interest.
References
- 1.Stass S, Mirro J, Melvin S, Pui CH, Murphy SB, Williams D. Lineage switch in acute leukemia. Blood. 1984;64:701–6. [PubMed] [Google Scholar]
- 2.Mirro J, Kitchingman G, Williams D, Lauzon GJ, Lin CC, Callihan T, et al. Clinical and laboratory characteristics of acute leukemia with the 4;11 translocation. Blood. 1986;67:689–97. [PubMed] [Google Scholar]
- 3.Mirro J, Kitchingman GR, Williams DL, Murphy SB, Zipf TF, Stass SA. Mixed lineage leukemia: the implications for hematopoietic differentiation. Blood. 1986;68:597–9. [PubMed] [Google Scholar]
- 4.Mirro J, Zipf TF, Pui CH, Kitchingman G, Williams D, Melvin S, et al. Acute mixed lineage leukemia: clinicopathologic correlations and prognostic significance. Blood. 1985;66:1115–23. [PubMed] [Google Scholar]
- 5.Morse HG, Heideman R, Hays T, Robinson A. 4;11 translocation in acute lymphoblastic leukemia: a specific syndrome. Cancer Genet Cytogenet. 1982;7:165–72. doi: 10.1016/0165-4608(82)90012-7. [DOI] [PubMed] [Google Scholar]
- 6.Prigogina EL, Fleischman EW, Puchkova GP, Kulagina OE, Majakova SA, Balakirev SA, et al. Chromosomes in acute leukemia. Hum Genet. 1979;53:5–16. doi: 10.1007/BF00289443. [DOI] [PubMed] [Google Scholar]
- 7.Van den Berghe H, David G, Broeckaert-Van Orshoven A, Louwagie A, Verwilghen R, Casteels-Van Daele M, et al. A new chromosome anomaly in acute lymphoblastic leukemia (ALL) Human Genet. 1979;46:173–80. doi: 10.1007/BF00291919. [DOI] [PubMed] [Google Scholar]
- 8.Pui CH, Sandlund JT, Pei D, Campana D, Rivera GK, Ribeiro RC, et al. Improved outcome for children with acute lymphoblastic leukemia: results of Total Therapy Study XIIIB at St Jude Children’s Research Hospital. Blood. 2004;104:2690–6. doi: 10.1182/blood-2004-04-1616. [DOI] [PubMed] [Google Scholar]
- 9.Hilden JM, Dinndorf PA, Meerbaum SO, Sather H, Villaluna D, Heerema NA, et al. Analysis of prognostic factors of acute lymphoblastic leukemia in infants: report on CCG 1953 from the Children’s Oncology Group. Blood. 2006;108:441–51. doi: 10.1182/blood-2005-07-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosaka Y, Koh K, Kinukawa N, Wakazono Y, Isoyama K, Oda T, et al. Infant acute lymphoblastic leukemia with MLL gene rearrangements: outcome following intensive chemotherapy and hematopoietic stem cell transplantation. Blood. 2004;104:3527–34. doi: 10.1182/blood-2004-04-1390. [DOI] [PubMed] [Google Scholar]
- 11.Tomizawa D, Koh K, Sato T, Kinukawa N, Morimoto A, Isoyama K, et al. Outcome of risk-based therapy for infant acute lymphoblastic leukemia with or without an MLL gene rearrangement, with emphasis on late effects: a final report of two consecutive studies, MLL96 and MLL98, of the Japan Infant Leukemia Study Group. Leukemia. 2007;21:2258–63. doi: 10.1038/sj.leu.2404903. [DOI] [PubMed] [Google Scholar]
- 12.Pui CH, Relling MV. Topoisomerase II inhibitor-related acute myeloid leukaemia. Br J Haematol. 2000;109:13–23. doi: 10.1046/j.1365-2141.2000.01843.x. [DOI] [PubMed] [Google Scholar]
- 13.Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23:6306–15. doi: 10.1200/JCO.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 14.Schoch C, Schnittger S, Klaus M, Kern W, Hiddemann W, Haferlach T. AML with 11q23/MLL abnormalities as defined by the WHO classification: incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood. 2003;102:2395–402. doi: 10.1182/blood-2003-02-0434. [DOI] [PubMed] [Google Scholar]
- 15.Basecke J, Whelan JT, Griesinger F, Bertrand FE. The MLL partial tandem duplication in acute myeloid leukaemia. Br J Haematol. 2006;135:438–49. doi: 10.1111/j.1365-2141.2006.06301.x. [DOI] [PubMed] [Google Scholar]
- 16.Djabali M, Selleri L, Parry P, Bower M, Young BD, Evans GA. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat Gen. 1992;2:113–8. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- 17.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, et al. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–8. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 18.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 19.Ziemin-van der Poel S, McCabe NR, Gill HJ, Espinosa R, III, Patel Y, Harden A, et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci USA. 1991;88:10735–9. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breen TR, Harte PJ. Trithorax regulates multiple homeotic genes in the bithorax and Antennapedia complexes and exerts different tissue-specific, parasegment-specific and promoter-specific effects on each. Development. 1993;117:119–34. doi: 10.1242/dev.117.1.119. [DOI] [PubMed] [Google Scholar]
- 21.Breen TR, Harte PJ. Molecular characterization of the trithorax gene, a positive regulator of homeotic gene expression in Drosophila. Mech Develop. 1991;35:113–27. doi: 10.1016/0925-4773(91)90062-b. [DOI] [PubMed] [Google Scholar]
- 22.Mazo AM, Huang DH, Mozer BA, Dawid IB. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc Natl Acad Sci USA. 1990;87:2112–6. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMahon KA, Hiew SY, Hadjur S, Veiga-Fernandes H, Menzel U, Price AJ, et al. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1:338–45. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Yagi H, Deguchi K, Aono A, Tani Y, Kishimoto T, Komori T. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood. 1998;92:108–17. [PubMed] [Google Scholar]
- 25.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–8. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 26.Kennison JA, Tamkun JW. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci USA. 1988;85:8136–40. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, et al. MLL targets SET domain methyl-transferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–17. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–28. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 29.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–49. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, et al. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277:10753–5. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 31.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA. 2001;98:12902–7. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsieh JJ, Cheng EH, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115:293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 33.Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol Cell Biol. 2003;23:186–94. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeda S, Chen DY, Westergard TD, Fisher JK, Rubens JA, Sasagawa S, et al. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Gene Develop. 2006;20:2397–409. doi: 10.1101/gad.1449406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood. 2002;100:3710–8. doi: 10.1182/blood-2002-04-1015. [DOI] [PubMed] [Google Scholar]
- 36.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–85. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 37.Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, Bochkarev A, et al. Structural basis for molecular recognition and presentation of histone H3 by WDR5. The EMBO J. 2006;25:4245–52. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–9. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 39.Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, et al. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat Struct Mol Biol. 2006;13:704–12. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Southall SM, Wong PS, Odho Z, Roe SM, Wilson JR. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell. 2009;33:181–91. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 41.Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol. 2001;21:2249–58. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, et al. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci USA. 1998;95:4152–7. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, Hess JL. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67:7275–83. doi: 10.1158/0008-5472.CAN-06-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–18. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 45.Milne TA, Hughes CM, Lloyd R, Yang Z, Rozenblatt-Rosen O, Dou Y, et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc Natl Acad Sci USA. 2005;102:749–54. doi: 10.1073/pnas.0408836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michel F, Crucifix C, Granger F, Eiler S, Mouscadet JF, Korolev S, et al. Structural basis for HIV-1 DNA integration in the human genome, role of the LEDGF/P75 cofactor. EMBO J. 2009;28:980–91. doi: 10.1038/emboj.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, et al. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–9. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 49.Zeleznik-Le NJ, Harden AM, Rowley JD. 11q23 translocations split the "AT-hook" cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc Natl Acad Sci USA. 1994;91:10610–4. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Birke M, Schreiner S, Garcia-Cuellar MP, Mahr K, Titgemeyer F, Slany RK. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res. 2002;30:958–65. doi: 10.1093/nar/30.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bach C, Mueller D, Buhl S, Garcia-Cuellar MP, Slany RK. Alterations of the CxxC domain preclude oncogenic activation of mixed-lineage leukemia 2. Oncogene. 2009;28:815–23. doi: 10.1038/onc.2008.443. [DOI] [PubMed] [Google Scholar]
- 52.Xia ZB, Anderson M, Diaz MO, Zeleznik-Le NJ. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc Natl Acad Sci USA. 2003;100:8342–7. doi: 10.1073/pnas.1436338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes. Genes Develop. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gregory GD, Vakoc CR, Rozovskaia T, Zheng X, Patel S, Nakamura T, et al. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol. 200:27, 8466–79. doi: 10.1128/MCB.00993-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scacheri PC, Davis S, Odom DT, Crawford GE, Perkins S, Halawi MJ, et al. Genome-wide analysis of menin binding provides insights into MEN1 tumorigenesis. PLoS Gen. 2006;2:e51. doi: 10.1371/journal.pgen.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hars ES, Lyu YL, Lin CP, Liu LF. Role of apoptotic nuclease caspase-activated DNase in etoposide-induced treatment-related acute myelogenous leukemia. Cancer Res. 2006;66:8975–9. doi: 10.1158/0008-5472.CAN-06-1724. [DOI] [PubMed] [Google Scholar]
- 57.Libura J, Slater DJ, Felix CA, Richardson C. Therapy-related acute myeloid leukemia-like MLL rearrangements are induced by etoposide in primary human CD34+ cells and remain stable after clonal expansion. Blood. 2005;105:2124–31. doi: 10.1182/blood-2004-07-2683. [DOI] [PubMed] [Google Scholar]
- 58.Betti CJ, Villalobos MJ, Diaz MO, Vaughan AT. Apoptotic triggers initiate translocations within the MLL gene involving the nonhomologous end joining repair system. Cancer Res. 2001;61:4550–5. [PubMed] [Google Scholar]
- 59.Gillert E, Leis T, Repp R, Reichel M, Hosch A, Breitenlohner I, et al. A DNA damage repair mechanism is involved in the origin of chromosomal translocations t(4;11) in primary leukemic cells. Oncogene. 1999;18:4663–71. doi: 10.1038/sj.onc.1202842. [DOI] [PubMed] [Google Scholar]
- 60.Nakada S, Katsuki Y, Imoto I, Yokoyama T, Nagasawa M, Inazawa J, et al. Early G2/M checkpoint failure as a molecular mechanism underlying etoposide-induced chromosomal aberrations. J Clin Invest. 2006;116:80–9. doi: 10.1172/JCI25716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102:2951–9. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- 62.Ferrando AA, Armstrong SA, Neuberg DS, Sallan SE, Silverman LB, Korsmeyer SJ, et al. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003;102:262–8. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- 63.Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–43. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 64.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–7. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 65.Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J, et al. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009 doi: 10.1038/leu.2009.33. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 66.Burmeister T, Meyer C, Schwartz S, Hofmann J, Molkentin M, Kowarz E, et al. The MLL recombinome of adult CD10-negative B-cell precursor acute lymphoblastic leukemia -results from the GMALL study group. Blood. 2009;113:4011–5. doi: 10.1182/blood-2008-10-183483. [DOI] [PubMed] [Google Scholar]
- 67.Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–37. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol. 2004;24:10470–8. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Slany RK, Lavau C, Cleary ML. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol Cell Biol. 1998;18:122–9. doi: 10.1128/mcb.18.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen J, Santillan DA, Koonce M, Wei W, Luo R, Thirman MJ, et al. Loss of MLL PHD finger 3 is necessary for MLL-ENL-induced hematopoietic stem cell immortalization. Cancer Res. 2008;68:6199–207. doi: 10.1158/0008-5472.CAN-07-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muntean AG, Giannola D, Udager AM, Hess JL. The PHD fingers of MLL block MLL fusion protein-mediated transformation. Blood. 2008;112:4690–3. doi: 10.1182/blood-2008-01-134056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schreiner SA, Garcia-Cuellar MP, Fey GH, Slany RK. The leukemogenic fusion of MLL with ENL creates a novel transcriptional transactivator. Leukemia. 1999;13:1525–33. doi: 10.1038/sj.leu.2401534. [DOI] [PubMed] [Google Scholar]
- 73.Zeisig DT, Bittner CB, Zeisig BB, Garcia-Cuellar MP, Hess JL, Slany RK. The eleven-nineteen-leukemia protein ENL connects nuclear MLL fusion partners with chromatin. Oncogene. 2005;24:5525–32. doi: 10.1038/sj.onc.1208699. [DOI] [PubMed] [Google Scholar]
- 74.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Gen. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 75.Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–54. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 77.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 008;28:2825–39. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–78. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 79.Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005;65:11367–74. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- 80.Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL, et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Develop. 2008;22:3403–8. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–68. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873–6. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 83.Luo RT, Lavau C, Du C, Simone F, Polak PE, Kawamata S, et al. The elongation domain of ELL is dispensable but its ELL-associated factor 1 interaction domain is essential for MLL-ELL-induced leukemogenesis. Mol Cell Biol. 2001;21:5678–87. doi: 10.1128/MCB.21.16.5678-5687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Simone F, Luo RT, Polak PE, Kaberlein JJ, Thirman MJ. ELL-associated factor 2 (EAF2), a functional homolog of EAF1 with alternative ELL binding properties. Blood. 2003;101:2355–62. doi: 10.1182/blood-2002-06-1664. [DOI] [PubMed] [Google Scholar]
- 85.Simone F, Polak PE, Kaberlein JJ, Luo RT, Levitan DA, Thirman MJ. EAF1, a novel ELL-associated factor that is delocalized by expression of the MLL-ELL fusion protein. Blood. 2001;98:201–9. doi: 10.1182/blood.v98.1.201. [DOI] [PubMed] [Google Scholar]
- 86.Palermo CM, Bennett CA, Winters AC, Hemenway CS. The AF4-mimetic peptide, PFWT, induces necrotic cell death in MV4–11 leukemia cells. Leuk Res. 2008;32:633–42. doi: 10.1016/j.leukres.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Srinivasan RS, Nesbit JB, Marrero L, Erfurth F, LaRussa VF, Hemenway CS. The synthetic peptide PFWT disrupts AF4-AF9 protein complexes and induces apoptosis in t(4;11) leukemia cells. Leukemia. 2004;18:1364–72. doi: 10.1038/sj.leu.2403415. [DOI] [PubMed] [Google Scholar]
- 88.Bennett CA, Winters AC, Barretto NN, Hemenway CS. Molecular targeting of MLL-rearranged leukemia cell lines with the synthetic peptide PFWT synergistically enhances the cytotoxic effect of established chemotherapeutic agents. Leuk Res. 2009;33:937–47. doi: 10.1016/j.leukres.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sobulo OM, Borrow J, Tomek R, Reshmi S, Harden A, Schlegelberger B, et al. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3) Proc Natl Acad Sci USA. 1997;94:8732–7. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rowley JD, Reshmi S, Sobulo O, Musvee T, Anastasi J, Raimondi S, et al. All patients with the T(11;16)(q23;p13.3) that involves MLL and CBP have treatment-related hematologic disorders. Blood. 1997;90:535–41. [PubMed] [Google Scholar]
- 91.Lavau C, Du C, Thirman M, Zeleznik-Le N. Chromatin-related properties of CBP fused to MLL generate a myelodysplastic-like syndrome that evolves into myeloid leukemia. EMBO J. 2000;19:4655–64. doi: 10.1093/emboj/19.17.4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cheung N, Chan LC, Thompson A, Cleary ML, So CW. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–15. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 93.Pal S, Sif S. Interplay between chromatin remodelers and protein arginine methyltransferases. J Cell Physiol. 2007;213:306–15. doi: 10.1002/jcp.21180. [DOI] [PubMed] [Google Scholar]
- 94.Strehl S, Borkhardt A, Slany R, Fuchs UE, Konig M, Haas OA. The human LASP1 gene is fused to MLL in an acute myeloid leukemia with t(11;17)(q23;q21) Oncogene. 2003;22:157–60. doi: 10.1038/sj.onc.1206042. [DOI] [PubMed] [Google Scholar]
- 95.Fuchs U, Rehkamp G, Haas OA, Slany R, Konig M, Bojesen S, et al. The human formin-binding protein 17 (FBP17) interacts with sorting nexin, SNX2, and is an MLL-fusion partner in acute myelogeneous leukemia. Proc Natl Acad Sci USA. 2001;98:8756–61. doi: 10.1073/pnas.121433898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.So CW, Lin M, Ayton PM, Chen EH, Cleary ML. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell. 2003;4:99–110. doi: 10.1016/s1535-6108(03)00188-0. [DOI] [PubMed] [Google Scholar]
- 97.Martin ME, Milne TA, Bloyer S, Galoian K, Shen W, Gibbs D, et al. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell. 2003;4:197–207. doi: 10.1016/s1535-6108(03)00214-9. [DOI] [PubMed] [Google Scholar]
- 98.Caslini C, Alarcon AS, Hess JL, Tanaka R, Murti KG, Biondi A. The amino terminus targets the mixed lineage leukemia (MLL) protein to the nucleolus, nuclear matrix and mitotic chromosomal scaffolds. Leukemia. 2000;14:1898–908. doi: 10.1038/sj.leu.2401933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garcia-Cuellar MP, Schreiner SA, Birke M, Hamacher M, Fey GH, Slany RK. ENL, the MLL fusion partner in t(11;19), binds to the cAbl interactor protein 1 (ABI1) that is fused to MLL in t(10;11)+ Oncogene. 2000;19:1744–51. doi: 10.1038/sj.onc.1203506. [DOI] [PubMed] [Google Scholar]
- 100.Zeisig BB, Milne T, Garcia-Cuellar MP, Schreiner S, Martin ME, Fuchs U, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–28. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Horton SJ, Grier DG, McGonigle GJ, Thompson A, Morrow M, De Silva I, et al. Continuous MLL-ENL expression is necessary to establish a "Hox Code" and maintain immortalization of hematopoietic progenitor cells. Cancer Res. 2005;65:9245–52. doi: 10.1158/0008-5472.CAN-05-1691. [DOI] [PubMed] [Google Scholar]
- 102.Price DH. Poised polymerases: on your mark...get set..go! . Molecular cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 103.Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–40. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li Z, Luo RT, Mi S, Sun M, Chen P, Bao J, et al. Consistent deregulation of gene expression between human and murine MLL rearrangement leukemias. Cancer Res. 2009;69:1109–16. doi: 10.1158/0008-5472.CAN-08-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Erfurth FE, Popovic R, Grembecka J, Cierpicki T, Theisler C, Xia ZB, et al. MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression. Proc Natl Acad Sci USA. 2008;105:7517–22. doi: 10.1073/pnas.0800090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–74. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci USA. 2005;102:14765–70. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, et al. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci USA. 2005;102:8603–8. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004;14:2063–9. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 110.Hess JL, Bittner CB, Zeisig DT, Bach C, Fuchs U, Borkhardt A, et al. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108:297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Horton SJ, Walf-Vorderwulbecke V, Chatters SJ, Sebire NJ, de Boer J, Williams O. Acute myeloid leukemia induced by MLL-ENL is cured by oncogene ablation despite acquisition of complex genetic abnormalities. Blood. 2009;113:4922–9. doi: 10.1182/blood-2008-07-170480. [DOI] [PubMed] [Google Scholar]
- 112.Stam RW, Schneider P, de Lorenzo P, Valsecchi MG, den Boer ML, Pieters R. Prognostic significance of high-level FLT3 expression in MLL-rearranged infant acute lymphoblastic leukemia. Blood. 2007;110:2774–5. doi: 10.1182/blood-2007-05-091934. [DOI] [PubMed] [Google Scholar]
- 113.Brown P, Levis M, Shurtleff S, Campana D, Downing J, Small D. FLT3 inhibition selectively kills childhood acute lymphoblastic leukemia cells with high levels of FLT3 expression. Blood. 2005;105:812–20. doi: 10.1182/blood-2004-06-2498. [DOI] [PubMed] [Google Scholar]
- 114.Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TC, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–9. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Eguchi M, Eguchi-Ishimae M, Knight D, Kearney L, Slany R, Greaves M. MLL chimeric protein activation renders cells vulnerable to chromosomal damage: an explanation for the very short latency of infant leukemia. Genes Chromosomes Cancer. 2006;45:754–60. doi: 10.1002/gcc.20338. [DOI] [PubMed] [Google Scholar]
- 116.Wiederschain D, Kawai H, Shilatifard A, Yuan ZM. Multiple mixed lineage leukemia (MLL) fusion proteins suppress p53-mediated response to DNA damage. J Biol Chem. 2005;280:24315–21. doi: 10.1074/jbc.M412237200. [DOI] [PubMed] [Google Scholar]
- 117.Wiederschain D, Kawai H, Gu J, Shilatifard A, Yuan ZM. Molecular basis of p53 functional inactivation by the leukemic protein MLL-ELL. Mol Cell Biol. 2003;23:4230–46. doi: 10.1128/MCB.23.12.4230-4246.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Megonigal MD, Rappaport EF, Nowell PC, Lange BJ, Felix CA. Potential role for wild-type p53 in leukemias with MLL gene translocations. Oncogene. 1998;16:1351–6. doi: 10.1038/sj.onc.1201637. [DOI] [PubMed] [Google Scholar]
- 119.Allen MD, Grummitt CG, Hilcenko C, Min SY, Tonkin LM, Johnson CM, et al. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. EMBO J. 2006;25:4503–12. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stam RW, den Boer ML, Schneider P, Nollau P, Horstmann M, Beverloo HB, et al. Targeting FLT3 in primary MLL-gene-rearranged infant acute lymphoblastic leukemia. Blood. 2005;106:2484–90. doi: 10.1182/blood-2004-09-3667. [DOI] [PubMed] [Google Scholar]
- 121.Armstrong SA, Kung AL, Mabon ME, Silverman LB, Stam RW, Den Boer ML, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3:173–83. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 122.Morgado E, Albouhair S, Lavau C. Flt3 is dispensable to the Hoxa9/Meis1 leukemogenic cooperation. Blood. 2007;109:4020–2. doi: 10.1182/blood-2006-01-039586. [DOI] [PubMed] [Google Scholar]