Molecular basis of the CRAC channel (original) (raw)
. Author manuscript; available in PMC: 2009 Aug 31.
Published in final edited form as: Cell Calcium. 2007 May 7;42(2):133–144. doi: 10.1016/j.ceca.2007.03.002
Abstract
Ca2+ release-activated Ca2+ (CRAC) channels, located in the plasma membrane, are opened upon release of Ca2+ from intracellular stores, permitting Ca2+ entry and sustained [Ca2+]i signaling that replenishes the store in numerous cell types. This mechanism is particularly important in T lymphocytes of the immune system, providing the missing link in the signal transduction cascade that is initiated by T cell receptor engagement and leads to altered expression of genes that results ultimately in the production of cytokines and cell proliferation. In the past three years, RNA interference screens together with over-expression and site-directed mutagenesis have identified the triggering molecule (Stim) that links store depletion to CRAC channel-mediated Ca2+ influx and the pore subunit (Orai) of the CRAC channel that allows highly selective entry of Ca2+ ions into cells.
Keywords: CRAC channel, T lymphocyte, Store-operated calcium, Stim, Orai
1. Introduction
This review will cover the discovery of Stim and Orai and their functional roles in store-operated Ca2+ influx, emphasizing our own contributions. Up front, we recognize that several other groups have been engaged in a similar quest; many of their key contributions are represented in accompanying articles in this series. We introduce the topic by first considering background information on calcium signaling in lymphocytes, the electrical description of CRAC current, and its physiological role in lymphocytes. Then we discuss how a candidate-based RNA interference (RNAi) screen led to the discovery of Drosophila Stim. In further work, we identified two functions of Stim and the mammalian homologue STIM1 (collectively referred to here as Stim1): the sensor function that initially detects the reduction of Ca2+ content in the lumen of the endoplasmic reticulum (ER); and the messenger function provided by Stim1 translocation to the plasma membrane to activate the CRAC channel. We further discuss our genome-wide RNAi screen that identified Drosophila Orai and additional genes that are required for CRAC channel activity; the recipe for greatly amplified CRAC current by co-expression of Stim + Orai; and the identification of amino acids that regulate Ca2+ selectivity, clinching the identification of Orai as the pore-forming component of the CRAC channel.
1.1. Lymphocytes and the early activation pathway of an immune response
T lymphocytes require direct contact with antigen-presenting cells (APC) such as dendritic cells to become activated. In parallel with work on the CRAC channel, the Cahalan lab group, in collaboration with Ian Parker at UCI, has used two-photon microscopy to image living T cells and dendritic cells interacting within intact mouse lymph nodes [1,2]. Under basal conditions without antigen, contacts between dendritic cells and T cells are initiated by virtue of robust T cell motility in combination with extension and retraction of dendritic cell dendrites, leading to a very high frequency of transient T cell/dendritic cell interactions [3]. Dendritic cells capture antigens in the periphery and bring them into the lymph node, serving as APC to initiate the immune response. When antigen is present, T cell–dendritic cell contact durations are prolonged as Ca2+ signaling is initiated; and after several hours, T cells are stably associated in clusters with individual dendritic cells [4]. Finally, the T cells let go and begin to divide about 24 h after initially encountering the antigen. Our story about the CRAC channels focuses on the T cell Ca2+ signal that is initiated by contact with APC and is crucial for initiation of an adaptive immune response.
1.2. Ion channels in human T cells
Ca2+ influx across the plasma membrane is primarily responsible for elevation of cytosolic free Ca2+ concentration ([Ca2+]i) following TCR engagement. But how does calcium enter the cell? Even earlier, nearly 25 years ago and shortly after the patch-clamp method was developed, researchers in the Cahalan lab initiated the study of ion channels in lymphocytes [5]. At that time, the importance of Ca2+ signaling to lymphocyte activation had already been proposed [6], and our initial expectation was that calcium channels might be detected electrically. At first, we investigated K+ channels that regulate the membrane potential of T cells (and thereby indirectly modulate Ca2+ signaling), but no Ca2+ current was detected. By combining single-cell Ca2+ monitoring with patch recording, Richard Lewis in the Cahalan lab found a tiny Ca2+-selective current with an inwardly rectifying I–V and a very positive reversal potential in Jurkat T cells [7]. The Ca2+ current, only a few picoamperes per cell, activated spontaneously during whole-cell recording with dialysis of a Ca2+ chelator. The same current could also be activated by T cell receptor stimulation during perforated-patch recording in a close temporal relationship with the upstroke of the cytosolic Ca2+ signal monitored simultaneously with a fluorescent Ca2+-indicator. The current could be blocked by external application of Ni2+ or Cd2+, agents that also blocked the mitogen-evoked Ca2+ signaling. During activation of the current, single-channel currents and even increased current noise were not evident, indicating a low single-channel conductance. At the time, a possible link of the Ca2+ current across the surface membrane to the internal stores was discussed, but the relation to store depletion and to models for “capacitative calcium entry” (CCE) [8] was unclear. The current was further studied by other groups, including Richard Lewis at Stanford and Reinhold Penner in Göttingen, and was shown to be activated by thapsigargin and by intracellular IP3 [9–11], strengthening its identity as a store-operated Ca2+ channel. It was given the name CRAC, for Ca2+ release-activated Ca2+ current, by Penner.
Fig. 1 highlights the function of ion channels during an encounter between a T cell and an APC that has processed the antigens specific for the T cell receptor expressed on the T cell. Very few channels are open initially; in fact only a few of the voltage-gated Kv1.3 channels in the cell suffice to maintain the resting membrane potential of about −50 mV [12]. Contact with peptide-MHC on the APC initiates the proximal signaling cascade involving adapter molecules and tyrosine kinases, resulting in phospholipase C activation and the formation of the two messengers, IP3 and DAG. IP3 opens the IP3 receptor-channels localized in the ER membrane, which in turn releases Ca2+ from the ER and begins to elevate cytosolic Ca2+. Depletion of the ER Ca2+ stores indirectly leads to Ca2+ influx through CRAC channels, driving [Ca2+]i into the micromolar range. As [Ca2+]i rises, Ca2+-activated K+ channels are opened and K+ leaves the cell, causing the membrane potential to become more negative to maintain the driving force for Ca2+ influx. The relative contribution of voltage-gated and Ca2+-activated K+ channels to regulate membrane potential and counterbalance the influx of Ca2+ depends upon the T cell subset and their activation state [13]. Ca2+ binds to calmodulin and activates calcineurin, a phosphatase that dephosphorylates a transcription factor NFAT, which then translocates to the nucleus where it binds to promoter regions to turn on expression of numerous genes, including the key cytokine, Interleukin-2 [14,15]. The pharmacological agent thapsigargin bypasses the upstream events in T cell signaling by inhibiting the SERCA pump in the ER membrane, which leads to depletion of the ER Ca2+ store and activation of CRAC channels that trigger subsequent events including gene expression. Thapsigargin was thus employed as a very useful tool in our RNAi screening effort, described in Sections 2 and 3 below.
Fig. 1.
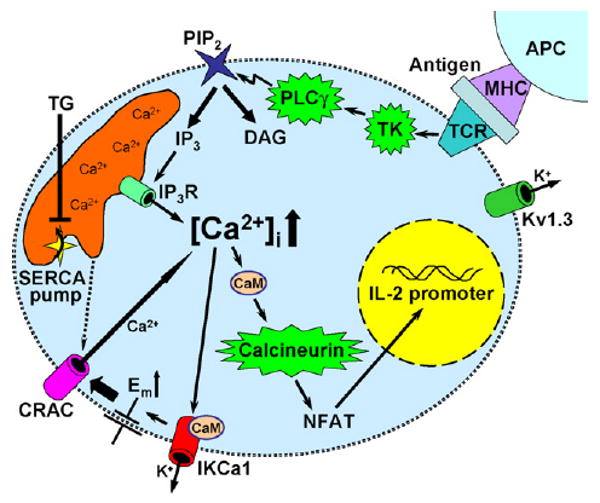
Ion channels and signaling pathways in T cells.
1.3. Ca2+ signaling and gene expression during T cell activation
By the mid-1990s, the calcium dependence of gene expression for T cell activation was determined quantitatively in single-cell Ca2+ assays combined with NFAT reporter gene monitoring [15,16]. To activate this key transcriptional regulatory pathway, at least 1–2 h of elevated [Ca2+]i is required. The Ca2+ signal may be oscillatory or sustained, and it is thought that oscillations in [Ca2+]i may provide a means to activate transcription of different genes selectively and without the requirement for continuous Ca2+ elevation, which could activate cell death pathways [17,18]. The content of the internal Ca2+ store is limited in T cells, and Ca2+ influx from outside of the cell is required to elevate Ca2+ to a level that is required for specific gene expression during T cell activation. CRAC channel activity and downstream Ca2+ effects via the calcineurin/NFAT pathway are thus critical for changes in gene expression elicited by TCR stimulation, as evidenced by altered gene expression in T cells that lack functional CRAC channels from patients with severe combined immune deficiency (SCID) disorder [14].
1.4. CRAC channels at the start of the millennium
As of 2003, then, the biophysical properties of CRAC current had been characterized, as summarized in Table 1. The immunological importance of the CRAC current to the NFAT pathway and to T cell activation was also clear. However, neither the channel itself nor the mechanism that links store depletion to CRAC channel opening were known. The major breakthroughs finally came in 2005 and 2006 when we and other groups used RNAi to identify genes that were required and site-directed mutagenesis to define the functional role of two proteins, Stim1 and Orai1, in turning on and physically embodying the CRAC channel, respectively.
Table 1.
Comparison of CRAC current biophysical properties
| Property | Native T cell CRAC [7,9–11,53–55] | Native S2 CRAC [26,33] | Stim + Orai [33,48] | Stim + E180D Orai [48] |
|---|---|---|---|---|
| Store dependence | Yes | Yes | Yes | Yes |
| Inactivation by: | ||||
| Store refilling | Yes | Yes | Not tested | Not tested |
| Local [Ca2+]i | Yes | Yes | Yes | Yes |
| Run-down | Yes | Yes | Yes | Yes |
| Fast inactivation (100 ms time scale) | Yes | No | No | No |
| Calcium selectivity | Yes | Yes | Yes | No |
| Permeant divalent cations | Ca2+ > Ba2+ > Sr2+» Mg2+ | Ba2+ > Ca2+ > Sr2+ » Mg2+ | Ca2+ ∼ Sr2+ ∼Ba2+ » Mg2+ | Negligible |
| Unitary channel conductance in 20 mM Ca2+ | 21 fS | 36 fS | Not tested | N/A |
| Monovalent current in zero divalent | Yes | Yes | Yes | Yes |
| Monovalent cation selectivity, _P_Cs/_P_Na | Na+ > Cs+, 0.09–0.13 | Na+ > Cs+, 0.17 | Na+ > Cs+, 0.13 | Na+ ∼ Cs+, 0.71 |
| Depotentiation | Yes | Yes | Yes | No |
| Pharmacology: | ||||
| Gd3+ or La3+ | nM (Gd3+), reversible | nM (Gd3+), reversible | nM (Gd3+), reversible | nM (Gd3+), reversible |
| SKF 96365 | μM, partially reversible | μM, partially reversible | Not tested | Not tested |
| 2-APB | Low μM – potentiation; high μM – inhibition | Low μM – potentiation; high μM – inhibition | Low μM – potentiation; high μM – inhibition | Low and high μM – complex behavior (potentiation and inhibition phases) |
2. RNAi candidate screen leads to identification of Stim, the Ca2+ sensor and messenger for CRAC channel activation
2.1. Choice of S2 cells for RNAi that led to the screening breakthrough
In the late summer of 2000, three of us (JR, KO and KS) joined a new company (now TorreyPines Therapeutics Inc.) and as part of an early project sought a novel way to identify the molecular components of store-operated Ca2+ entry (SOCE), especially the elusive CRAC channel. Previous work had suggested certain TRP channels might be the molecular counterparts of both store-operated Ca2+ channels and CRAC channels, but biophysical properties of TRP channels and CRAC channels did not match, and it seemed the field was nibbling around the edges because it lacked a more systematic and direct way to attack the problem. Then a colleague, Craig Montell, brought to our attention a newly developed technique for suppressing gene function called RNAi, which had shown utility in plants, worms, and flies. In these cases, introduction to cells of a double-stranded RNA (dsRNA) mimicking a specific mRNA sequence causes sequence-specific degradation of the corresponding endogenous mRNA, leading to the suppression of gene expression.
The Drosophila S2 cell line attracted our attention because its utility for studying Ca2+ signaling mechanisms was already known [19–24]. S2 cells were an attractive target for performing an RNAi-based screen to identify genes that controls SOCE because most fundamental cellular processes are conserved from Drosophila to mammals and the Drosophila genome had just been published earlier in 2000 [25]. Of great importance in the interpretation of such a screen, the TorreyPines group teamed-up with the Cahalan group at UC Irvine to show that S2 cells possess a store-operated Ca2+ current. A detailed patch-clamp analysis showed that the store-operated Ca2+ current in S2 cells was remarkably similar in biophysical characteristics to the CRAC current in human lymphocytes (Table 1) [26], so the choice of S2 cells turned out to be a good one. Both currents are highly Ca2+ selective (_P_Ca/_P_Na > 1000) and can be activated by maneuvers that deplete the ER Ca2+ store: cell dialysis with a Ca2+ chelator (passive store depletion); internal application of IP3; or external application of thapsigargin or ionomycin. Both exhibit the characteristic inwardly rectifying I_–_V shape with a reversal potential >50mV and an unusually low single-channel conductance in the femtosiemen range. In the absence of external divalent cations, both currents are carried by monovalent cations and sodium is more permeant than cesium. The monovalent Na+ current declines (depotentiation) over tens of seconds in the continued absence of external divalents. The pharmacological properties are also quite similar: currents can be suppressed by lanthanides (La3+, Gd3+), SKF96365 or 2-APB (high μM range); while the application of 2-APB at lower concentration potentiates both currents. A more detailed comparison of the biophysical properties of CRAC current in S2 and T cells is presented in Table 1, along with characteristics of CRAC current induced by expression of Stim + Orai. These initial findings validated the use of S2 cells for identifying SOC/CRAC channel components for us and other investigators. That S2 cells possess a CRAC-like channel may not be surprising, as the S2 cell line was originally derived from a primary culture of late stage (20–24 h old) Drosophila melanogaster embryos and, based upon gene expression patterns and phagocytic activity, is thought to represent a haemocyte or macrophage-like cell [27,28].
2.2. How RNAi works in S2 cells
The Drosophila S2 cell culture system is ideally suited for moderate- to high-throughput assays of gene function using RNAi-mediated gene silencing. This is because RNAi in S2 cells is simple, efficient, and long-lived. S2 cells can be treated with long, ∼500 base-pair dsRNAs, which are taken up easily from the medium by the cells [29]. An advantage of S2 cells is that they do not require transfection reagents for uptake of dsRNAs. The dsRNAs appear to enter the cells through receptor-mediated endocytosis [30]. Once inside the cells, dsRNAs are processed to 21–23 nucleotide fragments called small interfering RNAs (siRNAs) by the action of the enzyme Dicer. The use of long dsRNA probes increases the likelihood of producing siRNAs that efficiently bind to the RNA-inducing silencing complex (RISC), thereby reducing optimization steps necessary in designing siRNA probes for RNAi. Once bound to the RISC complex, siRNAs initiate degradation of the complementary mRNA sequences and subsequent suppression of protein translation. These considerations enhance the feasibility of conducting a high-throughput RNAi-based screen in S2 cells. Since the Drosophila genome is much smaller and was better examined at the time than the human genome, S2 cells provided the ideal model system to test the role of candidate genes in CRAC channel function by systematically suppressing functional expression through RNAi.
2.3. Choice of candidates by bioinformatics
At the time we initiated our first RNAi screen, genome-wide dsRNA libraries were not available, so we chose a bioinformatics approach to identify potential candidate genes. At the beginning, we tested about 20 genes that were either known TRP family members or TRP-related sequences. When suppression of these genes failed to affect store-operated Ca2+ entry in S2 cells, we searched the Drosophila genome for all genes annotated as ion channels in the GadFly database (release 2). In addition, we identified genes having an InterPro channel-like motif (e.g., PROSITE, PRINTS, PRODOM) and genes that shared a Multiple Expectation maximum for Motif Elicitation (MEME) profile built from members of the TRP family. From this list, we excluded genes we had already tested, well-characterized voltage-regulated channels, non-calcium channels, and other genes that had functional data published in the literature (e.g., cacophony, eag, Glu-RIIA, Glu-RIIB, para, Sh) retaining ∼80% of the candidates from the Gadfly-derived list and ∼90% of the motif-derived list. Finally, we selected other genes containing Ca2+ binding motifs, genes annotated as plasma membrane ATPases, or genes with putative roles in SOC influx, arriving at a final list of 150 genes that were then targeted by RNAi individually and tested. As it turned out, Stim was included in the list not only because of its putative Ca2+ binding domain, but also because a transient bug in the genome annotation identified Stim as having a potassium ion-channel motif. This highlights the fact that at the time of our initial screen the annotation of the fly genome was still changing. In addition, novel classes of genes with undefined sequence motifs and architectures could not be discovered by a bioinformatics approach. Thus, another key component of the CRAC channel was only discovered later using unbiased genome-wide RNAi screens (i.e., olf186-F, the Drosophila homolog of human Orai1 or CRACM1) [31–33].
2.4. How the initial candidate screen worked
The initial screen (Fig. 2) utilized a FluoroSkan single-channel fluorimeter to measure baseline and SOCE signals. S2 cells were treated with the appropriate dsRNAs in T-75 flasks for 5 days, harvested and plated in 96-well plates, and then loaded with the Ca2+ sensitive dye fluo-4 in buffer containing calcium. Cells were washed in calcium-free buffer and fluorescence levels were then monitored by the FluoroSkan. Initial fluorescence was recorded for a brief period to estimate basal Ca2+ levels, followed by addition of either vehicle or thapsigargin to discharge intracellular Ca2+ stores and activate store-operated Ca2+ channels. After a short incubation, CaCl2 was added to the cells and a single fluorescence reading was recorded three min after Ca2+ addition as an index of store-operated Ca2+ influx in each well. This single-point assay was possible because in our hands the Ca2+ entry signal became stable 2–4 min after Ca2+ addition, and it greatly facilitated the throughput of the RNAi screen. It was with this assay that we identified the activity of Stim [34] (first revealed in a patent application published in 2004 with priority to 2003 [35], and presented publicly at the Biophysical Society meeting in February 2005), a finding consistent with a subsequent, independent report by Liou et al. [36]. We later used this basic paradigm in a genome-wide RNAi screen to identify the role of olf186-F (Orai) [33].
Fig. 2.
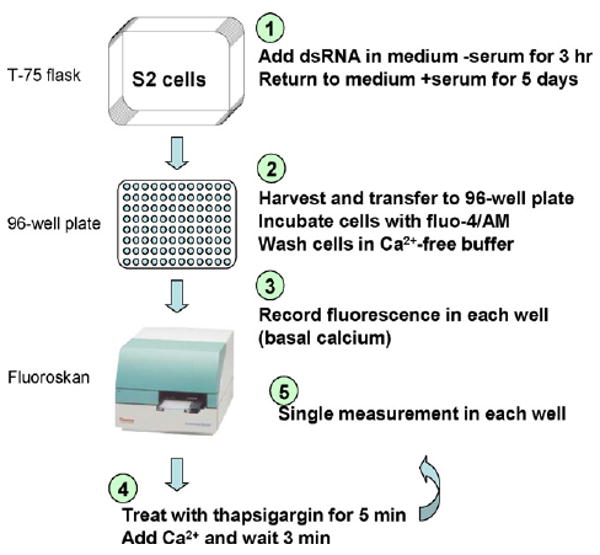
Candidate RNAi screening protocol.
2.5. Validation of Stim and STIM1 by single-cell Ca2+ imaging and patch-clamp
Our initial study validated the requirement for Stim by single-cell Ca2+ monitoring and by whole-cell recording in S2 cells pretreated with dsRNAs for Stim or control genes [34]. In Ca2+ imaging experiments, we used a thapsigargin and Ca2+ readdition protocol to assess SOCE in single cells. Following addition of thapsigargin in the absence of external Ca2+, a small transient rise in cytosolic free Ca2+ represents the release transient due to leak from the store into the cytoplasm. Subsequent readdition of external Ca2+ produces a very large Ca2+ signal that is due to Ca2+ influx through CRAC channels that are activated as a result of thagpsigargin-induced Ca2+ store depletion. In cells pretreated with dsRNAs for Stim, resting Ca2+ was unaltered and store content was normal, but Ca2+ influx upon readdition of external Ca2+ was profoundly inhibited. Patch-clamp experiments confirmed that knockdown of Stim by RNAi very effectively suppressed the CRAC current. There are two mammalian homologs of Drosophila Stim, STIM1 and STIM2. Suppression of STIM1 by treatment of HEK293 cells with STIM1-specific siRNAs inhibited thapsigargin-evoked Ca2+ entry and Ca2+ influx induced by muscarinic receptor stimulation, without altering resting Ca2+ levels, Ca2+ release transients, or membrane potentials [34]. In our hands, suppression of STIM2 had less of an effect on SOCE than did STIM1 knockdown. In Jurkat T cells, we again confirmed that suppression of STIM1 led to a profound reduction of the Ca2+ signal and CRAC current, this time using a short hairpin loop (shRNA) construct to reduce STIM1 expression selectively.
2.6. Dual functional role of Stim1: ER Ca2+ sensor and messenger to the plasma membrane
In the experiments described above, RNAi knockdown demonstrated a functional requirement for Stim1 expression in supporting CRAC channel activity. But how does Stim1 function, as a single span transmembrane protein that does not even remotely resemble any known ion channel? Drosophila Stim and its mammalian homologs, STIM1 and STIM2, are type 1 membrane proteins. The signal peptide sequence at the amino terminus and one predicted transmembrane segment indicated that the amino (N-) terminal portion of the protein faces the extracellular fluid or the lumen of the ER. The conserved N-terminal EF-hand motif suggested a possible role of Stim1 in Ca2+ signaling, and since Stim1 did not resemble any known ion channel we formulated the idea that it may serve as a Ca2+ sensor to trigger the process of CRAC channel activation. The approach we took was to overexpress the wild-type (WT) Stim1 protein or Stim1 mutant proteins in S2 and Jurkat T cells [37].
Our strategy for a series of point mutations was to disrupt Ca2+ binding to the EF-hand domain while preserving the overall protein structure, targeting different conserved residues that had been shown in other EF-hand proteins and by crystal structures to be essential for Ca2+ binding (Fig. 3). By making several different mutations on the same sites (D to N or D to A, and E to Q or E to A; Fig. 3 ovals) and corresponding mutations in different species (Stim and STIM1), we tried to avoid false-positive results. The mutants of Stim or STIM1 were expressed in S2 or Jurkat T cells, respectively, by transfection, and calcium signaling was compared in native cells and cells overexpressing either wild-type (WT) or mutant Stim/STIM1. Expression was confirmed by Western blot which showed similar expression levels of WT and mutants proteins. The S2 cells overexpressing the WT Stim had no phenotype, whereas expression of the EF-hand mutants produced an elevation of resting [Ca2+]i that we showed was due to constitutive activation of CRAC channels, since it was reduced to control levels by three inhibitors of CRAC current: 2-aminoethyldiphenyl borate (2-APB), SKF96365, and Gd3+. In Jurkat T cells, a modest enhancement of thapsigargin-evoked Ca2+ influx was produced by overexpressing the WT protein, and expression of the EF-hand mutant elevated cytosolic Ca2+ levels, as seen with S2 cells. This rise in Ca2+ was also completely blocked by CRAC channel inhibitors. Importantly, there was no change in ER store content or release kinetics, leading to the conclusion that expression of the EF-hand mutants bypassed the normal requirement for store depletion and activated CRAC channels even though the store was still replete with Ca2+. These findings suggested that Stim1 serves as an ER Ca2+ sensor that can trigger subsequent events when the Ca2+ store is depleted. According to this idea, Stim1 changes its state to Ca2+-unbound upon store depletion or by crippling the EF-hand and is the effector that initiates a sequence of events leading to activation of the CRAC channel in the plasma membrane.
Fig. 3.
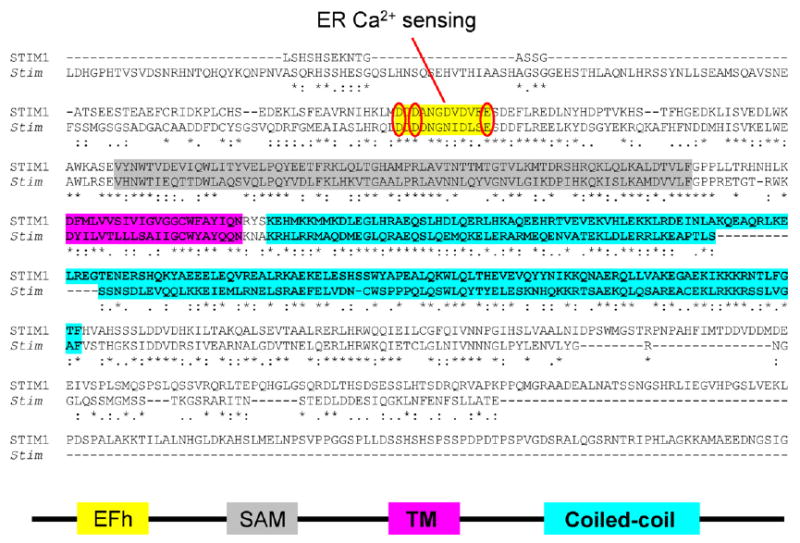
Stim1 sequences and motifs.
In parallel with these functional studies, we determined the distribution of the overexpressed STIM1 proteins by immunofluorescence using polyclonal antibodies specific to STIM1 and found a very different pattern of expression for WT and EF-hand mutant STIM1 [37]. The overexpressed WT STIM1 proteins were primarily localized in an ER-like pattern with some expression also visible at the plasma membrane, whereas the EF-hand mutant STIM1 proteins were predominantly seen at the plasma membrane in aggregations that we termed hotspots. Importantly, similar hotspots, or puncta, of native STIM1 were observed in several cell types, including Jurkat T cells, primary human T cells, RBL cells, and PC12 cells, using the same polyclonal antibodies shortly following store depletion. We used SERCA as a bona fide marker of the ER for comparison to the native STIM1 pattern of subcellular localization which changed following store depletion. In normal Ringer solution, there was extensive co-localization of both proteins. But when the store was depleted by thapsigargin, co-localization was reduced, because the STIM1 protein had migrated and was predominantly seen in hotspots on the cell surface. In addition, a time-course study showed that this redistribution of STIM1 to the plasma membrane was rapid, overlapping with the time-course of CRAC channel activation. After presenting these data at the annual Biophysical Society meeting (Feb. 2005), we decided to explore the subcellular localization of STIM1 with higher resolution by electron microscopy. The polyclonal antibodies were conjugated to quantum dots that are both highly fluorescent and visible as particles by electron microscopy. By this technique, native STIM1 in T cells was shown to redistribute to the plasma membrane following Ca2+ store depletion.
In summary, our results indicate that Stim1 has two functions: using the EF-hand to sense luminal ER Ca2+ content; and migrating to the plasma membrane when the EF-hand senses lowered Ca2+ in the ER. A similar conclusion was reached using an EF-hand YFP-tagged STIM1 mutant to activate SOCE and YFP-tagged STIM1 to track the migration of STIM1 to the cell surface following store depletion [36]. The Ca2+ sensor hypothesis was strengthened by a subsequent in vitro biochemical study using a core N-terminus STIM1 construct [38]. An apparent dissociation constant (_K_d) of 200–600 μM was measured, consistent with the functional range of Ca2+ levels within the ER upon store depletion.
2.7. A role for surface STIM1?
The presence of native STIM1 protein in both the plasma membrane and the ER was originally reported by using two different polyclonal antibodies against either N-terminus or C-terminus of STIM1 [39], and was confirmed by a later study [40]. By surface biotinylation, we found an increase in the surface-accessible native STIM1 protein upon store depletion in Jurkat T cells [37] that was confirmed in human platelets [41]. Yet, three reports showed that N-terminal YFP-, GFP-, and HA-tagged STIM1 is not extracellularly accessible following store depletion [36,42,43]. Results with tagged STIM1 constructs that fail to externalize indicate that acute surface exposure of STIM1 following Ca2+ store depletion may not be required for activation of the CRAC channel. Whether fluorescent protein or even smaller HA tags perturb STIM1 localization remains an issue. A recent study showed that N-terminal tagging of STIM1 with a small hexahistidine motif, but not with CFP, revealed cell surface exposure of STIM1 following thapsigargin treatment [44]. Thus, the presence of the larger fluorescent protein tag perturbed the re-localization of STIM1 into the plasma membrane. Several possibilities remain at this time, including a role for surface STIM1, in addition to STIM1 puncta next to the plasma membrane, in CRAC channel function. STIM1 on the plasma membrane was also reported to be essential for the activity of arachidonic acid-regulated Ca2+-selective (ARC) channels [45].
3. Genome-wide RNAi screen leads to the identification of olf186-F (Orai), the CRAC channel pore-forming subunit
3.1. Screening the fly genome with calcium signals
Because work on Stim1 made it clear that the actual CRAC channel had not yet been identified, we went back to RNAi screening in June 2005 and performed an unbiased and quantitative genome-wide screen at Harvard's Drosophila RNAi Screening Center (DRSC) to isolate additional molecules essential for SOC influx, with the primary goal of identifying the CRAC channel itself [33]. Fig. 4 illustrates some aspects of the screening protocol. At the DRSC, the entire Drosophila genome is represented in a set of 384-well plates, each well containing a dsRNA (amplicon) targeting a particular gene. Similar to the previous candidate screen that identified Stim, fluorescence from the Ca2+-sensitive dye fluo-4 was monitored systematically in each well at three time points, corresponding to “basal” (resting intracellular free Ca2+); “CCE” (TG-dependent Ca2+ influx) measured 4 min after adding thapsigargin; and “_F_max” (maximal fluorescence to normalize for cell number) after adding TritonX-100. The screen was finished in a four-week period and generated a large data set covering the entire genome [3 fluorescence measurement per well × 384 wells per plate × 63 plates × 2 duplicate experimental runs → >145,000 data points]. The overall high reproducibility was confirmed by a scatter plot with the two values derived from each amplicon.
Fig. 4.
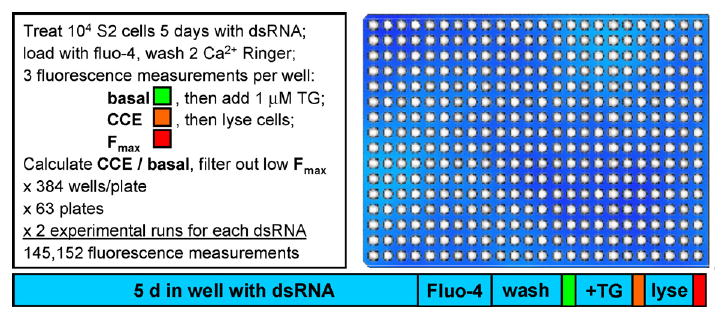
Genome-wide RNAi screening protocol.
3.2. Analyzing the screening data
Next, the screening results were processed. To assign statistical significance to the fluorescence values, _z_-scores for each well (calculated as the value of individual well minus the average, then divided by the standard deviation) were calculated. Candidate hits in four groups were selected if basal/_F_max or CCE/basal values were reduced or increased by more than three standard deviations from the mean. First, we computed the basal/_F_max for each well to provide a number for the normalized resting intracellular free Ca2+ level. Two groups (1 and 2) of candidates (hits) were defined by _z_-scores <−3 or >3, indicating abnormally low or high resting Ca2+, respectively. RNAi of these genes significantly influenced calcium homeostasis under resting conditions. Then, the CCE/basal ratio for each well was calculated to provide an index for the relative thapsigargin-evoked Ca2+ influx. This yielded an average value of about 2.1, and another two groups (3 and 4) were defined with _z_-scores <−3 or >3, indicating abnormally low or high store-operated Ca2+ entry, respectively. Assuming that RNAi of genes (e.g. Stim) that control SOCE do not influence the resting Ca2+ levels, overlapping candidates in groups 1 and 4 were removed from group 4, and overlapping candidates in groups 2 and 3 were removed from group 3 to avoid false-positive outcomes resulting from altered resting Ca2+ levels. The value of _F_max was used to filter out wells in which cells did not grow or failed to adhere, using a _z_-score of _F_max <−2 as a cutoff (confirmed by visual observation). In addition, we filtered out wells having more than five off-targets (non-specific RNAi effect). We then focused on filtered hits in group 3 with 75 remaining candidates, which should include the long-sought CRAC channel pore gene. The list was further narrowed down to those candidates with predicted transmembrane segments and with mammalian homologs; both are essential conditions for the CRAC pore subunit. In the end, a final list of 11 transmembrane protein hits was generated [33]. Gratifyingly, Stim was one of the strongest hits with a CCE/basal value of 1.26.
3.3. Olf186-F (Orai) as a conserved membrane protein with three human homologs
Uniquely among the filtered transmembrane protein hits, olf186-F (now renamed Orai), with a CCE/basal value of 1.31 and four predicted transmembrane segments, had no known function. Although only one member is represented in fly (dOrai, Fig. 5) and worm genomes, a conserved three-member gene family is present in all sequenced mammalian genomes. These are now renamed Orai1, Orai2, and Orai3 on human chromosomes 12, 7, and 16, respectively. Sequences and other key features are shown in Fig. 5. There are four predicted transmembrane segments in each Orai homolog which are all highly conserved, especially the first two transmembrane segments and the loop region in between. These provided hints for our follow-up mutagenesis strategy (see below).
Fig. 5.
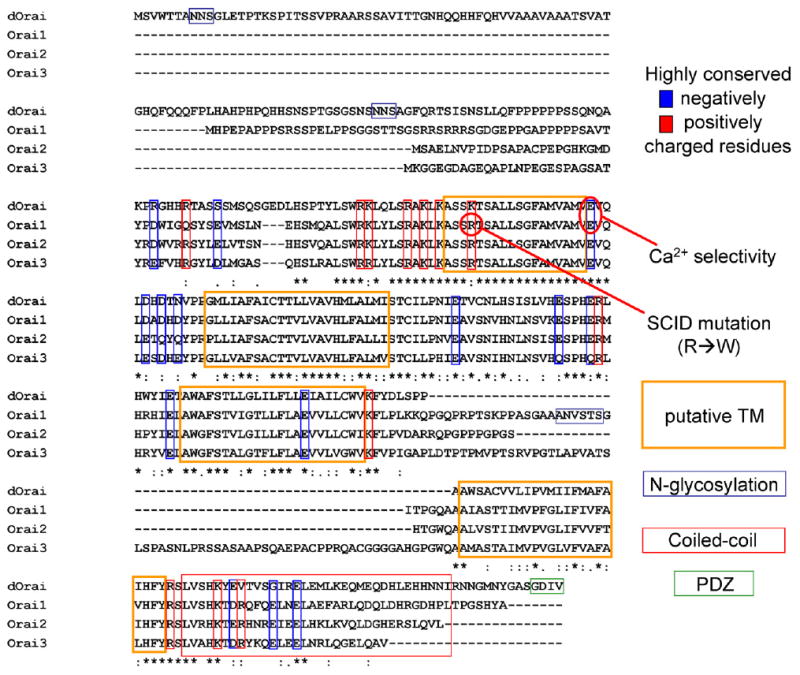
Fly and human Orai-related sequences and motifs.
3.4. Validation of hits by single-cell Ca2+ imaging and patch-clamp
Several final candidates were validated at the single-cell level by both Ca2+ imaging and patch-clamp analysis for direct RNAi effects on CRAC activity [33]. As for olf186-F (Orai), the vast majority of cells pretreated with dsRNA targeting olf186-F did not exhibit Ca2+ influx upon store depletion and had greatly reduced CRAC current densities, whereas both resting Ca2+ level and store content were the same as in control cells. We also validated the SERCA pump, which could have been a false-positive hit since the screen was based on thapsigargin. In single-cell Ca2+ assays and whole-cell recording from cells pretreated with dsRNA to knock down SERCA, there was a highly significant reduction in thapsigargin-evoked Ca2+ entry and in CRAC current densities recorded during passive store depletion. It remains to be determined whether this reduction resulted from abnormally low Ca2+ store content (assayed by ionomycin-evoked store release) or elevated [Ca2+]i levels. Syntaxin 5 was also validated as a genuine hit. Thus, by RNAi we confirmed with Ca2+ imaging and patch-clamp recording that Stim, Orai, SERCA, and Syntaxin 5 are each required for normal CRAC channel function.
An RNAi screen based on NF-AT translocation as a functional measurement was performed in DRSC at the same time [31]. Combined with genetic mapping of SCID patients which was initiated in 1996, Orai was also identified as an essential component for store-operated Ca2+ influx and the downstream NF-AT relocation into the nucleus. Moreover, a recessive point mutation in human Orai1 on chromosome 12 was isolated from SCID patients [31]. This channelopathy further reinforces the functional significance of CRAC channels (and Orai1) for the immune system. A third RNAi screen initiated several months later also identified the same gene (which was renamed as CRACM1 for CRAC modulator 1), together with another candidate (_dpr_3) called CRACM2 in the fly genome [32]. Remarkably, all three screens identified olf186-F (Orai, CRACM1) as an essential transmembrane protein.
3.5. Overexpression of Orai and Stim: amplified CRAC current
RNAi can test whether a particular molecule may be required for normal function. But in a complex multicomponent system, several required elements may be present. Thus, we sought to investigate effects of overexpression. We cloned the cDNA of Orai from S2 cells and expressed it with or without co-transfected Stim. When Orai was expressed alone, CRAC current density increased three-fold, a highly significant increase and in contrast to the lack of effect when Stim was overexpressed by itself. Importantly, when both genes were transfected, a huge CRAC current was recorded, and the kinetics of activation were accelerated compared to CRAC current induced by overexpression of Orai alone. The amplified CRAC current exhibited all the biophysical characteristics of native CRAC current in S2 cells (Table 1). These coexpression results provided further evidence that Orai may form the CRAC channel pore [33].
3.6. Mutations in Orai that affect CRAC channel Ca2+ selectivity
The ability of ion channels to select among various ions depends on interactions between amino acid side chains, ions and water molecules. Therefore, we sought to identify particular amino acids that, when mutated, resulted in altered ion selectivity of CRAC current following expression of Orai + Stim. Ion selectivity is usually accomplished by loop regions between transmembrane segments. It has been shown that negatively charged amino acids are the key elements for the Ca2+-selectivity filter of voltage-gated Ca2+ channels and TRPV6 [46,47]. The loop between transmembrane segments 1 and 2 of the fly Orai gene and mammalian Orai family members is the most conserved (Fig. 5). We focused on negatively charged residues and prepared a series of alanine substitutions. Mutation of the conservative glutamate to alanine at position 180 of Orai resulted in inhibition of the background S2 CRAC current, indicating a dominant-negative action. A conservative point mutation at this position to aspartate dramatically altered the ion selectivity properties of CRAC current. Whereas a large inward current developed during passive store depletion when WT Orai + Stim were co-transfected, an outward current developed when the point mutant E180D Orai was expressed together with Stim. This was because the CRAC channel I_–_V characteristic displayed pronounced outward rectification, rather than the normal inward rectification of native or amplified CRAC current. Ion substitution experiments clearly revealed a change from Ca2+ selective permeation to monovalent cation currents carried by Na+ or Cs+. Furthermore, several other properties related to ion selectivity were affected. In divalent-free solution, the relative Na+ to Cs+ permeability, _P_Na/_P_Cs, was reduced. The increase in Cs+ permeability is consistent with a slightly enlarged selectivity filter. Furthermore, the monovalent currents failed to depotentiate. The dramatic alterations of ion selectivity by a point mutation at position 180 of Orai indicate that Orai is the pore-forming subunit of the channel, responsible for Ca2+ selectivity. Interestingly, three other mutants near 180 had reduced sensitivity to the channel blocker Gd3+ possibly by reduced charge screening resulting in a reduced local concentration of the trivalent blocker. Taken together, these results lead to the conclusion that Orai is a bona fide CRAC channel pore subunit [48]. A corresponding point mutation in Orai1 at position 106 results in similar changes [49,50], although with some differences in rectification between the corresponding Orai and Orai1 mutants.
3.7. Interaction with Stim
So far, the CRAC channel gating mechanism has not been elucidated. The molecular events between Stim1 translocation and CRAC activation remain undefined. Both STIM1 and Orai1 accumulate at junctional sites where the ER and plasma membrane are closely apposed following store depletion [43,51]. In S2 cells, we found a greatly enhanced interaction between Orai and Stim upon store depletion, assessed by co-immunoprecipitation (co-IP), reflecting a dynamic interaction model for CRAC channel activation [48]. One possibility is that Stim delivers information about the store (empty) to Orai by direct physical contact, which in turn opens the channels. However, amino acid sequences required for the interaction and a possible requirement for other scaffolding proteins need to be further tested. As Drosophila Stim and Orai are both required to reconstitute CRAC channel activity in HEK cells (unpublished data), it will be interesting to see whether these two proteins alone form a minimal functional unit of CRAC channels. On the other hand, the redistribution of both STIM1 and Orai1 hint that there may be a finely tuned recruitment mechanism to generate a polarized activated CRAC channel distribution for Ca2+ signaling in microdomains. An interaction between mammalian STIM1 and Orai1 was also reported via co-IP assay without TG-treatment [50]. The interaction may be indirect or of low affinity as another co-IP based study showed that Orai1 did not associate with STIM1 in several detergent and salt conditions tested [52]. Further work is needed to reconcile these data and to determine how store depletion affects the interaction.
4. Overview and prospects
RNAi screening [31–34,36] and subsequent functional studies have clarified the molecular requirements and activation mechanisms of the CRAC channel, a sort of holy grail in the field of Ca2+ signaling. In human T cells, STIM1 senses ER Ca2+ content using an EF-hand motif near the N-terminus and translocates to the plasma membrane when the Ca2+ store is depleted [36,37]. The Ca2+ channel itself is formed from Orai1 in T cells, since a naturally occurring mutation within the first transmembrane segment abrogates channel activity in SCID patients [31], and a point mutant of a conserved glutamate residue between transmembrane segments 1 and 2 in Drosophila Orai or human Orai1 alters the Ca2+ ion selectivity of the channel [48–50].
The next challenge is to understand the detailed molecular composition and organization of CRAC channels, as a unique Ca2+ channel unrelated to other channel families. The subunit organization needs to be defined, and further site-directed mutagenesis with functional assays will provide more information. Ultimately a crystal or NMR structure will be essential to complete this mission. High-throughput screening may provide useful CRAC-specific blockers. Furthermore, the functional role of Stim and Orai homologs will need to be defined. The three human Orai homologs exhibit different tissue expression patterns. The fact that normal Orai2 and Orai3 genes cannot compensate the mutated Orai1 gene in SCID patients indicates that the three members may have different physiological roles [52]. Genetic targeting is sure to provide fresh insights, as will in vivo pharmacological targeting.
References
- 1.Cahalan MD, Parker I, Wei SH, Miller MJ. Two-photon tissue imaging: seeing the immune system in a fresh light. Nat Rev Immunol. 2002;2:872–880. doi: 10.1038/nri935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller MJ, Wei SH, Parker I, Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. 2002;296:1869–1873. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 3.Miller MJ, Hejazi AS, Wei SH, Cahalan MD, Parker I. T cell repertoire scanning is promoted by dynamic dendritic cell behavior and random T cell motility in the lymph node. Proc Natl Acad Sci USA. 2004;101:998–1003. doi: 10.1073/pnas.0306407101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller MJ, Safrina O, Parker I, Cahalan MD. Imaging the single cell dynamics of CD4+ T cell activation by dendritic cells in lymph nodes. J Exp Med. 2004;200:847–856. doi: 10.1084/jem.20041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeCoursey TE, Chandy KG, Gupta S, Cahalan MD. Voltage-gated K+ channels in human T lymphocytes: a role in mitogenesis? Nature. 1984;307:465–468. doi: 10.1038/307465a0. [DOI] [PubMed] [Google Scholar]
- 6.Metcalfe JC, Pozzan T, Smith GA, Hesketh TR. A calcium hypothesis for the control of cell growth. Biochem Soc Symp. 1980;45:1–26. [PubMed] [Google Scholar]
- 7.Lewis RS, Cahalan MD. Mitogen-induced oscillations of cytosolic Ca2+ and transmembrane Ca2+ current in human leukemic T cells. Cell Regul. 1989;1:99–112. doi: 10.1091/mbc.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Putney JW., Jr A model for receptor-regulated calcium entry. Cell Calcium. 1986;7:1–12. doi: 10.1016/0143-4160(86)90026-6. [DOI] [PubMed] [Google Scholar]
- 9.Hoth M, Penner R. Calcium release-activated calcium current in rat mast cells. J Physiol. 1993;465:359–386. doi: 10.1113/jphysiol.1993.sp019681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoth M, Penner R. Depletion of intracellular calcium stores activates a calcium current in mast cells. Nature. 1992;355:353–356. doi: 10.1038/355353a0. [DOI] [PubMed] [Google Scholar]
- 11.Zweifach A, Lewis RS. Mitogen-regulated Ca2+ current of T lymphocytes is activated by depletion of intracellular Ca2+ stores. Proc Natl Acad Sci USA. 1993;90:6295–6299. doi: 10.1073/pnas.90.13.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cahalan MD, Chandy KG, DeCoursey TE, Gupta S. A voltage-gated potassium channel in human T lymphocytes. J Physiol. 1985;358:197–237. doi: 10.1113/jphysiol.1985.sp015548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fanger CM, Rauer H, Neben AL, Miller MJ, Wulff H, Rosa JC, Ganellin CR, Chandy KG, Cahalan MD. Calcium-activated potassium channels sustain calcium signaling in T lymphocytes. Selective blockers and manipulated channel expression levels. J Biol Chem. 2001;276:12249–12256. doi: 10.1074/jbc.M011342200. [DOI] [PubMed] [Google Scholar]
- 14.Feske S, Giltnane J, Dolmetsch R, Staudt LM, Rao A. Gene regulation mediated by calcium signals in T lymphocytes. Nat Immunol. 2001;2:316–324. doi: 10.1038/86318. [DOI] [PubMed] [Google Scholar]
- 15.Negulescu PA, Shastri N, Cahalan MD. Intracellular calcium dependence of gene expression in single T lymphocytes. Proc Natl Acad Sci USA. 1994;91:2873–2877. doi: 10.1073/pnas.91.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fanger CM, Hoth M, Crabtree GR, Lewis RS. Characterization of T cell mutants with defects in capacitative calcium entry: genetic evidence for the physiological roles of CRAC channels. J Cell Biol. 1995;131:655–667. doi: 10.1083/jcb.131.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 18.Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392:933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 19.Hardie RC, Raghu P. Activation of heterologously expressed Drosophila TRPL channels: Ca2+ is not required and InsP3 is not sufficient. Cell Calcium. 1998;24:153–163. doi: 10.1016/s0143-4160(98)90125-7. [DOI] [PubMed] [Google Scholar]
- 20.Cordova D, Delpech VR, Sattelle DB, Rauh JJ. Spatiotemporal calcium signaling in a Drosophila melanogaster cell line stably expressing a Drosophila muscarinic acetylcholine receptor. Invert Neurosci. 2003;5:19–28. doi: 10.1007/s10158-003-0024-2. [DOI] [PubMed] [Google Scholar]
- 21.Swatton JE, Morris SA, Wissing F, Taylor CW. Functional properties of Drosophila inositol trisphosphate receptors. Biochem J. 2001;359:435–441. doi: 10.1042/0264-6021:3590435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Towers PR, Sattelle DB. A Drosophila melanogaster cell line (S2) facilitates post-genome functional analysis of receptors and ion channels. Bioessays. 2002;24:1066–1073. doi: 10.1002/bies.10178. [DOI] [PubMed] [Google Scholar]
- 23.Yagodin S, Hardie RC, Lansdell SJ, Millar NS, Mason WT, Sattelle DB. Thapsigargin and receptor-mediated activation of Drosophila TRPL channels stably expressed in a Drosophila S2 cell line. Cell Calcium. 1998;23:219–228. doi: 10.1016/s0143-4160(98)90120-8. [DOI] [PubMed] [Google Scholar]
- 24.Yagodin S, Pivovarova NB, Andrews SB, Sattelle DB. Functional characterization of thapsigargin and agonist-insensitive acidic Ca2+ stores in Drosophila melanogaster S2 cell lines. Cell Calcium. 1999;25:429–438. doi: 10.1054/ceca.1999.0043. [DOI] [PubMed] [Google Scholar]
- 25.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 26.Yeromin AV, Roos J, Stauderman KA, Cahalan MD. A store-operated calcium channel in Drosophila S2 cells. J Gen Physiol. 2004;123:167–182. doi: 10.1085/jgp.200308982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 28.Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- 29.Worby CA, Simonson-Leff N, Dixon JE. RNA interference of gene expression (RNAi) in cultured Drosophila cells. Sci STKE. 2001:PL1. doi: 10.1126/stke.2001.95.pl1. [DOI] [PubMed] [Google Scholar]
- 30.Ulvila J, Parikka M, Kleino A, Sormunen R, Ezekowitz RA, Kocks C, Ramet M. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J Biol Chem. 2006;281:14370–14375. doi: 10.1074/jbc.M513868200. [DOI] [PubMed] [Google Scholar]
- 31.Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 32.Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, Kraft S, Turner H, Fleig A, Penner R, Kinet JP. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang SL, Yeromin AV, Zhang XH, Yu Y, Safrina O, Penna A, Roos J, Stauderman KA, Cahalan MD. Genome-wide RNAi screen of Ca2+ influx identifies genes that regulate Ca2+ release-activated Ca2+ channel activity. Proc Natl Acad Sci USA. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, Velicelebi G, Stauderman KA. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roos J, Stauderman KA, Velicelebi G, Ohlsen K, DiGregorio PJ. Methods of modulating and of identifying agents that modulate intracellular calcium, in: WO2004/078995. 2003 [Google Scholar]
- 36.Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Jr, Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang SL, Yu Y, Roos J, Kozak JA, Deerinck TJ, Ellisman MH, Stauderman KA, Cahalan MD. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stathopulos PB, Li GY, Plevin MJ, Ames JB, Ikura M. Stored Ca2+ depletion-induced oligomerization of stromal interaction molecule 1 (STIM1) via the EF-SAM region: An initiation mechanism for capacitive Ca2+ entry. J Biol Chem. 2006;281:35855–35862. doi: 10.1074/jbc.M608247200. [DOI] [PubMed] [Google Scholar]
- 39.Manji SS, Parker NJ, Williams RT, Van Stekelenburg L, Pearson RB, Dziadek M, Smith PJ. STIM1: a novel phosphoprotein located at the cell surface. Biochim Biophys Acta. 2000;1481:147–155. doi: 10.1016/s0167-4838(00)00105-9. [DOI] [PubMed] [Google Scholar]
- 40.Spassova MA, Soboloff J, He LP, Xu W, Dziadek MA, Gill DL. STIM1 has a plasma membrane role in the activation of store-operated Ca2+ channels. Proc Natl Acad Sci USA. 2006;103:4040–4045. doi: 10.1073/pnas.0510050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez JJ, Salido GM, Pariente JA, Rosado JA. Interaction of STIM1 with endogenously expressed human canonical TRP1 upon depletion of intracellular Ca2+ stores. J Biol Chem. 2006;281:28254–28264. doi: 10.1074/jbc.M604272200. [DOI] [PubMed] [Google Scholar]
- 42.Mercer JC, Dehaven WI, Smyth JT, Wedel B, Boyles RR, Bird GS, Putney JW., Jr Large store-operated calcium selective currents due to co-expression of Orai1 or Orai2 with the intracellular calcium sensor, Stim1. J Biol Chem. 2006;281:24979–24990. doi: 10.1074/jbc.M604589200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hauser CT, Tsien RY. A hexahistidine-Zn2+-dye label reveals STIM1 surface exposure. Proc Natl Acad Sci USA. 2007;104:3693–3697. doi: 10.1073/pnas.0611713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mignen O, Thompson JL, Shuttleworth TJ. STIM1 regulates Ca2+ entry via arachidonate-regulated Ca2+-selective (ARC) channels without store-depletion or translocation to the plasma membrane. J Physiol. 2006 doi: 10.1113/jphysiol.2006.122432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ellinor PT, Yang J, Sather WA, Zhang JF, Tsien RW. Ca2+ channel selectivity at a single locus for high-affinity Ca2+ interactions. Neuron. 1995;15:1121–1132. doi: 10.1016/0896-6273(95)90100-0. [DOI] [PubMed] [Google Scholar]
- 47.Voets T, Janssens A, Droogmans G, Nilius B. Outer pore architecture of a Ca2+-selective TRP channel. J Biol Chem. 2004;279:15223–15230. doi: 10.1074/jbc.M312076200. [DOI] [PubMed] [Google Scholar]
- 48.Yeromin AV, Zhang SL, Jiang W, Yu Y, Safrina O, Cahalan MD. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature. 2006;443:226–229. doi: 10.1038/nature05108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prakriya M, Feske S, Gwack Y, Srikanth S, Rao A, Hogan PG. Orai1 is an essential pore subunit of the CRAC channel. Nature. 2006;443:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- 50.Vig M, Beck A, Billingsley JM, Lis A, Parvez S, Peinelt C, Koomoa DL, Soboloff J, Gill DL, Fleig A, Kinet JP, Penner R. CRACM1 multimers form the ion-selective pore of the CRAC channel. Curr Biol. 2006;16:2073–2079. doi: 10.1016/j.cub.2006.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luik RM, Wu MM, Buchanan J, Lewis RS. The elementary unit of store-operated Ca2+ entry: local activation of CRAC channels by STIM1 at ER-plasma membrane junctions. J Cell Biol. 2006;174:815–825. doi: 10.1083/jcb.200604015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gwack Y, Srikanth S, Feske S, Cruz-Guilloty F, Oh-Hora M, Neems DS, Hogan PG, Rao A. Biochemical and functional characterization of Orai family proteins. J Biol Chem. 2007 doi: 10.1074/jbc.M609630200. [DOI] [PubMed] [Google Scholar]
- 53.Lepple-Wienhues A, Cahalan MD. Conductance and permeation of monovalent cations through depletion-activated Ca2+ channels (ICRAC) in Jurkat T cells. Biophys J. 1996;71:787–794. doi: 10.1016/S0006-3495(96)79278-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zweifach A, Lewis RS. Calcium-dependent potentiation of store-operated calcium channels in T lymphocytes. J Gen Physiol. 1996;107:597–610. doi: 10.1085/jgp.107.5.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zweifach A, Lewis RS. Rapid inactivation of depletion-activated calcium current (ICRAC) due to local calcium feedback. J Gen Physiol. 1995;105:209–226. doi: 10.1085/jgp.105.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]