Recent advances in MeCP2 structure and function (original) (raw)
. Author manuscript; available in PMC: 2010 May 21.
Published in final edited form as: Biochem Cell Biol. 2009 Feb;87(1):219–227. doi: 10.1139/o08-115
Abstract
Mutations in methyl DNA binding protein 2 (MeCP2) cause the neurodevelopmental disorder Rett syndrome (RTT). The mechanism(s) by which the native MeCP2 protein operates in the cell are not well understood. Historically, MeCP2 has been characterized as a proximal gene silencer with 2 functional domains: a methyl DNA binding domain and a transcription repression domain. However, several lines of new data indicate that MeCP2 structure and function relationships are more complex. In this review, we first discuss recent studies that have advanced understanding of the basic structural biochemistry of MeCP2. This is followed by an analysis of cell-based experiments suggesting MeCP2 is a regulator, rather than a strict silencer, of transcription. The new data establish MeCP2 as a multifunctional nuclear protein, with potentially important roles in chromatin architecture, regulation of RNA splicing, and active transcription. We conclude by discussing clinical correlations between domain-specific mutations and RTT pathology to stress that all structural domains of MeCP2 are required to properly mediate cellular function of the intact protein.
Keywords: Rett syndrome, epigenomics, chromatin architecture, splice regulation, methyl DNA binding
Introduction: a historical perspective of MeCP2 structure and function
Methyl CpG binding protein (MeCP) 2 is a 53 kDa nuclear protein named for its ability to bind methylated DNA (Lewis et al. 1992; Meehan et al. 1992). In addition to preferentially binding methylated DNA, it was found that MeCP2 facilitated transcriptional repression in an in vitro transcription assay using native MeCP2 from rat brain nuclear extracts (Nan et al. 1997). These initial studies set a precedent for MeCP2 function to be predicated on methylated–DNA interactions. In this review, we will discuss the heuristic evolution of the view of MeCP2, from a single function protein to a multifunctional nuclear protein that directly affects chromatin architecture and is involved with gene expression in unanticipated ways.
A nuclear protein that preferentially bound methylated DNA in vitro without apparent regard for a consensus binding sequence was first described in 1989, and was named MeCP (Meehan et al. 1989, 1990). This was accomplished using synthetic double stranded (ds)DNA oligomers methylated by bacterial methyl transferases. Competition of the protein from synthetic DNA probes was observed only when the competitor DNA was methylated (Meehan et al. 1989). These assays selectively pulled down a protein into complexes from mouse brain, spleen, kidney, rat liver, and rabbit liver extracts. In each extract, MeCP preferentially bound the methylated DNA templates without sequence specificity. Further experiments revealed that MeCP was 2 distinguishable proteins — MeCP1 and MeCP2 (Meehan et al. 1992) — with MeCP1 requiring at least 12 symmetrically methylated CpGs and MeCP2 able to bind a single methylated CpG pair. MeCP2 was reported to be 100 times more abundant in adult somatic nuclei than MeCP1 (Meehan et al. 1992). When forming the initial hypothesis of MeCP2 function, it was proposed that MeCP2 normally binds methylated DNA in the context of chromatin, leading to long-term transcriptional repression. This hypothesis was corroborated by results showing that native MeCP2 purified from rat brain extracts was released upon micrococcol nuclease digestion of a methylated DNA probe, but not present on a nonmethylated DNA probe (Meehan et al. 1992). The complementary (c)DNA sequence for MeCP2 was also determined in that study by deriving amino acid sequences of digested native MeCP2 protein from rat brain extract, and subsequently designing oligonucleotide primers to amplify the messenger (m)RNA by the polymerase chain reaction. More evidence was provided when transiently transfected recombinant genes coding MeCP2 fused to the LacZ gene were expressed in mouse cell cultures, and similar localization to centromeric heterochomatin was observed, when compared with endogenous MeCP2 stained with anti-MeCP2 antibody. The inability of the MeCP2–LacZ fusion protein to localize to centromeric hereochomatin in methyl-transfererase-deficient mouse cells was provided as evidence that MeCP2 requires a methylated chromatin substrate for binding (Lewis et al. 1992).
The early driving hypothesis in MeCP2 research was methylated DNA binding preference. This hypothesis gave MeCP2 a name and also led to the isolation of the first functional domain: the methyl CpG binding domain (MBD) (Nan et al. 1993). By truncating either end of full-length MeCP2 and probing for methylated DNA binding preference, Nan et al. (1993) demonstrated that the MBD encompassed residues 78–163. This study was conducted using murine MeCP2 in mouse cell cultures. MeCP2 homologs in other mammals have not been fully characterized. Specifically, preferences for DNA methylation by MeCP2 homologs have not been thoroughly addressed in the available literature. The preference for methylated over unmethylated DNA for the Xenopus MeCP2 homolog is 20-fold, whereas the preference for murine (Fraga et al. 2003) and human (Nikitina et al. 2007_a_) MeCP2 for methylated, compared with unmethylated, DNA is 2–3-fold. A solution structure of the isolated MBD was solved by nuclear magnetic resonance (NMR) spectroscopy (Wakefield et al. 1999). The structured core appears as a wedge made up of a 3 stranded anti-parallel β-sheet on one side, with an α-helix on the C-terminal side. β-strands 1 and 2 are connected by a disordered loop of 5 residues, with 1 positively charged and 2 polar residues. The central wedge-shaped fold encompasses residues 103–145 of the MBD, and is flanked by 26 residues on the N-terminal side and 19 residues on the C-terminal side, with no detectable secondary structure. Recently, a high-resolution X-ray crystal structure of the MBD bound to DNA was published, showing that the hydration status of the methyl group on the 5′ carbon of cytosine stabilizes the MBD–DNA interface (Ho et al. 2008). The hydrophilic interaction between the MBD and stabilizing water molecules countered the previously proposed hypothesis that hydrophobic interactions stabilize the interaction. Of note, while the atomic structures have provided much detail about the MBD wedge motif, they do not explain the need for the flanking unstructured regions in recognizing methylated DNA.
The second MeCP2 domain to be characterized was the transcription repression domain (TRD). Using an in vitro β-actin transcription assay, different regions of MeCP2 were fused to the Gal 4 DNA binding domain. Results showed that residues 205–310 were required for transcriptional silencing, defining the TRD (Nan et al. 1997). Several mechanisms for the transcriptional repression by the TRD have been proposed. These mechanisms have recently been reviewed by Zlatanova (2005). Perhaps the most compelling hypothesis for transcriptional repression is based on the observation that the TRD binds the corepressor mSin3A, which is thought to recruit histone deacetylases (Jones et al. 1998; Nan et al. 1998; Wade et al. 1998). In this model, MeCP2 indirectly causes changes in chromatin architecture by mediating post-translational modifications of the histone tails. This model is not universally applicable to MeCP2 function, as histone-deacetylase-independent transcriptional repression has been observed (Yu et al. 2000). A lack of global histone tail modifications in MeCP2-null mice also argues against this model (Urdinguio et al. 2007). Since last reviewed, studies of transcriptional repression activity of MeCP2 have been relatively sparse, but a repression of tumor-associated genes by the TRD of MeCP2 has been observed in a chromatin immunoprecipitation (ChIP) assay (Wischnewski et al. 2007). This established a connection between MeCP2 and cancer-causing gene expression patterns.
The identification and characterization of the MBD and TRD, respectively, led to a more refined model, in which MeCP2 functions as a methyl DNA-specific proximal gene silencer that recruits corepressors and histone deacetylases (Meehan et al. 1992; Nan et al. 1997; Martinowich et al. 2003). In this regard, MeCP2 also binds to transcriptional silencing factors besides mSin3a, including N-CoR and c-Ski (Kokura et al. 2001).
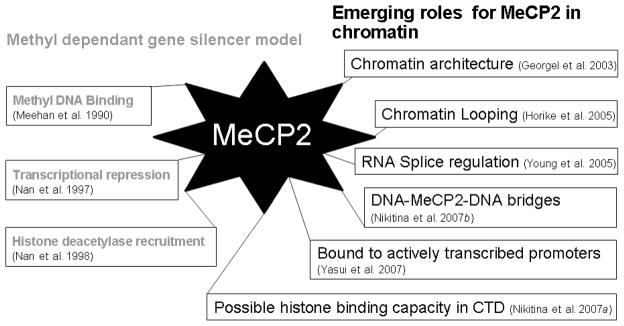
When the link between mutations in MeCP2 and Rett syndrome (RTT) was established in 1999, research on MeCP2 increased dramatically, and new studies were undertaken to determine how the protein functions on a global genomic scale (Amir et al. 1999). The first implication that MeCP2 has a histone-deacetylase-independent role in coordinating global chromatin architecture came from in vitro studies in 2003, which demonstrated that MeCP2 could directly compact chromatin without DNA methylation, ATP, or other proteins, such as mSin3A (Georgel et al. 2003). This study opened the door to understanding MeCP2 as a complex multifunctional nuclear protein with a prominent role in regulating global chromatin architecture. Since this observation, several other functions have been suggested for the protein. Some of these additional functions are depicted in Fig. 1.
Fig. 1.
Chronological representation of known MeCP2 functions. The progression of traditional understanding of MeCP2 as a methyl-dependent proximal gene silencer is shown in grey on the left, while studies implicating MeCP2 as having additional functions are listed on the right. CTD, carboxyl terminal domain.
In addition to the proposed roles in transcriptional repression and modulation of chromatin structure, a recent study potentially links MeCP2 function to mRNA splicing (Young et al. 2005). Using coimmunoprecipitation from HeLa cell extracts, MeCP2 was shown to interact with YB-1 (Young et al. 2005). The YB-1 protein is a highly conserved component of messenger ribonucleoprotein particles (mRNPs), and functions as the main mRNA packaging protein. The interaction between MeCP2 and YB-1 requires the presence of RNA, as coimmunoprecipitation treated with RNase failed to pull down YB-1 with MeCP2. It is unclear whether the RNA bridges MeCP2 and YB-1 or stabilizes a protein–protein interaction between the two. In the same study, it was also observed that MeCP2 affects the splicing of reporter mini-genes, and that a functional MBD was not required for the YB-1 interaction or splice regulation. These findings, in conjunction with the observation that there are aberrant alternative splicing patterns in a mouse model of RTT (Young et al. 2005), imply that MeCP2 has a previously uncharacterized function as a splice site regulator. Previously reported evidence that MeCP2 directly binds RNA with high affinity (0–10 nmol·L−1) (Jeffery and Nakielny 2004) is of renewed interest in light of the more recent observations of Young and co-workers (2005). RTT is considered a neural-specific phenotype caused by mutations in MeCP2; therefore, it is worth testing predictions made by the model that MeCP2 has a crucial role in splice site regulation and RNA binding. Predictions include neuronal-specific aberrant splice variants of the genes already observed to have dysregulated expression in MeCP2-null mouse models (Pelka et al. 2006).
New data about the MeCP2 gene itself have also been reported. The identification of a MeCP2 splice variant added an extra dimension to our understanding of how and where MeCP2 functions. This new splice variant was labeled the e1 isoform, and differs from the previously characterized e2 isoform only in the segments at their extreme amino termini. The e1 isoform has a 21 residue segment with an acidic pI of 4.25, while the e2 isoform N-terminal segment is 9 residues and has an basic pI of 9.5 (Mnatzakanian et al. 2004) (Fig. 2). These distinct structural differences, together with the differential distribution of the e1 and e2 isoforms between the dorsal thalamus and hypothalamus in developing postnatal mouse brains (Dragich et al. 2007), suggest that there are important undiscovered differences between the function of the 2 isoforms.
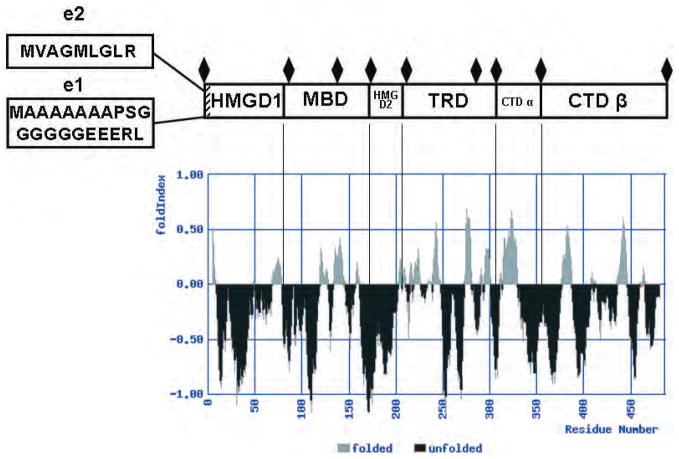
Fig. 2.
Diagram of MeCP2 domain organization, based on biochemical data aligned with a FoldIndex plot. Domains are labeled from amino to carboxyl terminal from left to right. The location and sequence of the alternatively spliced region of the e1 and e2 isoforms is shown at the left. The boundaries of trypsin-resistant bands are indicated by black diamonds. The full-length protein sequence was analyzed by the FoldIndex algorithm (http://bioportal.weizmann.ac.il/fldbin/findex), with a window setting of 10 and a step value of 1. Regions that are predicted to be disordered are indicated in black, while ordered regions are shown in gray. Note that under these settings, the first and last 10 amino acids are not represented in the plot.
Based on the observation that neurological defects in a mouse model of RTT can be reversed by re-expression of the wild-type protein, understanding the molecular mechanisms of native MeCP2 and how these mechanisms are malfunctioning in RTT has potential clinical application (Guy et al. 2007). This observation is significant in establishing RTT as a neurodevelopmental rather than a neurodegenerative disorder, and is consistent with a model for MeCP2 as a positive and negative regulator of transcription, a gene-specific splicing factor, and a chromatin architectural protein.
The structural and intrinsically disordered domains of MeCP2
Protein function is inexorably linked to structure; therefore, we will first consider recent advances in understanding the fundamental biochemistry of MeCP2. In particular, recent solution biophysical and protease digestion experiments have established that native MeCP2 is an intrinsically disordered protein composed of at least 6 distinct domains (Adams et al. 2007) (Fig. 2). Trypsin digestion of MeCP2 has the potential to occur throughout the length of the poly-peptide chain, as there are approximately 90 potential digestion sites dispersed evenly throughout the human MeCP2 sequence. The rate of fragment appearance is related to trypsin accessibility to the tertiary structure. N-terminal sequence analysis of kinetically stable tryptic bands identified 6 distinct MeCP2 domains (Adams et al. 2007). Listed from amino to carboxy termini, these are the HMGD1, MBD, HMGD2, TRD, carboxyl terminal domain (CTD)-α, and CTD-β (Fig. 2). Of note, the observed trypsin cleavage sites mapped to the boundaries of the 2 well-characterized functional domains (the MBD and TRD). There were also additional trypsin cleavage sites within both the MBD and TRD (Adams et al. 2007). The most N-terminal domain, named the HMGD1, shares amino acid composition similarity with the HMGA2 protein, and was rapidly digested to completion once released from the neighboring MBD domain. Likewise, the second HMG-like domain (HMGD2) was not detected as a kinetically stable band, suggesting that it is rapidly digested due to an unstructured conformation. The TRD sequence was recovered from the trypsin experiment, indicating that the availability of trypsin cleavage sites is more restricted in this domain. The experimental approach allowed the CTD to be divided into a CTD-α (residues 310–354) and a CTD-β (residues 355–486) (Adams et al. 2007). The CTD-β contains 2 identifiable sequence motifs: 7 consecutive histidines between residues 366–72 and a group 2 WW binding protein motif within a larger proline-rich region at residues 381–393. The His-rich motif in MeCP2 is highly conserved between species. The WW binding protein motif (residues 384–387) has been demonstrated to interact with splicing and transcription factors (Buschdorf and Stratling 2004; Kato et al. 2004). Interestingly, the proline-rich region in the CTD-β (residues 355–486) has been shown to bind HMGB1 (Dintilhac and Bernues 2002). At the level of chromatin structure, the CTD is required for MeCP2-mediated chromatin compaction. Nikitina et al. (2007 b) showed that the R294X MeCP2 mutant was able to bind naked, as well as wild-type, DNA, but could not condense chromatin into higher-order structures. This result is consistent with the early studies of Chandler et al. (1999), who showed that a region of the MeCP2 CTD contributed to the footprint of MeCP2 on nucleosomes.
While delineating the structural domains of MeCP2 was an important advance, understanding how these domains are organized into a functional tertiary structure is essential for understanding the normal cellular functions of the protein, and why certain mutations lead to an RTT phenotype. The next section discusses recent studies indicating that MeCP2 has an atypical tertiary structure permeated with an unusual amount of disorder.
MeCP2 tertiary structure
Due to the lack of NMR or X-ray crystal structure data, characterization of the tertiary structure of full-length MeCP2 has been accomplished by solution biochemical and biophysical methods. Circular dichroism (CD) of recombinant human MeCP2 showed that full-length protein was approximately ~35% β-strand/turn, 5% α-helix, and almost 60% unstructured. CD further indicated that the isolated MBD fragment (residues 78–168) was ~10% α-helix, 51% β-strand/turn, and 38% unstructured (Adams et al. 2007), levels that approximated the amount of secondary structure seen with NMR (Wakefield et al. 1999). A recombinant fragment of MeCP2 comprising the TRD (residues 198–305) was 85% unstructured, according to CD. When studied by analytical ultracentrifugation, MeCP2 behaved as a monomer over a wide range of ionic conditions and molar concentrations, and had an unusually low sedimentation coefficient (2.2 S) and a correspondingly high frictional coefficient ratio (f/_f_o = 2.4). These results are in agreement with sucrose gradient results obtained from a single set of experimental conditions (Klose and Bird 2004). Importantly, the CD data and the high f/_f_o value indicate that MeCP2 has a coil-like tertiary structure similar to that of a partially denatured protein (Adams et al. 2007). Based on these observations, it is not surprising that MeCP2 has been reported to have certain anomalous physicochemical properties. For example, the 53 kDa MeCP2 monomer yields an apparent molecular mass of 500 kDa, according to gel filtration, and migrates at an apparent mass of 75–80 kDa on SDS gels (Klose and Bird 2004).
The CD and sedimentation results, together with the anomalous behavior in gel matrices, indicate that the MeCP2 tertiary structure possesses the features of an intrinsically disordered protein (Adams et al. 2007). The concept of intrinsic or native disorder in proteins has recently gained much attention (Dunker et al. 2001, 2005; Uversky et al. 2005; Chen et al. 2006; Hansen et al. 2006). Prediction algorithms indicate that there is a preponderance of intrinsic disorder in proteins such as transcription factors (Haynes et al. 2006; Liu et al. 2006). It has been hypothesized that the presence of intrinsic disorder permits transient, low-affinity protein–protein and protein–nucleic acid interactions (Uversky et al. 2005; Liu et al. 2006). In the case of MeCP2, the location of order and disorder can be reproducibly predicted by several programs, such as PONDR (Li et al. 1999) and FoldIndex (Prilusky et al. 2005). The FoldIndex prediction plot for MeCP2 (Fig. 2) aligned with the domains of MeCP2. Unlike the core and linker histones, which are predicted to have disorder at their terminal domains, MeCP2 is predicted to have short stretches of order interspersed between long stretches of internal disorder over the length of the entire peptide chain. Of note, many of the predicted order–disorder boundaries coincide with the boundaries of the structural domains identified by proteolysis. Also, the known functional domains (i.e., the MBD and TRD) are predicted to be significantly disordered, consistent with the experimental data. Taken together, all available evidence suggests that intrinsic disorder is a key determinant of MeCP2 tertiary structure, and is likely to be an important constituent of all 6 MeCP2 structural domains (Fig. 2).
Two conceptual models of MeCP2 tertiary structure can be imagined to fit the data. In the first model, several β-sheet/turn structural motifs are interspersed along the length of the MeCP2 amino acid chain, connected by disordered regions, and with no intermotif interactions. This could be viewed as an inchworm model. However, electron micrographs of MeCP2 do not support this model (Nikitina et al. 2007_b_). The second model is based on the CD finding that β strands/turns are the predominant type of classical secondary structure in MeCP2. In this model, MeCP2 strand-forming regions that are separated by tens or possibly hundreds of amino acids apart on the linear polypeptide chain are connected by β sheets or other forms of β structures. For example, one can picture a small half β barrel with large regions of intrinsic disorder making up loops on either side of the β sheet stacks. Although the new studies represent significant progress, a better understanding of the unusual tertiary structure of MeCP2 will be necessary to decipher the molecular links between MeCP2 domain organization, the multifunctionality of the protein, and the cellular pathogenesis of RTT.
Evidence for global genomic functions of MeCP2
A recent important genomics paper has raised questions about the current paradigms of how MeCP2 acts at the cellular level. Using a ChIP-chip approach, Yasui et al. (2007) performed an overall epigenomic binding analysis of MeCP2. Two important aspects of MeCP2 function were revealed by this work. First, MeCP2 is not always associated with transcriptionally repressed genes and, instead, is often associated with actively transcribed genes. Second, the majority (59%) of dsDNA binding sites for MeCP2 within the genome are thousands of base pairs away from intragenic regions, let alone methylated promoters. These new results suggest that the fundamental question of what MeCP2 is doing in the nucleus remains to be answered. Clearly, the finding that a majority of MeCP2 molecules bind actively transcribed promoters indicates that this protein has additional functions that are not explained by the traditional proximal gene silencer model.
This ChIP-chip analysis consisted of scanning 26.3 Mb of imprinted and nonimprinted chromosomal loci of known or suspected genes targeted by MeCP2. A custom high-density gene chip or oligonucleotide (oligo) microarray was constructed with 50 mer oligos attached with a step of 32 base pairs per 50 mer, such that each successive oligo contained 18 bp in the 5′ end of the oligo, matching the last 18 bp of the 3′ end of the previous oligo. Repetitive sequences were removed prior to gene chip construction, so only unique sequences were assayed for binding in the selected chromosomal regions. Human neurons from the SH-SY5Y cell line were used as the source of chromatin for immunoprecipitation of MeCP2. The SH-SY5Y cell line was selected based on its ability to doubly express MeCP2 during differentiation (Jung et al. 2003). The high-affinity immunoglobulin Y-specific antibody used in this study recognized the C-terminus of MeCP2. The C-terminus is identical in both e1 and e2 MeCP2 splice variants; therefore, the antibody did not distinguish between isoforms. This yielded an averaged binding along the assayed chromosomal cross-section. Further understanding of the difference between the 2 isoforms would come with a similar epigenomic analysis using isoform specific antibodies.
In the CHiP-chip experiments, MeCP2 was observed to occupy many active promoters and to bind mostly to nonmethylated sites along intergenic spaces (Yasui et al. 2007). Of those intergenic binding sites, 58.4% were 10 kb or more away from transcription start sites or transcription end sites. Interestingly, the majority of the MeCP2 intragenic binding sites were intronic, which is consistent with a potential functional role for MeCP2 in pre-mRNA splicing (Young et al. 2005). The other main conclusion gathered by the epigenomic ChIP-chip analysis was that the majority (62.6%) of MeCP2-bound promoters are actively expressed genes, including, for example, the immediate early response gene JUNB (Yasui et al. 2007).
Perhaps the crux of this ChIP-chip analysis, and the most contradictory to the initial paradigm of MeCP2 function, was the finding that MeCP2 is not concentrated at densely methylated promoters. To determine which of the assayed promoters had the highest methylation levels, genome-wide promoter methylation analysis by methylated DNA immunoprecipitation (MeDIP) was performed on a microarray containing 24 275 presumed human promoters (Yasui et al. 2007). This methylation seeking immunoprecipitation technique was applied to identically differentiated SH-SY5Y cells. Comparing the results from the promoter methylation-dependant immunoprecipitation assay with those from the MeCP2 binding assay revealed that only 2.2% of the top 4062 promoters with highest measurable levels of methylation were bound by MeCP2. The finding that MeCP2 is not concentrated at densely methylated promoters contradicts the original paradigm of MeCP2 function. The hypothesis that MeCP2 promoter binding is coupled to transcriptional repression is further compromised by the finding that, with certain promoter occupancy, expression levels of the target protein actually increase. The observation that the RNA-SEH2A gene expression decreases with MeCP2 deficiency is another specific example implicating MeCP2 as a regulator rather than strictly a repressor of targeted genes associated with neuronal development (Yasui et al. 2007).
There are caveats to the applicability of results generated by in vitro ChIP-chip techniques to an in vivo model of RTT. Using neuroblastoma cells as a source of chromatin may not reflect tissue-specific chromatin binding profiles for MeCP2 in normal neuronal tissue, and low-affinity DNA binding events may be overexaggerated on the gene chip results. However, the model of MeCP2 as being associated with actively transcribed genes and a majority of nonmethylated CpG sequences is reinforced by more recent literature. Gene expression patterns studied using in the hypothalamus of mice reveal that nearly 85% of genes bound by MeCP2 are actively transcribed (Chahrour et al. 2008). This study adds to the list of MeCP2 interaction partners by demonstrating that CREB1 protein is pulled down in ChIP with MeCP2. The specificity of the interaction is further established by the interaction of MeCP2 with CREB1 at activated promoters, but not at repressed targets.
Recent in vivo work also corroborates the conclusions of the CHiP-chip epigenomic MeCP2 binding analysis. For example, MeCP2 knockout mice and RTT patients do not aberrantly express genes regulated by promoter methylation (Traynor et al. 2002; Tudor et al. 2002). These results, together with the evidence that MeCP2 binds a majority of actively transcribing promoters, argues for MeCP2 having a role as both a positive and negative regulator of transcription (Yasui et al. 2007; Chahrour et al. 2008). This evidence further supports the view that MeCP2 uses something in addition to, or other than, methylation for certain cellular functions, and suggests a model in which MeCP2 functions as an architectural chromatin protein and both a positive and negative regulator of transcription, rather than a gene-specific methylation-directed silencer.
As mentioned previously, in vitro studies are also consistent with a role for MeCP2 in regulating global chromatin architecture, independent of methylation status. Genomic dsDNA is packed in chromatin by wrapping around octamers of basic histone proteins, which form nucleosomes (Luger et al. 1997). Nucleosomes are interspersed at semi-regular intervals along the DNA strand, creating arrays of nucleosomes, termed chromatin. MeCP2 has been shown to bind to chromatin fibers and directly compact them into folded and oligomeric structures when bound to model nucleosomal arrays (Georgel et al. 2003; Nikitina et al. 2007a, 2007b). The arrays used in these studies were unmethylated, implicating a role for MeCP2 in modulating chromatin architecture, independent of methylation status. Although it is unequivocal that MeCP2 preferentially binds methylated DNA, and even more specifically the methylated linker DNA of mononucleosomes and nucleosomal arrays (Nikitina et al. 2007_a_; Ishibashi et al. 2008), the fact remains that MeCP2 is able to significantly alter higher-order chromatin independent of methylation remains a clearly documented property of the protein, and is likely due, in part, to the presence of multiple MeCP2 DNA and (or) chromatin binding domains besides the MBD (Nikitina et al. 2007_a_).
The abundance of MeCP2 in adult somatic nuclei (1–5 × 105 molecules/nucleus) implies that there is a global genomic role for MeCP2. The in vitro studies of chromatin compaction are currently the best explanation for the genomic occupancy of MeCP2. Even though MeCP2 may function as a global chromatin architectural protein, no change in histone modification profiles were observed, compared with wild-type in MeCP2-null mice (Urdinguio et al. 2007). This finding can be reconciled by considering that MeCP2 can modify genomic chromatin architecture directly and independently of histone modifier proteins (Georgel et al. 2003; Nikitina et al. 2007a, 2007b). Considering that specific genes, such as BDNF and TRKB, are down- or upregulated, respectively, in the MeCP2 knockout mouse model leaves open the prospect of unidentified factors directing MeCP2 localization (Abuhatzira et al. 2007).
Potential clinical correlation between MeCP2 domains and RTT
Missense and nonsense mutations that cause RTT are found in all 6 structural domains of MeCP2 (Moore et al. 2005). This argues that all domains are required for proper function of the protein. Toward this end, recent studies have attempted to correlate the location of the mutation with specific facets of RTT pathogenesis. Though the majority of RTT cases are caused by either single point mutations or truncation mutations in MeCP2, the diagnosis of RTT is defined via clinical observation, not genetically (Glaze 2004).
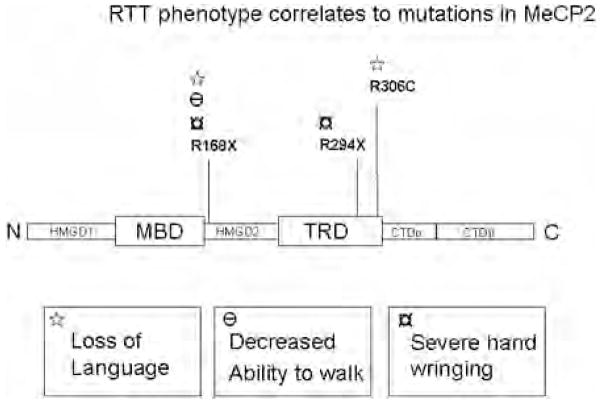
Perhaps the most influential factor in disease severity is the type of X-chromosome inactivation (XCI). Females with favorably skewed XCI may have very mild learning disabilities, while a spectrum of severity correlates with increasingly unfavorable XCI to autism, and on to latter-onset RTT (Glaze 2004). Some attempts have been made to correlate MeCP2 mutations to disease severity by comparing data in the international RTT database with clinically reported severity. Two common RTT mutations in MeCP2, the R168X and T158M mutations, were chosen for a study in which the degree of XCI skewing and direction were observed and correlated with RTT severity (Archer et al. 2007). That study reports that there is a statistically significant correlation between unfavorable XCI and measured clinical severity. Although the correlation appears to be statistically significant, it does not appear very robust. One MeCP2 mutation that has been directly implicated in elevated severity, regardless of XCI skew, is the R270X mutant. This truncation mutant decreases life expectancy in RTT patients, compared with all other recorded mutations (Jian et al. 2005). Recently, Neul et al. (2008) have established that individuals with the R168X mutation are more severely affected than those with R294X and other late carboxy-terminal truncating mutations, highlighting the importance of domains that are C-terminal to the MBD. These researchers have established that different point mutations affect the severity of the 3 main pathological problems associated with RTT: loss of language, walking, and hand use. The correlation between domain mutation and disease phenotype is depicted in Fig. 3. The most severe mutation observed was the R168X. Individuals with this truncation mutation lose the ability to walk, use their hands properly, and, more frequently, loose their entire vocabulary. The truncation mutations in the C-terminal are less severe. These patients have a higher probability of walking and retaining vocabulary, although they exhibit other significant problems. The R306C point mutation only affects language skills. Taken together, the clinical studies and the distribution of mutations in the RTT database suggest that all 6 MeCP2 structural domains shown in Fig. 2 must function together to mediate the normal cellular actions of MeCP2.
Fig. 3.
Schematic depicting MeCP2 with contingent domains labeled. Two common Rett syndrome (RTT) truncation mutations (R168X and R294X) and an RTT point mutation (R306C) are indicated along the peptide. The phenotypic pathology or combinations are correlated to each mutation with symbols.
In closing, the fundamental question of how MeCP2 functions in the nucleus of nearly all vertebrate tissues should be approached in the future from the new structural and functional perspectives discussed above. Specifically, at the molecular level, what is the relationship between intrinsic disorder, MeCP2 domain organization, and protein function? How do different mutations in the highly disordered MeCP2 tertiary structure cause different neurodevelopmental RTT symptoms? How does MeCP2 affect chromatin architecture in vivo? Does MeCP2 have to bind to currently unknown proteins that modulate its capacity to regulate transcription? What, exactly, is the role of MeCP2 in pre-mRNA splicing? Does MeCP2 directly bind to RNA molecules? Are there cellular functions of MeCP2 that have yet to be discovered? The new data require that we step back and ask these and many other related questions.
Acknowledgments
This work was supported by National Institutes of Health grant GM66834.
Abbreviations
RTT
Rett syndrome
CTD
carboxyl terminal domain
Footnotes
1
This paper is one of a selection of papers published in this Special Issue, entitled 29th Annual International Asilomar Chromatin and Chromosomes Conference, and has undergone the Journal’s usual peer review process.
References
- Abuhatzira L, Makedonski K, Kaufman Y, Razin A, Shemer R. MeCP2 deficiency in the brain decreases BDNF levels by REST/CoREST-mediated repression and increases TRKB production. Epigenetics. 2007;2:214–222. doi: 10.4161/epi.2.4.5212. [DOI] [PubMed] [Google Scholar]
- Adams VH, McBryant SJ, Wade PA, Woodcock CL, Hansen JC. Intrinsic disorder and autonomous domain function in the multifunctional nuclear protein, MeCP2. J Biol Chem. 2007;282:15057–15064. doi: 10.1074/jbc.M700855200. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Archer H, Evans J, Leonard H, Colvin L, Ravine D, Christodoulou J, et al. Correlation between clinical severity in patients with Rett syndrome with a p.R168X or p.T158M MECP2 mutation, and the direction and degree of skewing of X-chromosome inactivation. J Med Genet. 2007;44:148–152. doi: 10.1136/jmg.2006.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschdorf JP, Stratling WH. A WW domain binding region in methyl-CpG-binding protein MeCP2: impact on Rett syndrome. J Mol Med. 2004;82:135–143. doi: 10.1007/s00109-003-0497-9. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler SP, Guschin D, Landsberger N, Wolffe AP. The methyl-CpG binding transcriptional repressor MeCP2 stably associates with nucleosomal DNA. Biochemistry. 1999;38:7008–7018. doi: 10.1021/bi990224y. [DOI] [PubMed] [Google Scholar]
- Chen JW, Romero P, Uversky VN, Dunker AK. Conservation of intrinsic disorder in protein domains and families: II. functions of conserved disorder. J Proteome Res. 2006;5:888–898. doi: 10.1021/pr060049p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dintilhac A, Bernues J. HMGB1 interacts with many apparently unrelated proteins by recognizing short amino acid sequences. J Biol Chem. 2002;277:7021–7028. doi: 10.1074/jbc. M108417200. [DOI] [PubMed] [Google Scholar]
- Dragich JM, Kim YH, Arnold AP, Schanen NC. Differential distribution of the MeCP2 splice variants in the postnatal mouse brain. J Comp Neurol. 2007;501:526–542. doi: 10.1002/cne.21264. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/S1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Montoya G, Taysavang P, Wade PA, Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J Biol Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- Glaze DG. Rett syndrome: of girls and mice – lessons for regression in autism. Ment Retard Dev Disabil Res Rev. 2004;10:154–158. doi: 10.1002/mrdd.20030. [DOI] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JC, Lu X, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J Biol Chem. 2006;281:1853–1856. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]
- Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, et al. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLOS Comput Biol. 2006;2:e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol Cell. 2008;29:525–531. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Thambirajah AA, Ausio J. MeCP2 preferentially binds to methylated linker DNA in the absence of the terminal tail of histone H3 and independently of histone acetylation. FEBS Lett. 2008;582:1157–1162. doi: 10.1016/j.febslet.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Jeffery L, Nakielny S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J Biol Chem. 2004;279:49479–49487. doi: 10.1074/jbc.M409070200. [DOI] [PubMed] [Google Scholar]
- Jian L, Archer HL, Ravine D, Kerr A, de Klerk N, Christodoulou J, et al. p.R270X MECP2 mutation and mortality in Rett syndrome. Eur J Hum Genet. 2005;13:1235–1238. doi: 10.1038/sj.ejhg.5201479. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Jung BP, Jugloff DG, Zhang G, Logan R, Brown S, Eubanks JH. The expression of methyl CpG binding factor MeCP2 correlates with cellular differentiation in the developing rat brain and in cultured cells. J Neurobiol. 2003;55:86–96. doi: 10.1002/neu.10201. [DOI] [PubMed] [Google Scholar]
- Kato Y, Nagata K, Takahashi M, Lian L, Herrero JJ, Sudol M, Tanokura M. Common mechanism of ligand recognition by group II/III WW domains: redefining their functional classification. J Biol Chem. 2004;279:31833–31841. doi: 10.1074/jbc.M404719200. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. MeCP2 behaves as an elongated monomer that does not stably associate with the Sin3a chromatin remodeling complex. J Biol Chem. 2004;279:46490–46496. doi: 10.1074/jbc.M408284200. [DOI] [PubMed] [Google Scholar]
- Kokura K, Kaul SC, Wadhwa R, Nomura T, Khan MM, Shi-nagawa T, et al. The Ski protein family is required for MeCP2-mediated transcriptional repression. J Biol Chem. 2001;276:34115–34121. doi: 10.1074/jbc.M105747200. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-O. [DOI] [PubMed] [Google Scholar]
- Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting protein disorder for N-, C-, and internal regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Meehan R, Antequera F, Lewis J, MacLeod D, McKay S, Kleiner E, Bird AP. A nuclear protein that binds preferentially to methylated DNA in vitro may play a role in the inaccessibility of methylated CpGs in mammalian nuclei. Philos Trans R Soc Lond B Biol Sci. 1990;326:199–205. doi: 10. 1098/rstb.1990.0004. [DOI] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnatzakanian GN, Lohi H, Munteanu I, Alfred SE, Yamada T, MacLeod PJ, et al. A previously unidentified MECP2 open reading frame defines a new protein isoform relevant to Rett syndrome. Nat Genet. 2004;36:339–341. doi: 10.1038/ng1327. [DOI] [PubMed] [Google Scholar]
- Moore H, Leonard H, Fyfe S, De Klerk N, Leonard N. InterRett – The application of bioinformatics to international Rett syndrome research. Ann Hum Biol. 2005;32:228–236. doi: 10.1080/03014460500075068. [DOI] [PubMed] [Google Scholar]
- Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/S0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Neul JL, Fang P, Barrish J, Lane J, Caeg EB, Smith EO, et al. Specific mutations in Methyl-CpG-Binding Protein 2 confer different severity in Rett syndrome. Neurology. 2008;70:1313–1321. doi: 10.1212/01.wnl.0000291011.54508.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitina T, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Grigoryev SA, Woodcock CL. MeCP2-chromatin interactions include the formation of chromatosome-like structures and are altered in mutations causing Rett syndrome. J Biol Chem. 2007a;282:28237–28245. doi: 10.1074/jbc.M704304200. [DOI] [PubMed] [Google Scholar]
- Nikitina T, Shi X, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Woodcock CL. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol Cell Biol. 2007b;27:864–877. doi: 10.1128/MCB.01593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelka GJ, Watson CM, Radziewic T, Hayward M, Lahooti H, Christodoulou J, Tam PP. Mecp2 deficiency is associated with learning and cognitive deficits and altered gene activity in the hippocampal region of mice. Brain. 2006;129:887–898. doi: 10.1093/brain/awl022. [DOI] [PubMed] [Google Scholar]
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, et al. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- Traynor J, Agarwal P, Lazzeroni L, Francke U. Gene expression patterns vary in clonal cell cultures from Rett syndrome females with eight different MECP2 mutations. BMC Med Genet. 2002;3:12. doi: 10.1186/1471-2350-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor M, Akbarian S, Chen RZ, Jaenisch R. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci USA. 2002;99:15536–15541. doi: 10.1073/pnas.242566899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdinguio RG, Pino I, Ropero S, Fraga MF, Esteller M. Histone H3 and H4 modification profiles in a Rett syndrome mouse model. Epigenetics. 2007;2:11–14. doi: 10.4161/epi.2.1.3698. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Veenstra GJ, Imhof A, Sera T, et al. Histone deacetylase directs the dominant silencing of transcription in chromatin: association with MeCP2 and the Mi-2 chromodomain SWI/SNF ATPase. Cold Spring Harb Symp Quant Biol. 1998;63:435–445. doi: 10.1101/sqb.1998.63.435. [DOI] [PubMed] [Google Scholar]
- Wakefield RI, Smith BO, Nan X, Free A, Soteriou A, Uhrin D, et al. The solution structure of the domain from MeCP2 that binds to methylated DNA. J Mol Biol. 1999;291:1055–1065. doi: 10.1006/jmbi.1999.3023. [DOI] [PubMed] [Google Scholar]
- Wischnewski F, Friese O, Pantel K, Schwarzenbach H. Methyl-CpG binding domain proteins and their involvement in the regulation of the MAGE-A1, MAGE-A2, MAGE-A3, and MAGE-A12 gene promoters. Mol Cancer Res. 2007;5:749–759. doi: 10.1158/1541-7786.MCR-06-0364. [DOI] [PubMed] [Google Scholar]
- Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, et al. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci USA. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci USA. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Thiesen J, Stratling WH. Histone deacetylase-independent transcriptional repression by methyl-CpG-binding protein 2. Nucleic Acids Res. 2000;28:2201–2206. doi: 10.1093/nar/28.10.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J. MeCP2: the chromatin connection and beyond. Biochem Cell Biol. 2005;83:251–262. doi: 10.1139/o05-048. [DOI] [PubMed] [Google Scholar]