dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties (original) (raw)
Abstract
Mi-2 and ISWI, two members of the Snf2 superfamily of ATPases, reside in separate ATP-dependent chromatin remodelling complexes. These complexes differ in their biochemical properties and are believed to perform distinct functions in the cell. We have compared the remodelling activity of recombinant Drosophila Mi-2 (dMi-2) with that of recombinant ISWI. Both proteins are nucleosome-stimulated ATPases and promote nucleosome mobilization. However, dMi-2 and ISWI differ in their interaction with nucleosome core particles, in their substrate requirements and in the direction of nucleosome mobilization. We have used antibodies to immobilize a complex containing dMi-2 and the dRPD3 histone deacetylase from Drosophila embryo extracts. This complex shares the nucleosome-stimulated ATPase and nucleosome mobilization properties of recombinant dMi-2, demonstrating that these activities are maintained in a physiological context. Its functional properties distinguish dMi-2 from both SWI2/SNF2 and ISWI, defining a new family of ATP-dependent remodelling machines.
Keywords: ATPase/chromatin/Mi-2/nucleosome remodelling
Introduction
Molecular machines that carry out the fundamental processes of DNA replication, recombination, repair and transcription need to recognize and bind to their DNA substrate. The packaging of DNA into chromatin, however, hinders their access to DNA. Chromatin remodelling factors facilitate DNA binding by creating a more dynamic chromatin structure (reviewed in Cairns, 1998; Kingston and Narlikar, 1999). Two broad classes of remodelling activities have received much attention in recent years. The first class is comprised of ATP-dependent chromatin remodelling complexes containing ATPases related to SWI2/SNF2 or ISWI (Kingston and Narlikar, 1999). These remodel interactions between histones and DNA in a poorly defined reaction requiring ATP hydrolysis. The second class consists of complexes that acetylate or deacetylate specific lysine residues in the N-terminal tails of histones (Kouzarides, 1999). Acetylation is believed to lead to a more open chromatin structure by changing the nucleosome–nucleosome interactions involved in the folding of the nucleosomal fibre (Luger et al., 1997a; Tse et al., 1998). Alternatively, acetylated lysine tails could serve as recognition sites for chromatin binding factors, which in turn elicit changes in chromatin structure.
The view that ATP-dependent chromatin remodelling and histone acetylation/deacetylation are carried out by separate entities has recently been challenged. Complexes containing the ATPases Mi-2α and/or Mi-2β as well as histone deacetylases HDAC1 and HDAC2 have been identified in human and frog (Tong et al., 1998; Wade et al., 1998; Xue et al., 1998; Zhang et al., 1998). These complexes combine ATP-dependent chromatin remodelling and histone deacetylase (HDAC) activities, and have accordingly been named NuRD or NRD [nucleosome remodelling and deacetylation (Tong et al., 1998; Xue et al., 1998; Zhang et al., 1998)]. The Mi-2 subunits of these complexes belong to the CHD (_c_hromo-_h_elicase/ATPase-_D_NA binding) family of proteins (Woodage et al., 1997). Four CHD proteins have been identified in vertebrates: CHD1, CHD2, CHD3 (Mi-2α) and CHD4 (Mi-2β). CHD proteins share two conserved chromo domains and one ATPase domain. In addition, CHD1 and CHD2 have a DNA-binding domain whereas Mi-2α and Mi-2β contain two PHD fingers (Stokes and Perry, 1995; Woodage et al., 1997). The physiological function of CHD proteins is not clear. CHD1 has been localized to decompacted interphase chromosomes in mammalian cells and to regions of high transcriptional activity on Drosophila polytene chromosomes, implicating this family member in transcriptional control (Stokes et al., 1996; Kelley et al., 1999).
Histone deacetylation of nucleosomal substrates by Mi-2 complexes is stimulated by ATP hydrolysis, arguing that energy-dependent remodelling of the nucleosome is required for the deacetylases to gain access to histone tails (Tong et al., 1998; Xue et al., 1998; Zhang et al., 1998). The Mi-2 complexes are believed to repress transcription through their remodelling and deacetylation activities in a targeted manner. Two ways of recruiting Mi-2 complexes to promoter regions have been proposed. First, targeting could be mediated via the interaction between the Mi-2 complex and DNA-bound transcriptional repressors. Indeed, the Mi-2 complex copurifies with the Ikaros repressor and functionally interacts with thyroid hormone receptor in vitro (Xue et al., 1998; Kim et al., 1999). Furthermore, a Drosophila homologue of Mi-2 (dMi-2) physically and genetically interacts with the hunchback repressor (Kehle et al., 1998). Secondly, the Mi-2 complex could be targeted to methylated DNA either directly via its MBD3 subunit or indirectly via association with the MBD2A-methylated DNA-binding protein (Wade et al., 1999; Zhang et al., 1999).
Several observations suggest that Mi-2 complexes are involved in cell cycle regulation and that their deregulation contributes to cancer. Mi-2 itself was originally identified as an autoimmune antigen in patients suffering from dermatomyositis. These patients suffer an increased risk of developing cancer (Seelig et al., 1995, 1996). Furthermore, Mi-2 is targeted by the E7 oncoprotein of human papilloma virus 16 (Brehm et al., 1999). Finally, the Mi-2 complex subunit MTA2 is closely related to MTA1, which has been found to be overexpressed in cancer cells with a high potential for metastasis (Toh et al., 1994, 1997, 1999).
Mi-2 and other CHD proteins are members of the growing SNF2 superfamily of ATPases (Eisen et al., 1995). The founding member of this family, yeast SWI2/SNF2, is a well characterized ATP-dependent chromatin remodelling enzyme. However, since the similarity between members of the Snf2 superfamily is restricted to the ATPase domain it is unclear whether all family members are nucleosome remodelling factors. SWI/SNF, Sth1 and ISWI complexes can facilitate factor binding to chromatin and activate transcription from a chromatin template in vitro (for review see Kingston and Narlikar, 1999). Furthermore, both SWI/SNF and ISWI complexes are able to promote movement of a nucleosome along DNA (Hamiche et al., 1999; Längst et al., 1999; Whitehouse et al., 1999). Both types of ATPase also display a series of distinct biochemical properties. Whereas BRG1 and hBRM ATPase activities are stimulated to the same extent by nucleosomes and naked DNA, the ISWI ATPase is preferentially stimulated by nucleosomes (Corona et al., 1999; Phelan et al., 1999). We have recently demonstrated that ATPase activity and nucleosome mobilization by ISWI depends on an intact histone H4 tail (C.R.Clapier, G.Längst, D.F.V.Corona, P.B.Becker and K.P.Nightingale, manuscript in preparation). In contrast, remodelling of nucleosomal arrays by SWI/SNF is variably affected by the removal of all four histone tails depending on the precise assay conditions (Logie and Peterson, 1997; Guyon et al., 1999). These findings show that SWI/SNF and ISWI complexes differ in the way they interact with the nucleosome to promote chromatin remodelling.
The Mi-2 complexes can also facilitate transcription factor binding to a nucleosomal template and increase restriction enzyme access to nucleosomal DNA (Tong et al., 1998; Xue et al., 1998; Zhang et al., 1998). It is unclear to what extent their activities resemble those of SWI2/SNF2, Sth1 and ISWI-containing complexes. The Mi-2 ATPases have variably been suggested to comprise a family of their own (Tyler and Kadonaga, 1999) or to belong to the SWI/SNF family based on the comparison of amino acid sequences of their ATPase domains (Kingston and Narlikar, 1999). It is assumed that the Mi-2 ATPase is responsible for chromatin remodelling by the Mi-2 complex. However, the remodelling activites of recombinant Mi-2 protein have not been analysed and we do not know what role other complex subunits play in chromatin remodelling.
We have expressed recombinant dMi-2 in order to study its chromatin remodelling activities. We find that dMi-2 is a nucleosome-stimulated ATPase. Furthermore, dMi-2 is stimulated by nucleosomes that lack all four histone tails. Recombinant dMi-2 binds nucleosomes and promotes nucleosome mobilization in an ATP-dependent manner. Remarkably, the direction of nucleosome mobilization promoted by dMi-2 and ISWI is different in our assay. Moreover, unlike ISWI, dMi-2 can mobilize nucleosomes lacking the H4 N-terminal tail. We have partially purified a dMi-2-containing complex from Drosophila embryo extracts (here referred to as ‘dMi-2 complex’) with ATPase, HDAC and nucleosome mobilization activities. Recombinant dMi-2 and dMi-2 from Drosophila embryos share the same ATPase and nucleosome mobilization properties. We propose that dMi-2 defines a novel family of chromatin remodelling complexes that is mechanistically distinct from both ISWI and SWI/SNF complexes.
Results
A dMi-2-containing complex in Drosophila
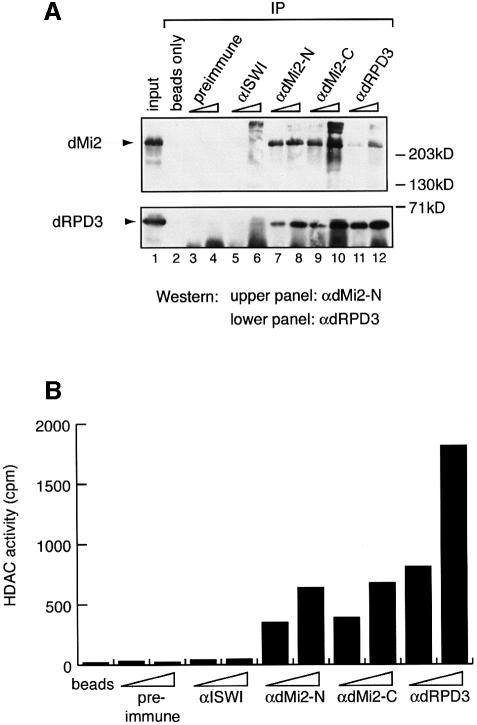
In order to study the chromatin remodelling activities of a putative Drosophila Mi-2 complex we raised two antibodies directed against the N-terminus (αdMi-2-N) and the C-terminus of dMi-2 (αdMi-2-C), respectively, as well as an antibody directed against the dRPD3 (αdRPD3) HDAC. We used these antibodies for immunoprecipitation from a crude nuclear extract prepared from Drosophila embryos and for detection of dMi-2 and dRPD3 in the immunoprecipitates by western blot analysis. The αdMi-2-N antibody recognized a protein of the expected molecular weight (220 kDa) in the extract (Figure 1A, lane 1, upper panel). Both dMi-2 antisera as well as the αdRPD3 antiserum immunoprecipitated the 220 kDa αdMi-2-N-reactive protein. No 220 kDa band was detected when antiserum was omitted from the immunoprecipitation (beads only) or when preimmune or αISWI serum was used. We conclude that the 220 kDa protein is dMi-2. The αdRPD3 antiserum recognizes a protein of ∼60 kDa in nuclear extract, which we will refer to as dRPD3. αdRPD3 as well as both αdMi-2 antisera immunoprecipitated dRPD3 (Figure 1A, lower panel). Again, omission of antiserum or use of preimmune or αISWI serum failed to precipitate significant amounts of dRPD3. These results argue that dMi-2 and dRPD3 form a stable complex in Drosophila embryos.
Fig. 1. (A) dMi-2 and dRPD3 coimmunoprecipitate. Drosophila embryo nuclear extract was immunoprecipitated with 1 and 5 µl of various antisera as indicated. dMi-2 and dRPD3 were detected by western analysis using the αdMi-2-N (upper panel) or the αdRPD3 antiserum (lower panel). Lane 1: 20% input. Positions of dMi-2 and dRPD3 bands are indicated on the left, molecular weights are indicated on the right. (B) αdMi-2 antibodies coprecipitate histone deacetylase activity. Immunoprecipitations were performed as in (A) using 1 and 5 µl of antisera. HDAC activity was measured using a radioactive histone H4 peptide as a substrate. (C) αdMi-2 and αdRPD3 antibodies coprecipitate nucleosome-stimulated ATPase activity. Immunoprecipitations were performed as in (A). Antisera used are indicated at the top. Each immunoprecipitate was split into three and used for ATPase assays in the absence or presence of 50 ng of free or nucleosomal DNA. ATPase activity values are expressed as percentage conversion of ATP.
We next tested whether the immunoprecipitates were active in an in vitro HDAC assay (Figure 1B). αdRPD3 and both αdMi-2 antisera precipitated HDAC activity whereas the control precipitates contained only background levels of activity.
We next investigated the ATPase activity of the immunoprecipitated dMi-2 complex. The immunoprecipitates were split into three, and ATPase activity of each aliquot was tested in the absence (control) or presence of naked plasmid DNA or nucleosomes produced from plasmid DNA and purified Drosophila histones by salt gradient dialysis, respectively. As shown in Figure 1C, low levels of ATPase activity were detected when no antiserum (beads) or preimmune serum was used for immunoprecipitation irrespective of the presence of naked DNA or nucleosomes in the ATPase reaction. The αdMi-2 antisera also precipitated a low level of ATPase activity that was not significantly increased in the presence of naked DNA. ATPase activity was strongly enhanced, however, if nucleosomes were present in the reaction, demonstrating the presence of a nucleosome-stimulated ATPase activity in these immunoprecipitates. We also detected nucleosome-stimulated ATPase activity when αdRPD3 antiserum was used for immunoprecipitation, albeit at a lower level. These findings suggest that the immunoprecipitated dMi-2 complex possesses ATPase activity that is stimulated by nucleosomes but not by naked DNA.
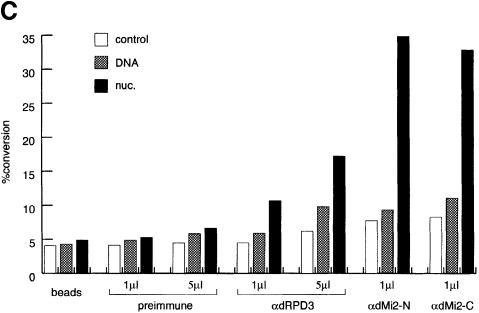
To estimate the size of the dMi-2 complex we subjected it to Superose 6 gel filtration chromatography (Figure 2). Aliquots of Superose 6 fractions were immunoprecipitated with αdRPD3 and αdMi-2-C antisera, and the precipitates were subjected to HDAC and ATPase assays, respectively. dMi-2-associated nucleosome-stimulated ATPase activity was found to elute between fractions 20 and 24. The same fractions also contained the main peak of dRPD3-associated HDAC activity. The molecular weight corresponding to fractions 20–24 is ∼1.0 MDa. Taken together our results suggest that, like its vertebrate counterparts, dMi-2 resides in a high molecular weight complex that possesses HDAC and nucleosome-stimulated ATPase activity.
Fig. 2. dMi-2 resides in a large complex. Partially purified dMi-2-containing fractions (see Materials and methods) were fractionated over a Superose 6 sizing column. Aliquots of fractions were immunoprecipitated with 1 µl of αdMi-2-C and subjected to ATPase assays, or immunoprecipitated with 3 µl of αdRPD3 antiserum and subjected to the HDAC assay as indicated. Molecular weights of fractions eluting from the Superose 6 column were determined with molecular weight standards (Pharmacia) and are indicated below fraction numbers.
Recombinant dMi-2 is a nucleosome-stimulated ATPase
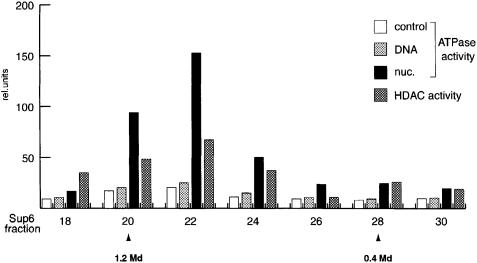
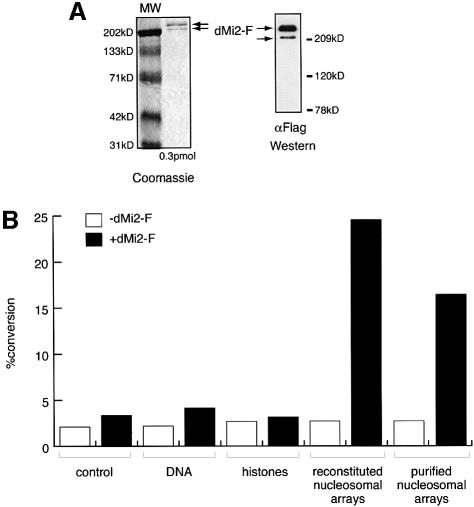
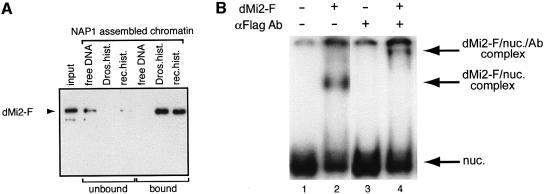
Although it has not yet been formally demonstrated, it is widely assumed that dMi-2 and its vertebrate homologues confer ATPase activity to Mi-2-containing complexes. It has been proposed that the non-enzymatic subunits of chromatin remodelling complexes regulate the activity and specificity of the ATPases (Kingston and Narlikar, 1999). For example, it is conceivable that nucleosome stimulation of the ATPase requires a non-enzymatic subunit of the dMi-2 complex. To establish the intrinsic properties of dMi-2 we expressed flag epitope-tagged full length dMi-2 (dMi-2-F) using the baculovirus system and purified the recombinant protein to apparent homogeneity by immunoaffinity chromatography. The purified recombinant protein gave rise to two closely migrating bands on SDS–PAGE gels. Both bands were recognized by α-Flag antibody in a western blot analysis (Figure 3A). We do not know at present whether these bands represent degradation products or post-translationally modified forms of dMi-2. As expected, recombinant dMi-2 did not display HDAC activity (data not shown). Recombinant dMi-2 had weak ATPase activity in the absence of DNA or nucleosomes, which was not significantly stimulated by the addition of DNA or core histones (Figure 3B). However, addition of nucleosomal arrays isolated from Drosophila embryos or nucleosomes reconstituted from plasmid DNA and purified Drosophila histones greatly stimulated the ATPase activity of the recombinant protein. These results verify that the nucleosome-stimulated ATPase activity is an intrinsic feature of dMi-2 and is not conferred by other subunits of the dMi-2 complex.
Fig. 3. Recombinant dMi-2 is a nucleosome-stimulated ATPase. (A) Recombinant dMi-2-F (0.3 pmol) was applied to SDS–PAGE and either stained with Coomassie Blue (left panel) or analysed by western blotting using α-Flag antibody (right panel). The position of dMi-2-F-derived bands is indicated by arrows. MW: molecular weight marker. (B) Recombinant dMi-2-F (200 fmol) was used for ATPase assays in the absence (control) or presence of 50 ng of naked DNA (DNA), 50 ng of core histones purified from Drosophila embryos (histones), 50 ng of nucleosomal arrays reconstituted by salt gradient dialysis (reconstituted nucleosomal arrays) and 50 ng of nucleosomal arrays purified from Drosophila embryos as indicated.
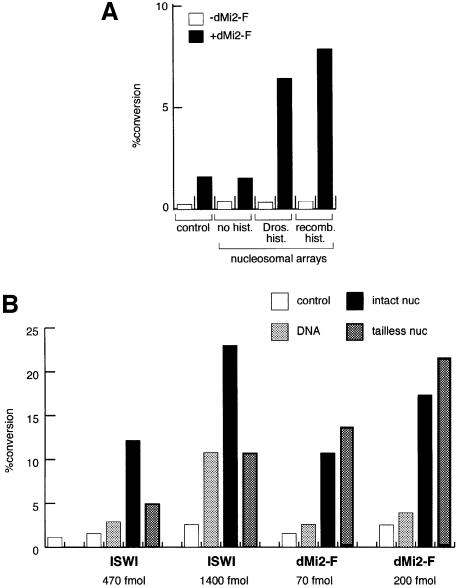
The Drosophila histones used for reconstitution of nucleosomes represent a mixture of the different post-translationally modified forms that exist in vivo. It is conceivable that the dMi-2 ATPase does not respond to the nucleosome structure as such but rather to a particular histone isoform. To address this issue we reconstituted nucleosomes from recombinant histones expressed in Escherichia coli, which lack post-translational modifications, using yNAP1 as an assembly vehicle (Akthar and Becker, 2000). Nucleosomes assembled on plasmid DNA either with purified or recombinant histones activated dMi-2 to a similar extent (Figure 4A). The assembly vehicle, yNAP1, by itself had no effect. Thus, nucleosomal arrays consisting of fully defined components (DNA and four recombinant core histones) suffice to stimulate the dMi-2 ATPase.
Fig. 4. Activation of dMi-2 by recombinant nucleosomes. (A) Recombinant dMi-2-F (200 fmol) was used for ATPase assays in the absence (control) or presence of 50 ng of nucleosomal arrays assembled with yNAP1 as a histone deposition vehicle. To the yNAP1 assembly reactions either no histones (no hist.), histones purified from Drosophila embryos (Dros. hist.) or recombinant histones expressed in E.coli (recomb. hist.) were added. (B and C) Nucleosomal arrays were assembled with yNAP1 using either no histones (DNA), recombinant intact histones (intact nuc) or recombinant histones lacking all four tails (tailless nuc) and used to stimulate ATPase activities of recombinant ISWI and dMi-2-F as indicated. The amounts of ISWI and dMi-2-F in (B) were chosen to give similar activation by intact nucleosomes. ATPase assay and quantification were done as described above.
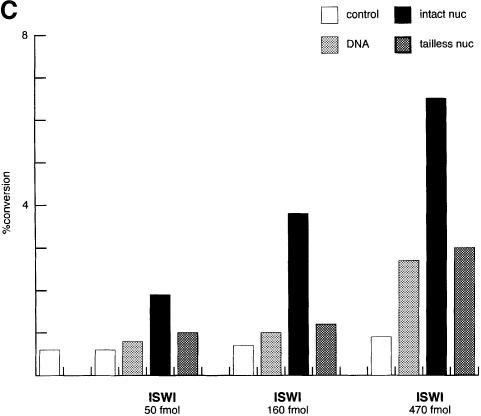
Stimulation of the dMi-2 ATPase does not require histone tails
The activity of the better characterized ATPases ISWI and SWI2/SNF2 depends on the presence of histone N-terminal domains to a varying degree (Georgel et al., 1997; Corona et al., 1999; Guyon et al., 1999; Logie et al., 1999). In order to monitor the effect of N-terminal tail sequences we reconstituted nucleosomes from recombinant Xenopus histones lacking N-termini (Luger et al., 1997b) and compared intact and tailless nucleosomes for stimulation of the ATPase activities of recombinant ISWI and dMi-2 (Figure 4B). We chose the amounts of ISWI and dMi-2 used in this experiment to give comparable activation by intact nucleosomes. The ATPase activity of ISWI was strongly stimulated by intact nucleosomes reconstituted from recombinant histones in agreement with our previous findings (Corona et al., 1999). Nucleosomes lacking all four histone tails did not stimulate ISWI beyond the levels observed with naked DNA. We were unable to detect significant activation of the ISWI ATPase by tailless nucleosomes at several enzyme concentrations tested (Figure 4C). In sharp contrast to ISWI, dMi-2 was equally well stimulated by intact and tailless nucleosomes (Figure 4B), indicating that nucleosomal activation of the dMi-2 ATPase is histone tail independent.
dMi-2 binds nucleosomes
The observations that dMi-2 ATPase activity is stimulated by nucleosomes suggest that dMi-2 can physically associate with nucleosomes. To address this question we immobilized biotinylated DNA on streptavidin-coated paramagnetic beads, and reconstituted nucleosomes on this DNA using yNAP1 as an assembly vehicle. To test for dMi-2 binding the nucleosomes on beads were incubated with recombinant dMi-2 and then stringently washed to remove loosely bound proteins. Western analysis revealed that dMi-2 bound strongly to nucleosomal arrays assembled from either purified or recombinant histones (Figure 5A). No binding to naked DNA was detected under these conditions. We conclude that dMi-2 binds preferentially to nucleosomal arrays over naked DNA. We also investigated whether dMi-2 can form stable complexes with mononucleosomes in a bandshift assay (Figure 5B). For this assay mononucleosomes were generated from purified Drosophila histones by polyglutamic acid-mediated assembly (Längst et al., 1999). First, we tested binding to a mononucleosome assembled on a 248 bp DNA fragment. Incubation with recombinant dMi-2 resulted in the formation of a slower migrating complex (Figure 5B, lane 2), which was supershifted upon addition of α-Flag antibody to the binding reaction (lane 4). We conclude that dMi-2 can associate with a mononucleosome to form a complex that is sufficiently stable to withstand electrophoresis through polyacrylamide gels.
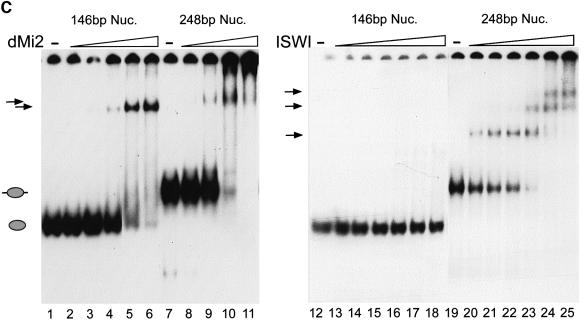
Fig. 5. Recombinant dMi-2 binds to nucleosomes. (A) Biotinylated linearized DNA was bound to streptavidin-coated paramagnetic beads and assembled into nucleosomal arrays using yNAP1 as a histone deposition vehicle. Assembly reactions contained either no histones (free DNA), Drosophila histones (Dros.hist.) or recombinant histones (rec.hist.) as indicated. Bound and unbound materials were subjected to SDS–PAGE and western blot analysis using the M2 antibody (anti-Flag; Sigma). Thirty percent input was loaded as a control (input). (B) Recombinant dMi-2-F (240 fmol, lanes 2 and 4) was incubated with mononucleosomes containing a 248 bp radioactively labelled DNA fragment in the absence or presence of α-Flag antibody as shown. Resulting complexes were separated by native gel electrophoresis and visualized following autoradiography. Free mononucleosomes (nuc.) and dMi-2-F′–nucleosome complexes are indicated by arrows. (C) Recombinant dMi-2-F (lanes 2 and 8: 40 fmol; lanes 3 and 9: 80 fmol; lanes 4 and 10: 120 fmol; lanes 5 and 11: 240 fmol; lane 6: 360 fmol) and ISWI (lanes 13 and 20: 5 fmol; lanes 14 and 21: 10 fmol; lanes 15 and 22: 15 fmol; lanes 16 and 23: 25 fmol; lanes 17 and 24: 50 fmol; lanes 18 and 25: 75 fmol) were incubated with mononucleosomes containing 146 or 248 bp of radioactively labelled DNA as indicated. The positions of the 146 bp (grey ellipse) and 248 bp nucleosome (grey ellipse with protruding DNA ends) are shown on the left. The positions of nucleosome complexes are indicated by arrows.
Next we compared the ability of recombinant dMi-2 and ISWI to bind two different mononucleosome species in the bandshift assay (Figure 5C). We compared binding to a mononucleosome (core particle) containing 146 bp of DNA with binding to a nucleosome assembled on the 248 bp fragment. The primary difference between these substrates is that in the 146 bp nucleosome all DNA is entirely in contact with the histone octamer, whereas the 248 bp nucleosome contains ∼100 bp of naked DNA extending from the octamer. Incubation with increasing amounts of dMi-2 resulted in the formation of a dMi-2–core particle complex (Figure 5C, lanes 1–6) or a complex of dMi-2 with the 248 bp nucleosome (lanes 7–11). In contrast, we failed to detect stable ISWI–core particle complexes (Figure 5C, lanes 12–18), even if higher amounts of ISWI were used (data not shown). Incubation of increasing amounts of ISWI with the 248 bp nucleosome produced up to three ISWI–nucleosome complexes, suggesting that more than one ISWI molecule can simultaneously associate with this substrate (Figure 5C, lanes 19–25).
We conclude that dMi-2 can stably associate with a mononucleosomal substrate lacking free DNA, in agreement with our finding that free DNA does not stimulate the dMi-2 ATPase. In marked contrast, ISWI only binds the nucleosome when free DNA is protruding from the histone octamer. These results strongly suggest that dMi-2 and ISWI recognize different features of the nucleosome.
dMi-2 promotes nucleosome mobilization
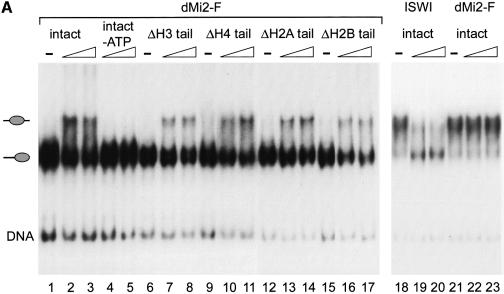
ATP hydrolysis by Mi-2 is believed to drive remodelling of chromatin by the Mi-2 complex. In order to characterize nucleosome remodelling directly, we made use of a recently established ‘nucleosome sliding’ assay. Previously, we have shown that the CHRAC complex and recombinant ISWI can mobilize mononucleosomes to change their position on short DNA fragments (Längst et al., 1999). Nucleosome reconstitution on a 248 bp fragment derived from the mouse rDNA promoter results in the formation of two main species, which differ in the position of the nucleosome relative to the DNA ends and which can be separated by gel electrophoresis under non-denaturing conditions (Längst et al., 1999). Mononucleosomes positioned near the ends and at the centre of the DNA fragment were used as substrates for mobilization by dMi-2 and ISWI (Figure 6A). To avoid loss of labelled nucleosomes due to formation of stable enzyme–substrate complexes (as in the bandshift assay, Figure 5B) the reaction was stopped by addition of excess unlabelled nucleosomes. As reported previously, recombinant ISWI moved the centrally positioned nucleosome to the ends of the fragment (Figure 6A, lanes 18–20) but not vice versa (Längst et al., 1999 and data not shown). Remarkably, recombinant dMi-2 behaved in exactly the opposite way: dMi-2 failed to move the central nucleosome (Figure 6A, lanes 21–23) but mobilized the end-positioned nucleosome (lanes 1–3). dMi-2-promoted nucleosome mobilization was ATP dependent (Figure 6A, lanes 4 and 5). We also tested the effect of removal of individual histone tails on nucleosome mobilization by dMi-2. dMi-2 was able to mobilize nucleosomes lacking individual histone N-termini (Figure 6A, lanes 6–17). We were not able to assay the effect of the simultaneous removal of all four tails on nucleosome sliding by dMi-2 because tailless nucleosomes failed to position correctly on the DNA fragment (data not shown). We conclude that not only are histone tails dispensable for stimulation of the dMi-2 ATPase but also that no single tail is required for nucleosome mobilization.
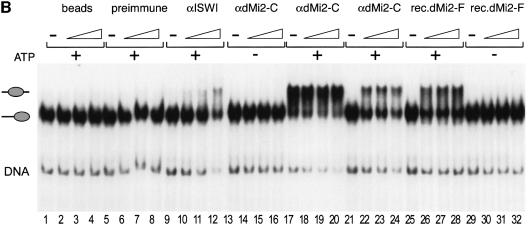
Fig. 6. dMi-2 mobilizes mononucleosomes. (A) Recombinant dMi-2 and ISWI were incubated with positioned mononucleosomes reconstituted with either four wild-type recombinant histones (intact) or three wild-type histones and one lacking the N-terminal tail (Δ) as indicated. Nucleosomes were then separated by native gel electrophoresis. The positions of free DNA, end-positioned mononucleosomes and centre-positioned mononucleosomes are shown on the left. (B) Recombinant dMi-2 and immunoprecipitates (see Materials and methods) were incubated with positioned nucleosomes as described above. The antisera used and presence of ATP in the reactions are indicated on the top (beads: no antibody control). rec.dMi2-F: recombinant dMi-2-F. The positions of free DNA and the two nucleosome species are shown on the left.
We have previously shown that the presence of the CHRAC subunits affects the direction of nucleosome sliding by ISWI (Längst et al., 1999). Like recombinant dMi-2, the CHRAC complex moves the mononucleosome from the end towards the centre of the DNA fragment but fails to mobilize the nucleosome in the opposite direction. In order to investigate whether the presence of dMi-2-associated proteins affects the direction of dMi-2- mediated nucleosome mobilization we tested immunoprecipitated dMi-2 complexes in the mobilization assay (Figure 6B). The αdMi-2-C antiserum precipitated an activity that mobilized the end-positioned nucleosome (Figure 6B, lanes 21–24) but not the centrally positioned nucleosome (lanes 17–20). This mobilization activity was ATP dependent (Figure 6B, lanes 13–16). Immunoprecipitated dMi-2 displayed the same nucleosome mobilization activity as recombinant dMi-2 (Figure 6B, lanes 25–32). Material precipitated with protein A agarose beads alone and preimmune serum had no nucleosome mobilization activity (Figure 6B, lanes 1–8). αISWI antiserum precipitated an activity that mobilized the mononucleosome from the ends towards the centre of the DNA fragment (Figure 6B, lanes 9–12), which could be due to CHRAC or any other ISWI-containing complex (Längst et al., 1999). Taken together, our analysis of ATPase and nucleosome mobilization activity suggests that the properties of recombinant dMi-2 are fully preserved in a native dMi-2 complex.
Discussion
We have for the first time directly compared two unrelated recombinant chromatin remodelling ATPases, dMi-2 and ISWI, in nucleosome binding, nucleosome mobilization and ATPase assays. This comparative analysis has uncovered a number of fundamental differences between these two remodelling enzymes, as described below.
Importance of free DNA for nucleosome binding and ATPase activity of dMi-2 and ISWI
Both recombinant ATPases can be stimulated by nucleosomes assembled from recombinant histones. Whereas the dMi-2 ATPase does not respond to free DNA, the ISWI ATPase is stimulated by free DNA to some extent. Free DNA also appears to play a role in the interaction of ISWI with the nucleosome: ISWI binds to a nucleosome reconstituted on 248 bp of DNA, which displays free DNA, but not to the core particle consisting entirely of 146 bp of nucleosomal DNA under the stringent conditions of our bandshift assay. In striking contrast, dMi-2 interacts with both nucleosomes equally well, demonstrating that it does not require free DNA for interaction. Taken together, these observations suggest that ISWI, but not dMi-2, recognizes its chromatin substrate in part through an interaction with free DNA.
Nucleosome mobilization by dMi-2 and ISWI
The ability to promote the movement of a nucleosome along DNA has been demonstrated for the ISWI-containing NURF and CHRAC complexes, recombinant ISWI and the SWI/SNF complex (Hamiche et al., 1999; Längst et al., 1999; Whitehouse et al., 1999). It is conceivable that all remodelling ATPases mobilize nucleosomes in the same way in our assay and that this simply reflects a common chromatin remodelling activity. Indeed, ISWI and dMi-2 share the intrinsic capacity to promote nucleosome mobilization in an ATP-dependent manner. Surprisingly, however, ISWI and dMi-2 move the nucleosome in opposite directions in our assay. ISWI moves nucleosomes prepositioned at the centre to the ends of a DNA fragment but is not able to mobilize end-positioned nucleosomes (Längst et al., 1999). dMi-2 moves nucleosomes positioned at the end of the DNA fragment to a central position but fails to mobilize the central mononucleosome. Whether this observed directionality of nucleosome movement translates to a regulatory difference in a physiological chromatin context is unclear at present. Nevertheless, the observed difference in the direction of nucleosome movement within the constraints of our assay system suggests that ISWI and dMi-2 interact differently with nucleosomes and that they employ different mechanisms to mobilize them. At present we do not understand the mechanisms of nucleosome mobilization. ISWI only moves the central nucleosome, which is flanked on either side by free DNA. Given that free DNA plays a role in substrate recognition by ISWI we speculate that ISWI needs to interact with two DNA segments extending from the nucleosome in order to mobilize it. This scenario would predict that at least two ISWI molecules simultaneously interact with the central mononucleosome. In agreement with this hypothesis we have observed the formation of multiple ISWI–nucleosome complexes in the bandshift assay, most likely reflecting the binding of multiple ISWI molecules to the 248 bp nucleosome. In contrast, dMi-2 nucleosome binding and ATPase activity is not influenced by free DNA, pointing to a different mechanism of nucleosome mobilization. Interestingly, nucleosome mobilization by dMi-2 in our assay is similar to that of ISWI in the con text of the CHRAC complex (Längst et al., 1999). Identification of the principle that modulates ISWI activity to change the direction of nucleosome mobilization should shed light on the mechanism of nucleosome mobilization by dMi-2 as well.
While this manuscript was under revision, Guschin and coworkers reported that the highly purified Xenopus Mi-2 complex can redistribute a nucleosome positioned near the end of a fragment derived from the Xenopus thyroid hormone receptor βA gene towards more central positions in an ATP-dependent manner (Guschin et al., 2000). It appears that the directionality of Mi2-mediated nucleosome mobilization has been conserved across species.
Importance of histone tails for dMi-2 and ISWI
Nucleosomal stimulation of ISWI ATPase activity and nucleosome mobilization by ISWI are sensitive to the removal of specific histone tails (C.R.Clapier, G.Längst, D.F.V.Corona, P.B.Becker and K.P.Nightingale, manuscript in preparation). In contrast, dMi-2 ATPase activity and nucleosome mobilization by dMi-2 are unaffected, suggesting that these are tail-independent processes. The substrate requirements for ATPase and chromatin remodelling activity of dMi-2 reside within the globular domain of the core nucleosome. The differences in dependence on the histone N-termini reinforces the notion that ISWI and dMi-2 approach the nucleosome in fundamentally different ways.
dMi-2 and other remodelling enzymes
A corresponding comparison between SWI2/SNF2 or Sth1 ATPases and dMi-2 has not been carried out to date. It is not known in which way SWI2/SNF2 and Sth1 ATPases mobilize nucleosomes and whether or not histone tails are required for this activity. It is clear, however, that important differences exist between SWI2/SNF2 or Sth1 and dMi-2 activities. First, SWI2/SNF2 and Sth1 ATPases are both activated to the same extent by nucleosomes and free DNA (Cairns, 1998; Kingston and Narlikar, 1999). ISWI is preferentially stimulated by nucleosomes but also shows some activation by naked DNA (Corona et al., 1999; see also Figure 4B). This contrasts sharply with the exclusive stimulation of recombinant dMi-2 and the dMi-2 complex by nucleosomes. In agreement with our results, ATPase activity of the Xenopus dMi-2 complex is stimulated by chicken erythrocyte mononucleosomes but not by salmon sperm DNA (Wade et al., 1998). In contrast, the ATPase activity of the human NuRD complex is strongly stimulated by naked DNA (Zhang et al., 1998). It is possible that this discrepancy is due to species-specific differences between different Mi-2 complexes or different experimental setups.
Based on our biochemical analysis we propose that dMi-2 defines a new class of nucleosome remodelling ATPases. It will be important to determine the molecular basis for the observed differences between dMi-2 and other ATPases in order to gain a better understanding of how chromatin remodelling machines work.
The dMi-2 complex
By biochemical fractionation of Drosophila embryo extracts we have demonstrated that dMi-2 resides in a large complex (∼1.0 MDa) that also contains the HDAC dRPD3. The estimated size of the complex suggests that it contains further subunits in addition to dMi-2 and dRPD3. Most likely some of the additional dMi-2-associated subunits will correspond to proteins identified in vertebrate Mi-2 complexes. Indeed, several of these homologous sequences can be found in Drosophila EST databases (Wade et al., 1999). The vertebrate Mi-2 complexes have been implicated in mediating transcriptional repression by binding to methylated DNA. In contrast to its vertebrate counterparts, the Drosophila genome is not methylated to any appreciable extent. It will be fascinating to determine whether the dMi-2 complex contains subunits related to the methylated DNA-binding proteins present in the vertebrate complexes and what their role might be in the absence of DNA methylation. A major challenge now is to understand how the different enzymatic subunits of the dMi-2 complex cooperate to regulate chromatin.
Materials and methods
Preparation of antisera
dMi-2 (aa 4–518), dMi-2 (aa 1223–1982) and dRPD3 (aa 337–521) were expressed from pQE His-tag expression vectors in E.coli and purified over a Ni-NTA column. The recombinant protein fragments were further purified by SDS–PAGE and used to raise rabbit polyclonal antisera αdMi-2-N, αdMi-2-C and αdRPD3, respectively.
Fractionation of Drosphila embryo extracts
Crude nuclear extract from Drosophila embryos (Nightingale et al., 1998) was separated by FPLC over a Biorex70 (Amersham-Pharmacia) cation exchange column using stepwise salt elution. dMi-2-containing fractions were pooled, dialysed against HEMG100 [25 mM HEPES pH 7.6, 100 mM KCl, 12.5 mM MgCl2, 0.5 mM EDTA, 0.5 mM EGTA, 1 mM dithiothreitol (DTT), 0.2 mM phenylmethylsulfonyl fluoride, 10% glycerol] and applied to a ResourceQ (Amersham-Pharmacia) anion exchange column. dMi-2 was eluted from this column using a salt gradient. dMi-2-containing fractions from the ResourceQ column were pooled, concentrated with a centricon concentrator and applied to a Superose 6 sizing column.
Immunoprecipitations and western analysis
Immunoprecipitations were carried out in HEMG200 (200 mM KCl) supplemented with 0.1% NP-40. Antisera were added to 75–200 µl of nuclear extract prepared from 0–24 h old Drosophila embryos (Nightingale et al., 1998) and incubated for 3 h at 4°C. Protein A beads (Roche) were added and incubation was continued for 1 h. Protein A–antibody complexes were collected by centrifugation and washed four times in 1 ml of HEMG100/0.1% NP-40. Protein A–antibody complexes were either resuspended in loading buffer for SDS–PAGE or used immediately for ATPase and nucleosome mobilization assays. For western blot analysis immunoprecipitates were separated by SDS–PAGE, transferred to Hybond membrane (Amersham) and probed with the relevant antibodies as described (Brehm et al., 1999).
For nucleosome mobility assays, 250 µl of a dMi-2-containing Biorex 70 fraction were precipitated with 10 µl of preimmune, 3 µl of αdMi-2-C or 10 µl of αISWI antiserum. Protein A agarose–antibody complexes were extensively washed and resuspended in 10 µl of HEMG100/0.1% NP-40. Volumes of 0.2, 0.5 and 1.0 µl were added to the nucleosome mobilization assay.
Expression of recombinant dMi-2 and histones
Recombinant ISWI was prepared as described previously (Corona et al., 1999). The dMi-2 coding sequence was flag-tagged at the C-terminus using PCR and appropriate primers, and cloned into the pVL1393 expression vector. Sf9 cells were transfected and recombinant baculovirus was produced using the Invitrogen MaxBac system according to the manufacturer’s instructions. For expression, exponentially growing Sf9 cells were infected with recombinant virus and harvested 48 h post-infection. Infected cells were collected by centrifugation, resuspended in HEMG500 containing 0.1% NP-40. Cells were lysed by two cycles of freeze–thaw in liquid nitrogen followed by sonification (2× 15 s bursts) in a Branson sonifier. Lysates were cleared by centrifugation and filtration (0.45 µm). M2 affinity gel (Sigma) was added to the lysate. After incubation for 12 h at 4°C the affinity gel was recovered by centrifugation and washed twice with 1 ml of HEMG500/0.1% NP-40, twice with 1 ml of HEMG1000/0.1% NP-40 and twice with 1 ml of HEMG100/0.1% NP-40. Bound protein was eluted with 0.25 mg/ml flag peptide (Sigma) in HEMG100/0.1% NP-40, aliquotted and stored at –70°C. Recombinant wild-type and tailless histones were expressed in E.coli, purified and reconstituted into nucleosomes as described (Luger et al., 1997b).
ATPase and HDAC assays
ATPase assays were performed essentially as described elsewhere (Corona et al., 1999). HDAC assays were performed as described elsewhere (Brehm et al., 1998).
yNAP1 assembly and binding assay
yNAP1 assisted nucleosome assembly on linear DNA immobilized on paramagnetic beads, and MNase analysis was done as described in Akthar and Becker (2000). Binding reactions were performed with 40–200 fmol dMi-2-F and 50–200 ng of freshly assembled chromatin in 30 µl of HEMG100 containing 0.1% NP-40 and 0.1 mg/ml bovine serum albumin (BSA) as described (Akthar and Becker, 2000). Beads were washed with HEMG containing increasing concentrations of KCl and NP-40. Residual binding of dMi-2 to DNA was observed with less stringent wash conditions (HEMG100/0.1% NP-40). Binding to chromatin was found to be resistant to washes in HEMG1000/1% NP-40.
Isolation of nucleosomal arrays from embryo extracts
Nucleosomal arrays were prepared from isolated Drosophila embryo essentially as described in Kornberg et al. (1989).
Nucleosome reconstitution
Salt gradient dialysis. Nucleosomes were assembled from DNA and either purified or recombinant histones by salt gradient dialysis essentially as described previously (Längst et al., 1999). Mononucleosomes were reconstituted on a 248 bp DNA fragment representing sequences between –232 and +16 relative to the mouse rDNA transcription start site (+1), or on a 146 bp DNA fragment derived from pUC9. DNA fragments were synthesized by PCR and end-labelled with [γ-32P]ATP by polynucleotide kinase. Salt dialysis was performed in Sartorius collodion bags that were rinsed with water and blocked with HI salt buffer (10 mM Tris–HCl pH 7.6, 2 M NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 0.05% NP-40), containing 200 ng/ml BSA. A typical assembly reaction (500 µl) contained 60 µg of DNA, 40 µg of BSA and 60 µg of histones in HI salt buffer, adjusted to a final concentration of 2.5 M NaCl. The salt was continuously reduced to 50 mM NaCl during 16–24 h. The efficiency of chromatin assembly was monitored by measuring the restrained superhelicity after complete relaxation with topoisomerase I.
Nucleosome assembly with polyglutamic acid. Mononucleosomal particles were assembled using purified histones and polyglutamic acid (PGA, Sigma P4886) according to Stein et al. (1979). Histones and PGA were mixed in a 2:1 mass ratio in 0.15 M KCl and incubated for 1 h at room temperature. Precipitates were removed by centrifugation and the supernatant (HP-Mix) was stored in aliquots at –20°C. Different ratios of DNA to HP-Mix were incubated for 3 h at room temperature and analysed by electromobility shift assay to reveal optimal conditions for nucleosome assembly.
Nucleosome shift and mobility assay
Nucleosome shift and mobility assays were essentially carried out as described previously (Längst et al., 1999). Nucleosomes were assembled with PGA on the 248 bp rDNA fragment. Nucleosomes were separated by electrophoresis on 5% polyacrylamide gels in 0.5× TBE. Gel slices containing the separated nucleosomes were crushed and the nucleosomes were eluted for 1 h at 4°C in EX100 buffer containing 200 ng/µl BSA.
Isolated mononucleosomes (60 fmol) of defined positions on the 248 bp rDNA fragment were incubated with ISWI (2–5 fmol), dMi-2 (25–50 fmol) or affinity purified dMi-2 complex as indicated in the figure legends for 90 min at room temperature in 8 µl of EX100 buffer containing 1 mM ATP, 1 mM DTT and 400 ng/µl BSA. The reaction was stopped by the addition of 100 ng of salt-dialysed nucleosomes and further incubation for 5 min. Nucleosomes were then separated by native gel electrophoresis in 5% polyacrylamide gels in 0.5× TBE.
Acknowledgments
Acknowledgements
We are indebted to Asifa Akthar for providing advice and reagents. We would like to thank Christa Schwarzlose and Irene Vetter for expert assistance. C.R.C. is supported by the EMBL International PhD Programme.
References
- Akthar A. and Becker,P.B. (2000) Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell, 5, 1–9. [DOI] [PubMed] [Google Scholar]
- Brehm A., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. [DOI] [PubMed] [Google Scholar]
- Brehm A., Nielsen,S.J., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1999) The E7 oncoprotein associates with Mi-2 and histone deacetylase activity to promote cell growth. EMBO J., 18, 2449–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B.R. (1998) Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem. Sci., 23, 20–25. [DOI] [PubMed] [Google Scholar]
- Corona D.F., Langst,G., Clapier,C.R., Bonte,E.J., Ferrari,S., Tamkun,J.W. and Becker,P.B. (1999) ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell, 3, 239–245. [DOI] [PubMed] [Google Scholar]
- Eisen J.A., Sweder,K.S. and Hanawalt,P.C. (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res., 23, 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel P.T., Tsukiyama,T. and Wu,C. (1997) Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J., 16, 4717–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin D., Wade,P.A., Kikyo,N. and Wolffe,A.P. (2000) ATP-dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry, 39, 5238–5245. [DOI] [PubMed] [Google Scholar]
- Guyon J.R., Narlikar,G.J., Sif,S. and Kingston,R.E. (1999) Stable remodeling of tailless nucleosomes by the human SWI–SNF complex. Mol. Cell. Biol., 19, 2088–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiche A., Sandaltzopoulos,R., Gdula,D.A. and Wu,C. (1999) ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell, 97, 833–842. [DOI] [PubMed] [Google Scholar]
- Kehle J., Beuchle,D., Treuheit,S., Christen,B., Kennison,J.A., Bienz,M. and Muller,J. (1998) dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science, 282, 1897–1900. [DOI] [PubMed] [Google Scholar]
- Kelley D.E., Stokes,D.G. and Perry,R.P. (1999) CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma, 108, 10–25. [DOI] [PubMed] [Google Scholar]
- Kim J. et al. (1999) Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity, 10, 345–355. [DOI] [PubMed] [Google Scholar]
- Kingston R.E. and Narlikar,G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- Kornberg R.D., LaPointe,J.W. and Lorch,Y. (1989) Preparation of nucleosomes and chromosomes. Methods Enzymol., 170, 3–14. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (1999) Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev., 9, 40–48. [DOI] [PubMed] [Google Scholar]
- Längst G., Bonte,E.J., Corona,D.F. and Becker,P.B. (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell, 97, 843–852. [DOI] [PubMed] [Google Scholar]
- Logie C. and Peterson,C.L. (1997) Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J., 16, 6772–6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logie C., Tse,C., Hansen,J.C. and Peterson,C.L. (1999) The core histone N-terminal domains are required for multiple rounds of catalytic chromatin remodeling by the SWI/SNF and RSC complexes. Biochemistry, 38, 2514–2522. [DOI] [PubMed] [Google Scholar]
- Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997a) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Luger K., Rechsteiner,T.J., Flaus,A.J., Waye,M.M. and Richmond,T.J. (1997b) Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol., 272, 301–311. [DOI] [PubMed] [Google Scholar]
- Nightingale K.P., Wellinger,R.E., Sogo,J.M. and Becker,P.B. (1998) Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. EMBO J., 17, 2865–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan M.L., Sif,S., Narlikar,G.J. and Kingston,R.E. (1999) Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell, 3, 247–253. [DOI] [PubMed] [Google Scholar]
- Seelig H.P., Moosbrugger,I., Ehrfeld,H., Fink,T., Renz,M. and Genth,E. (1995) The major dermatomyositis-specific Mi-2 autoantigen is a presumed helicase involved in transcriptional activation. Arthritis Rheum., 38, 1389–1399. [DOI] [PubMed] [Google Scholar]
- Seelig H.P., Renz,M., Targoff,I.N., Ge,Q. and Frank,M.B. (1996) Two forms of the major antigenic protein of the dermatomyositis-specific Mi-2 autoantigen. Arthritis Rheum., 39, 1769–1771. [DOI] [PubMed] [Google Scholar]
- Stein A., Whitlock,J.P. and Bina,M. (1979) Acidic polypeptides can assemble both histones and chromatin in vitro at physiological ionic strength. Proc. Natl Acad. Sci. USA, 76, 5000–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes D.G. and Perry,R.P. (1995) DNA-binding and chromatin localization properties of CHD1. Mol. Cell. Biol., 15, 2745–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes D.G., Tartof,K.D. and Perry,R.P. (1996) CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc. Natl Acad. Sci. USA, 93, 7137–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh Y., Pencil,S.D. and Nicolson,G.L. (1994) A novel candidate metastasis-associated gene, mta1, differentially expressed in highly metastatic mammary adenocarcinoma cell lines. cDNA cloning, expression and protein analyses. J. Biol. Chem., 269, 22958–22963. [PubMed] [Google Scholar]
- Toh Y., Oki,E., Oda,S., Tokunaga,E., Ohno,S., Maehara,Y., Nicolson,G.L. and Sugimachi,K. (1997) Overexpression of the MTA1 gene in gastrointestinal carcinomas: correlation with invasion and metastasis. Int. J. Cancer, 74, 459–463. [DOI] [PubMed] [Google Scholar]
- Toh Y., Kuwano,H., Mori,M., Nicolson,G.L. and Sugimachi,K. (1999) Overexpression of metastasis-associated MTA1 mRNA in invasive oesophageal carcinomas. Br. J. Cancer, 79, 1723–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J.K., Hassig,C.A., Schnitzler,G.R., Kingston,R.E. and Schreiber,S.L. (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature, 395, 917–921. [DOI] [PubMed] [Google Scholar]
- Tse C., Sera,T., Wolffe,A.P. and Hansen,J.C. (1998) Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol., 18, 4629–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler J.K. and Kadonaga,J.T. (1999) The ‘dark side’ of chromatin remodeling: repressive effects on transcription. Cell, 99, 443–446. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Jones,P.L., Vermaak,D. and Wolffe,A.P. (1998) A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol., 8, 843–846. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Gegonne,A., Jones,P.L., Ballestar,E., Aubry,F. and Wolffe,A.P. (1999) Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nature Genet., 23, 62–66. [DOI] [PubMed] [Google Scholar]
- Whitehouse I., Flaus,A., Cairns,B.R., White,M.F., Workman,J.L. and Owen-Hughes,T. (1999) Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature, 400, 784–787. [DOI] [PubMed] [Google Scholar]
- Woodage T., Basrai,M.A., Baxevanis,A.D., Hieter,P. and Collins,F.S. (1997) Characterization of the CHD family of proteins. Proc. Natl Acad. Sci. USA, 94, 11472–11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Wong,J., Moreno,G.T., Young,M.K., Cote,J. and Wang,W. (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell, 2, 851–861. [DOI] [PubMed] [Google Scholar]
- Zhang Y., LeRoy,G., Seelig,H.P., Lane,W.S. and Reinberg,D. (1998) The dermatomyositis-specific autoantigen Mi-2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell, 95, 279–289. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ng,H.H., Erdjument-Bromage,H., Tempst,P., Bird,A. and Reinberg,D. (1999) Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev., 13, 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]