Detection of Protein S-Nitrosylation with the Biotin Switch Technique (original) (raw)
. Author manuscript; available in PMC: 2011 Jun 22.
Abstract
Protein _S_-nitrosylation, the post-translational modification of cysteine thiols to form _S_-nitrosothiols, is a principle mechanism of nitric oxide-based signaling. Studies have demonstrated myriad roles for _S_-nitrosylation in organisms from bacteria to humans, and recent efforts have greatly advanced our scientific understanding of how this redox-based modification is dynamically regulated during physiological and pathophysiological conditions. The focus of the current review is the biotin switch technique (BST), which has become a mainstay assay for detecting _S_-nitrosylated proteins in complex biological systems. Potential pitfalls and modern adaptations of the BST are discussed, as are future directions for this assay in the burgeoning field of protein _S_-nitrosylation.
Keywords: Biotin switch technique, _S_-nitrosylation, _S_-nitrosothiol, redox, cysteine, thiol, nitric oxide, nitrosative stress
BASICS OF S-NITROSYLATION
Protein _S_-nitrosylation—the covalent adduction of a nitroso group to a cysteine thiol side chain—has recently emerged as a principle mechanism by which nitric oxide (NO) mediates a wide range of cellular functions and phenotypes [1, 2]. _S_-nitrosylation regulates diverse pathways such as G-protein-coupled receptor signaling [3–5], death receptor-mediated apoptosis [6–11], glutamate-dependent neurotransmission [12–15], vesicular trafficking [16–19], stimulation of prostaglandin synthesis [20–22], and the unfolded protein response [23]. In addition, aberrant _S_-nitrosylation is implicated in disease states such as tumor initiation and growth [24–28], neurodegeneration [23, 29–32] and malignant hyperthermia [33]. Consequently, much effort is focused on understanding the role of _S_-nitrosylation in normal physiology and its contribution to pathophysiology. For example, several recent studies have shown that dysregulated _S_-nitrosylation of the ryanodine receptor (Ca2+-release channel) may contribute to cardiac arrhythmias [34], heat stroke [33] and impaired exercise capacity [35]. As scientific interest in protein _S_-nitrosylation continues to intensify, an increasing number of studies are relying on the biotin switch technique (BST) for the detection of endogenously _S_-nitrosylated proteins (protein-SNOs). The introduction of this assay by Jaffrey et al. in 2001 [36] has served as an impetus for studies probing _S_-nitrosylation in vivo, largely due to its superb compatibility with ubiquitous molecular methods (e.g. SDS-PAGE, immunodetection, mass spectrometry).
NO- VS. SULFUR-BASED ASSAYS OF S-NITROSYLATION
The sulfur-nitrogen bond of an SNO is particularly labile and can undergo both homolytic and heterolytic cleavage reactions [37, 38]. The lability of the S-NO bond has served as the cornerstone for numerous SNO detection strategies, though the chemistries employed following SNO cleavage differ greatly between assays (Fig. 1). Most techniques detect the NO or nitrite (NO2−) liberated upon S-NO cleavage, and hence can be considered “NO-based” strategies. In these assays, divalent mercury (e.g. HgCl2) is often employed to heterolytically cleave the S-NO bond, producing a mercury-thiol complex and nitrosonium ion (NO+); the latter is a potent nitrosant and undergoes rapid hydration to NO2− at neutral pH. Techniques (spectrophotometric or fluorescent) that detect the NO2− product include the Saville [39–41], diaminonapthalene [39, 42] and diaminofluorescein assays [42–45].
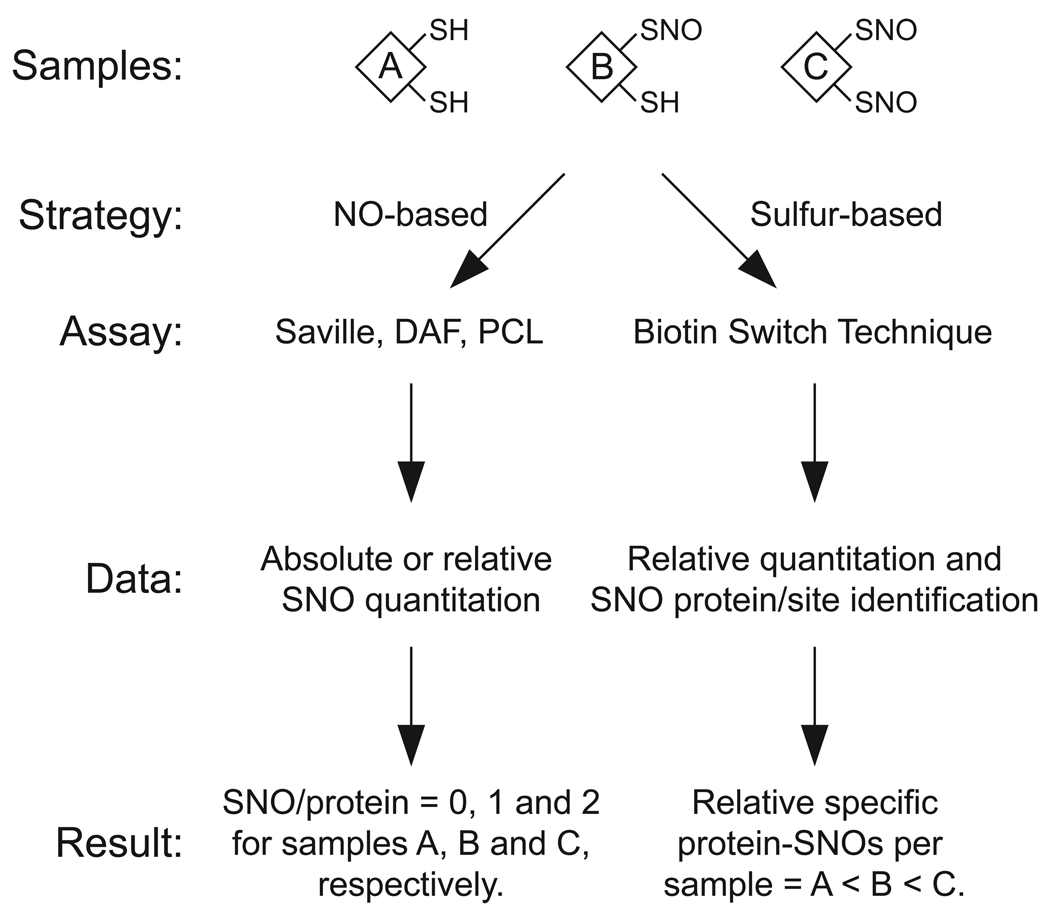
Fig. 1.
A general comparison of NO- and sulfur-based strategies for detecting protein _S_-nitrosylation. As an example, three lysates containing various amounts of protein _S_-nitrosylation are subjected to both NO- and sulfur-based assays. NO-based strategies include the Saville and diaminofluorescein (DAF) assays, which employ a chemical probe, and Hg-coupled photolysis-chemiluminescence (PCL), which detects NO gas liberated by SNO homolysis and can differentiate SNO from metal-NO. Importantly, this assay is highly sensitive (low nanomolar SNO concentrations can be detected) and has been well-validated with genetic models of disrupted NO/SNO metabolism [108, 109]. It therefore serves as a standard method for probing _S_-nitrosylation in vivo. With a complex biological sample (e.g. a lysate), these NO-based strategies can readily determine the absolute amount of SNO per sample, but cannot readily detect an individual protein-SNO. A sulfur-based strategy, such as the biotin switch technique (BST), employs covalent “tagging” at the sulfur atom of each SNO, thus facilitating relative quantitation and protein-SNO identification.
Another common NO-based technique employs homolytic or reductive conditions to cleave the S-NO bond, followed by chemiluminescent detection of the liberated NO via reaction with ozone. Such methods include Hg-coupled photolysis-chemiluminescence [46, 47] and the copper-cysteine-carbon monoxide (3C) assay [48–50]. Though each of these NO-based methods is well suited for SNO quantitation (relative to SNO standards), they have limited use in functional studies of _S_-nitrosylated proteins within complex mixtures because the proteins of interest must be purified (e.g. by immunoprecipitation) prior to SNO measurement. While this method has been applied successfully in a number of cases—including _S_-nitrosylated hemoglobin [51–53], caspase-3 [11, 54], thioredoxin-1 [55], c-Jun N-terminal kinase [56], G-protein-coupled receptor kinase 2 [5], ryanodine receptor [57, 58] and prokaryotic OxyR [59]—the arduous nature of the approach has limited its application.
In contrast to NO-based assays, the BST is unique in that it targets the sulfur atom of an SNO without regard for the fate of any liberated NO species; it can thus be considered a “sulfur-based” strategy. As the BST employs covalent “tagging” of protein-SNOs, it can detect individual protein-SNOs in a complex mixture (since the tag is added to the protein of interest). For studies of a specific protein or class of proteins, the BST has therefore proved to be of great utility (> 200 publications to date).
As described above, the major differences between NO- and sulfur-based strategies for SNO detection are summarized in Fig. 1. Importantly, the type of information gained from either approach is different. NO-based assays are capable of absolute quantitation (e.g. 10 nmol SNO per 1 mg protein), though they do not generally discriminate the source of each protein-SNO. In contrast, the BST tends to be more qualitative (relatively quantitative); its great advantage is its ability to detect individual protein-SNOs in a complex mixture, as well as to identify novel protein-SNOs.
WORKFLOW OF THE BIOTIN SWITCH TECHNIQUE
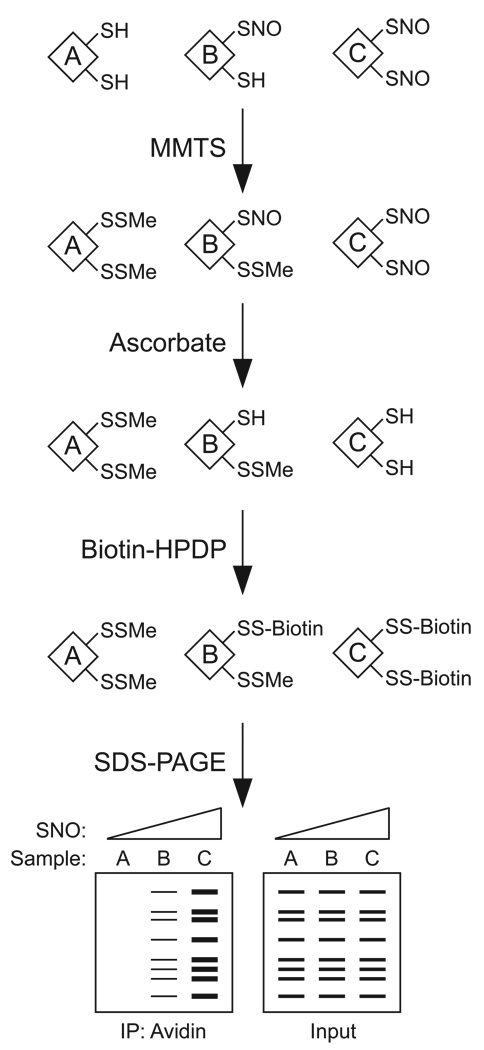
As illustrated by Fig. 2, the BST consists of three principal steps: 1) “blocking” of free cysteine thiols by _S_-methylthiolation with MMTS (a reactive thiosulfonate); 2) conversion of SNOs to thiols via transnitrosation with ascorbate; and 3) in situ “labeling” by _S_-biotinylation of the nascent thiols with biotin-HPDP, a reactive mixed disulfide of biotin. The degree of biotinylation (and thus _S_-nitrosylation) is determined by either anti-biotin immunoblotting or streptavidin pull-down followed by immunoblotting for the protein(s) of interest.
Fig. 2.
Overview of the biotin switch technique. In the example shown, three lysates with various degrees of _S_-nitrosylation are subjected to the assay. The “blocking” step involves _S_-methylthiolation of each cysteine thiol with _S_-methylmethanethiosulfonate (MMTS). Next, ascorbate is employed to convert each SNO to a free thiol via a transnitrosation reaction to generate _O_-nitrosoascorbate. In the “labeling” step, each nascent free thiol (previously an SNO site) is biotinylated with biotin-HPDP. Biotinylated proteins are enriched by avidin affinity media, and analyzed by SDS-PAGE/immunoblotting. Total protein-SNOs or an individual protein-SNO can be detected with avidin-HRP or with an antibody against a protein of interest, respectively. As illustrated, the degree of pulldown correlates with protein _S_-nitrosylation. Prior to avidin pulldown, a small fraction of each sample is analyzed to determine protein “input.”
The blocking step is typically initiated by the addition of SDS and MMTS, followed by heating at 50 °C and separation of proteins from excess MMTS by acetone precipitation. The combination of heat and SDS functions to fully denature proteins, thus granting MMTS optimal access to natively buried protein thiols. Effective blocking of free thiols is required to minimize “background” biotinylation (i.e. biotinylation that results from incomplete blocking) and to maximize assay sensitivity.
The second step in the BST converts the SNO to a free thiol so that it can be biotinylated. This is achieved by allowing ascorbate—a di-enol antioxidant with unique reactivity towards SNOs—to undergo a transnitrosation reaction with the protein-SNO. As initially reported by Holmes and Williams [60], this reaction proceeds via a concerted nitroso transfer mechanism whereby the SNO and ascorbate are converted to thiol and _O_-nitrosoascorbate. Notably, ascorbate is several orders of magnitude more reactive than other _N_- or _O_-based nucleophiles (e.g. amines, phenols) [61, 62]. The reaction between SNOs and ascorbate also exhibits a critical pH-dependence and is favored by transition metal chelation: alkaline conditions promote the nitroso transfer reaction, likely by increasing the fraction of mono-deprotonated (i.e. nucleophilic) ascorbate, whereas transition metals may enable alternative chemistry (see below). The specificity of the BST for protein-SNOs is predicated on the fact that ascorbate will convert SNOs to free thiols without reducing other cysteine-based oxidations such as _S_-glutathionylation or _S_-oxides (sulfenic, sulfinic and sulfonic acids). This specificity is supported by thermodynamic measurements (discussed below) and by several experimental validations [36, 63].
The third step of the BST involves biotinylation of the nascent thiol (i.e. previous SNO site). This is performed concomitantly with the ascorbate reaction, such that the newly liberated free thiols are immediately biotinylated. Excess biotin-HPDP is removed by acetone precipitation, and protein biotinylation (i.e. previous _S_-nitrosylation) is assessed by one of multiple routes. Avidin-agarose is frequently employed to enrich the biotinylated proteins, followed by elution and SDS-PAGE. It is important to note that the biotin tag is attached via a disulfide linkage, and therefore reductants (e.g. DTT) will remove the tag. This reductive elution strategy is frequently employed to elute biotinylated proteins from avidin-agarose prior to SDS-PAGE, especially when the biotin-tag is not employed for immunodetection (e.g. when blotting an individual protein). As demonstrated in Fig. 3A, the BST readily detects _S_-nitrosylated GAPDH in cytokine-stimulated macrophages, in which inducible nitric oxide synthase (iNOS) is upregulated [64–66]. Alternatively, total protein-SNOs can also be assessed by non-reducing SDS-PAGE and immuno-detection of the biotin tag. As shown in Fig. 3B, this approach easily detects total protein-SNOs in HEK293 cells following treatment with _S_-nitrosocysteine (CysNO).
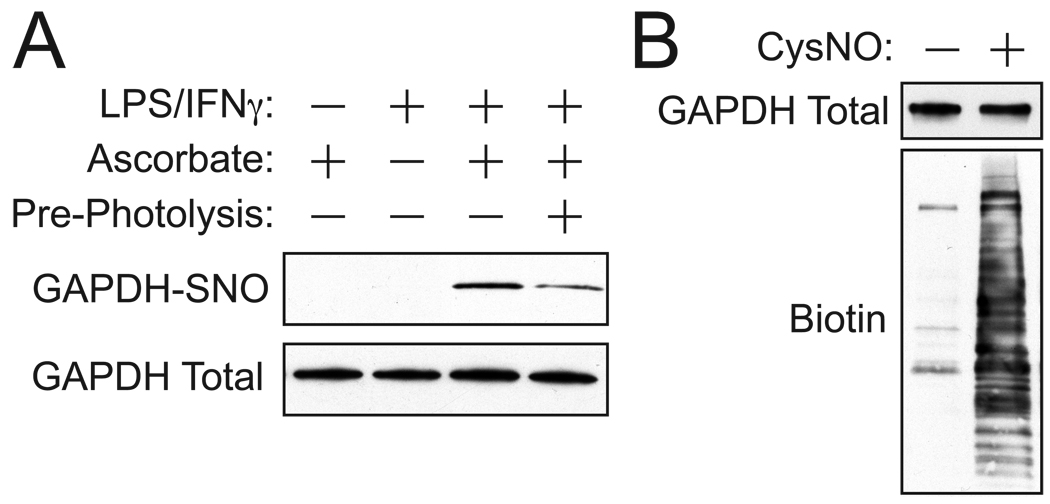
Fig. 3.
A typical BST detects both endogenous and exogenous _S_-nitrosylation in cultured mammalian cells. A) Murine RAW264.7 macrophages were either untreated or cytokine-stimulated with lipopolysaccharide (500 ng/ml) and IFN-γ (100 U/ml) for 16 h, which drives NO production. Cellular extracts were subjected to the BST and probed for _S_-nitrosylated GAPDH (GAPDH-SNO), along with ascorbate and pre-photolysis controls. Notably, omission of ascorbate leads to nearly complete loss of biotinylation and pre-photolysis with a Hg vapor lamp greatly attenuates the same signal. B) Whole cellular protein-SNOs are detected by treating HEK293 cells with 200 µM _S_-nitrosocysteine (CysNO) for 10 min. Cellular extracts were subjected to the BST and 5% of each biotinylation reaction (~ 40 µg) was analyzed by immunoblotting with avidin-HRP and anti-GAPDH antibody (for “input”).
It is important to note that the BST can be used to assay _S_-nitrosylation by constitutive NOS isoforms (nNOS and eNOS; Fig. 4A) in addition to iNOS, and to examine _S_-nitrosylation dynamics in the context of signal transduction, as demonstrated for G-protein-coupled receptor kinase 2 (GRK2) following stimulation of multiple G-protein-coupled receptors (Fig. 4B). Thus, when performed appropriately it is highly sensitive as well as specific. Further, the BST can be used to identify the NOS isoform(s) involved (Fig. 4A and C), and to reveal the participation of denitrosylase activities or endogenous _S_-nitrosylating agents (Fig. 4D) across a wide range of cellular stimuli. Collectively, these examples illustrate the utility of the BST for assessing _S_-nitrosylation in vivo, particularly when used alongside the methodological, pharmacological and genetic tools demonstrated in Figs. 3 and 4.
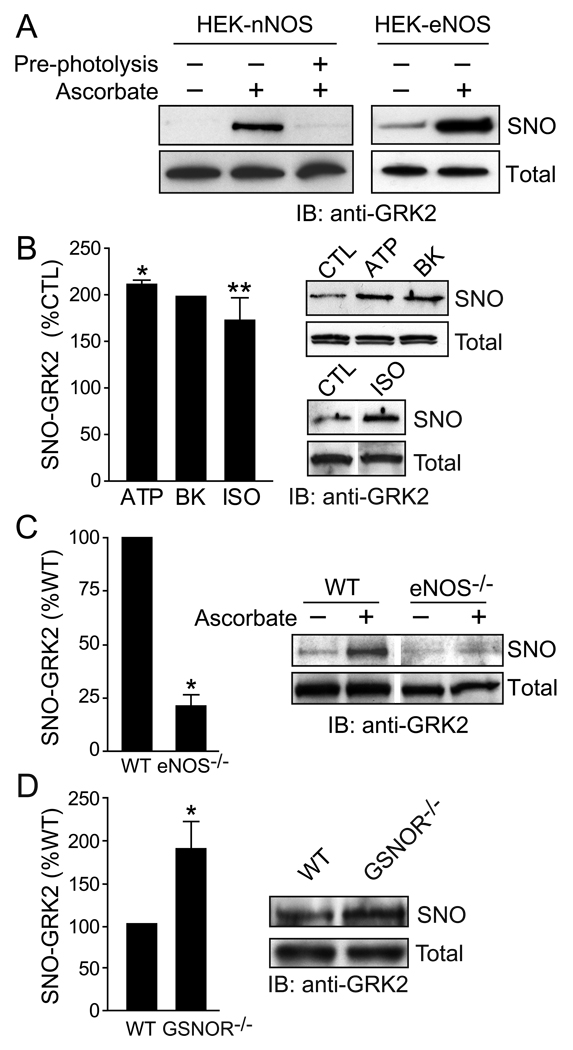
Fig. 4.
The BST can be combined with pharmacological and genetic tools to study _S_-nitrosylation in primary cells and tissues in vivo. A) GRK2 is _S_-nitrosylated in HEK293 cells that stably overexpress the constitutive NOS isoforms (eNOS and nNOS). B) G protein coupled receptor (GPCR)-specific agonists (ATP, adenosine triphosphate; BK, bradykinin; ISO, isoproteronol; CTL, control), each of which leads to eNOS activation, increase GRK2 _S_-nitrosylation in human umbilical vein endothelial cells. C) GRK2 _S_-nitrosylation is diminished in lungs from eNOS−/− mice relative to wild type (WT) mice. D) GRK2 _S_-nitrosylation is increased in lungs from mice lacking a major SNO-metabolizing enzyme, GSNO reductase (GSNOR−/−). Figure adapted from reference [5].
PRE-PHOTOLYSIS AS AN INDEPENDENT STRATEGY IN THE BST
There is little doubt that the signals arising from a BST following treatment with CysNO (Fig. 3B) represent bona fide _S_-nitrosylated proteins. Like all other biological assays, however, internal and external controls should be employed to verify data obtained with the BST, particularly when assaying endogenously _S_-nitrosylated proteins. These approaches frequently include: 1) omission of ascorbate during the labeling step of the BST; 2) overexpression or knockdown of a particular NOS isoform; and/or 3) pharmacological activation or inhibition of NOS. Though each of these approaches has been consistently applied to assay SNOs by the BST, we recently introduced an independent technique that involves photolysis of the SNO prior to performing the BST (dubbed “pre-photolysis”) [5, 63]. This methodology—a UV-based homolysis of the S-NO bond—is a simple and specific tool for assaying SNOs with the BST, particularly under conditions where NOS activation or inhibition are either unfeasible (e.g. human patient samples) or ineffective (e.g. NOS activity-independent, a characteristic of signaling activated by denitrosylation). This approach is based on Hg-coupled photolysis-chemiluminescence, in which the Hg-displaceable pool of NO groups (i.e. SNO) is photolyzed and detected via reaction with ozone. Photolysis eliminates SNO. Consequently, pre-photolysis serves as a complementary approach to BST. As shown in Fig. 3A, the signal from endogenously _S_-nitrosylated GAPDH is attenuated by a brief exposure to high intensity UV light. A schematic illustration in Fig. 4 shows how pre-photolysis and ascorbate can be combined to rigorously assess a complex sample for protein _S_-nitrosylation.
DESIGN OF THE PRE-PHOTOLYSIS APPARATUS
Pre-photolysis is performed by exposing the sample(s) to a strong UV radiation source within a light-sealed reaction chamber. Overall such an apparatus should: 1) deliver high intensity radiation at 335 nm (the ideal wavelength for S-NO photolysis [47, 67, 68]); 2) prevent heating of the sample to avoid thermolytic reactions; 3) allow multiple samples to be photolyzed simultaneously; and 4) protect the operator from any harmful UV exposure. We employ a 200 watt, 1.9 amp Hg vapor lamp (Ace Glass, catalog # 7825-32), which delivers approximately 50% of its energy in the ultraviolet region. This lamp, along with an appropriate power supply, can be fitted into a sealed photochemical reaction cabinet (e.g. Ace Glass, catalog # 7836-20). Samples are generally photolyzed in borosilicate glass vessels (Pierce catalog # 13504). Typically a two-minute exposure approximately 3 cm from the lamp is sufficient to photolyze at least 50% of a protein-SNO.
Several more economical sources of UV radiation may also suffice for pre-photolysis, though they are likely less efficacious than a Hg vapor lamp. These include a UV transilluminator (often used to visualize DNA with ethidium bromide staining) and a UV crosslinker that provides radiation with wavelengths greater than 300 nm. In the latter case, a suitable apparatus would be the Ultraviolet Products (UVP) CL-1000 crosslinker (UVP, catalog # 81-0112-01) fitted with five long-range UV lamps (UVP, catalog # 34-0006-01).
METHODOLOGICAL ISSUES AND CONCERNS RAISED ABOUT THE BST
Despite its widespread use, the BST is technically challenging and labor intensive. Since each step contains potential sources error, a rigorous experimentalist will include proper negative and positive controls to add confidence to their results. In the case of _S_-nitrosylation, numerous tools can be employed to check assay efficiency and to manipulate the stability or formation of protein-SNOs, thus adding great confidence to results obtained with the BST.
When comparing protein-SNOs across multiple conditions, it is imperative that each sample contains equivalent protein abundance, or “inputs”. While some degree of normalization can be allowed, linearity (with respect to protein input and biotinylation) of the BST has not been demonstrated. Since proteins are subjected to multiple acetone precipitation steps and pellet washes, SDS-PAGE analysis should be performed on the biotinylated material prior to avidin pulldown. The final step of the BST is, for all intents and purposes, an affinity pulldown, which are almost always reported in the literature as both an “input” and a “pulldown.” The BST should be held to the same standards.
The blocking step of the BST can also present a technical challenge, as some protein thiols can be resistant to complete blocking, resulting in high levels of SNO-independent biotinylation. In the absence of proper controls, incomplete blocking can be problematic since it may lead to misinterpretation of the data (i.e. “false-positive” signal). However, if performed correctly (e.g. use of ascorbate), ineffective blocking will lower assay sensitivity (increase signal to noise) rather than specificity. Conversly, reverse strategies such as omitting ascorbate or employing pre-photolysis can be used to determine the degree of blocking. Both of these treatments will attenuate a true SNO signal. Further, the blocking reaction can be lengthened to improve efficiency, though SNO stability may be compromised at 50 °C due to thermolysis.
The specificity of the BST for SNOs (versus other cysteine-based modifications) had been questioned in several works. Here we review the actual data in these publications, and explain why observations are more likely artifact than source of real concern. Landino et al. suggested that ascorbate reduces tubulin disulfides, a claim the authors use to challenge the validity of the BST [69]. However, as discussed below, reduction of alkyl (biological) disulfides by ascorbate is highly unfavorable thermodynamically, and an explanation for how “disulfides” were purportedly reduced was not provided. These authors in fact employed high concentrations of peroxynitrite to “oxidize” tubulin, though peroxynitrite also nitrosates thiols (albeit at low yields) thereby generating SNOs [70–72]; it is likely that _S_-nitrosylated tubulin was generated under these conditions. A later report by Huang et al. argued that the BST yields “artifactual” ascorbate-dependent biotinylation of native reduced BSA [73] (i.e. SNO-independent). The authors did not provide a mechanistic basis for their observation, and subsequent work has reproduced the findings of Huang, which was caused by a window light artifact (BST should be performed in the dark) [63].
Most recently Guisitarini et al. have challenged the BST mainly on the grounds that ascorbate reduces DTNB [74]. However, this well known reaction has no bearing on reduction of biological (alkyl) disulfides: DTNB is a highly electrophilic, aryl disulfide that readily undergoes both hydrolysis and homolysis, the latter facilitated by ascorbate [75, 76]. DTNB is therefore not representative of biological disulfides or in vivo Cys-based oxidation products. More perplexing, however, is these authors’ suggestion that ascorbate reduces low molecular weight biological disulfides (e.g. glutathione, homocysteine and cysteine disulfide) and protein mixed disulfides, but not intramolecular protein disulfides (under denaturing conditions). In fact, none of these reactions are favored thermodynmically, and there is no kinetic or thermodynamic basis for the difference between mixed and intramolecular protein disufides under denaturing conditions. On closer inspection, yield of reaction with cystine and glutathione disulfide was extremely low and appeared to saturate at a few percent over several hours, consistent with the presence of a contaminant (e.g. contaminant metals) or other artifact. Further, the authors measured disulfide reduction in the continuous presence of fluorescent thiol alkylators (bromobimanes), which irreversibly pull reactions by Le Chatelier’s principle, and treated other samples with DTT to prevent “possible re-oxidation of thiols after their reduction by ascorbate” [74]. These approaches, which would artifactually generate thiols, cast doubt on the conclusions. These authors also do not provide a mechanistic explanation for their results or reference prior work to the contrary [77–81]. But most important of all, they do not perform the BST, whereas a comparative analysis of the BST across cell lysates derivatized by _S_-glutathionylation, _S_-oxidation and _S_-nitrosylation revealed that only the _S_-nitroylated proteins were detected [63].
Shared between all these claims of “artifactual signals” is the notion that ascorbate can reduce alkyl disulfides. However, this concept is both unsupported and difficult to reconcile experimentally with the use of MMTS as a blocking agent, which converts protein thiols to mixed methyl disulfides. Clearly the BST would never work if ascorbate removed these “blocked” methyl disulfides. Further, the two electron standard reduction potential of cysteine disulfides is −170 to −320 mV [77], indicating that this endergonic reaction will not likely couple to the two electron oxidation of ascorbate, which is also unfavorable (the standard reduction potential of dehydroascorbate to ascorbate is +70 mV [82]). Thus, one would not expect ascorbate to directly reduce any biological protein cysteine oxidation products (except highly transient and unstable thiyl or sulfinyl radicals). In contrast, these electrochemical measurements favor the reverse reaction—thiol-dependent reduction of dehydroascorbate to ascorbate—a scenario supported by extensive in vitro and in vivo experimentation [78, 80, 81]. The same thermodynamic arguments hold for higher _S_-oxides as well [77].
Although the BST is highly specific for protein SNOs [63], we have noted that the presence of indirect sunlight (from an adjacent window) during the labeling step closely recapitulates the artifactual results of Huang et al. [73]. This SNO-independent biotinylation reaction is fully reversed by DTT, confirming that indirect sunlight indeed promotes an artifactual ascorbate-dependent protein-biotin disulfide during the labeling step. Under these conditions, biotin-HPDP (a synthetic aryl disulfide) undergoes quantitative reduction to biotin-thiol, though ascorbate exhibited no measureable reactivity in the absence of sunlight [63]. This production of high micromolar biotin-thiol during the BST would lead to artifactual protein biotinylation via thiol/disulfide exchange with “blocked” (i.e. _S_-methylthiolated) proteins. The mechanism of this side reaction may involve homolysis of biotin-HPDP to generate an unstable alkyl thiyl radical of the biotin group and a resonance-delocalized aryl radical of 2-thiopyridine. In the presence of ascorbate, these thiyl radicals would be reduced to thiol [77] and therefore contribute to the observed artifact. Importantly, similar reactions between light, aryl disulfides and ascorbate have been reported [75, 83]. These issues underscore the critical need to perform the labeling step of the BST in a light-free environment, and to employ metal chelators (more below).
POTENTIAL PROBLEMS ASSOCIATED WITH EXOGENOUS METALS IN THE BST
Ascorbate will drive one-electron reductions of Cu2+ and Fe3+ ions, which may result in reactions that compromise BST specificity, including production of ascorbate and hydroxyl radicals. These reactions are readily mitigated through the use of metal chelators (e.g. EDTA, DTPA, neocuproine) [84–87]. Despite the widespread application of metal chelation with the BST, one recent report suggests a metal-dependence for the BST to efficiently detect SNOs [88]. However, this conclusion overlooks the different chemistry operative in the presence and absence of transition metals: transition metals favor reductive chemistry by ascorbate over transnitrosation. Thus at low concentrations of ascorbate (<1 mM) as employed by Wang et al. [88], transition metals promote direct SNO reduction (metals serve as catalysts). Conversely, transnitrosation reactions, which form the basis of BST specificity are favored by metal sequestration and higher ascorbate concentrations [60]. Increasing the amount of ascorbate (5 – 50 mM ascorbate) should remedy the reported “requirement” for exogenous metals that may increase assay sensitivity, but at the expense of specificity. In particular, redox active metals are known to promote manifold reactions (both ascorbate dependent and independent), such as Cu2+-mediated thiol oxidation [89–91] and Fenton chemistry [92–94]. Given the potential for such side reactions to compromise assay specificity, the addition of exogenous redox active metals in the BST cannot be advocated without more rigorous validation in complex biological systems.
Given the known reactivity of Hg towards thiols and SNOs, numerous studies have attempted to employ Hg salts (e.g. HgCl2) to quench SNOs prior to performing the BST (and thus serve as an internal assay control in lieu of omitting ascorbate). Despite the frequent application of Hg salts in NO-based assays (described above), the high affinity of Hg salts for free thiols [95–97] makes this strategy inherently problematic for the BST. As nicely demonstrated by Zhang et al. [98], HgCl2 prevents biotinylation of free thiols and lowers any signal irrespective of _S_-nitrosylation status. Since all sources of biotinylation (i.e. signals) in the BST are attenuated by Hg salts, it is unclear how such a strategy could be used to distinguish an SNO from a false positive signal due to incomplete blocking; therefore, Hg salts should not be used in lieu of other well-validated controls (e.g. pre-photolysis, NOS inhibition, omitting ascorbate). By contrast, in an NO-based assay such as photolysis-chemiluminesence, Hg-thiol reactions identify the source of NO with Cys thiol by quenching the signal.
MODERN PERMUTATIONS AND ADAPTATIONS OF THE BST
Various groups have adapted the BST to suit specific experimental goals. For example, several labs have trypsinized the biotinylated material prior to avidin pulldown to allow mass spectrometry-based determination of protein-SNO sites (i.e. specific cysteine residues targeted by _S_-nitrosylation) [5, 99–101]. In addition, a thiol-reactive hexahistidine tag has been used in lieu of biotin-HPDP to further facilitate mass spectrometric analysis [102]. These studies have further demonstrated the utility of the BST for SNO-oriented proteomics and bioinformatics. However, some of these strategies rely on a single peptide (containing the SNO site) for the identification of an entire protein, and thus stringent mass spectrometric criteria must be applied to these approaches to avoid incorrect protein (and SNO site) identification.
In another novel approach, Kettenhofen et al. substituted biotin-HPDP with thiol-reactive cyanine dyes to combine in-gel fluorescent detection strategies with the BST [103, 104]. Han et al. similarly employed coumarin-based fluorophores to the same end [105]. A major advantage of this approach is that it is readily adaptable to 2-dimensional differential in-gel electrophoresis (2D-DIGE) techniques where multiple differentially fluorescent-labeled samples are co-analyzed on a single 2D electrophoresis gel [106, 107].
TYPICAL PROTOCOL FOR THE BIOTIN SWITCH TECHNIQUE
- Overall steps of the BST:
- Blocking
- Labeling
- Pulldown
- SDS-PAGE and/or immunodetection
- General considerations for the BST:
- A wide range of protein amount can be utilized (generally 0.3 to 5 mg of total protein per sample).
- A sensitive antibody with low background signal is critical for detecting an individual protein-SNO within a lysate.
- As discussed above, positive and negative controls should be included in every BST. For example, addition of 0.1 mM CysNO to one sample (prior to the BST) is an optimal positive control. A non-ascorbate or photolyzed sample serves as a convenient negative control.
- Acetone precipitations are typically used to remove excess reagents and isolate proteins. However, for small sample volumes (< 100 µl) or protein quantities (< 300 µg), acetone precipitation is often inefficient or inconsistent. Under these conditions, P-6 desalting gel (Bio-Rad) or G-25 sephadex (Amersham) can be substituted for acetone precipitations.
- Necessary reagents and notes:
- Methyl methanethiosulfonate (MMTS) – Fluka, catalog # 64306.
- Biotin-HPDP – Pierce, catalog # 21341.
- Streptavidin agarose – Fluka, catalog # 85881.
- Sodium ascorbate – Fluka, catalog # 11140. This ascorbate has been assayed for metal content, which is reported on the bottle. We have noted that this ascorbate is considerably more stable in aqueous solution than other preparations of ascorbate (i.e. yellowing due to slow oxidation to dehydroascorbate is undetectable).
- Stock solutions:
- HEN buffer (100 mM HEPES, 1 mM EDTA, 0.1 mM neocuproine pH 8.0).
- Alternatively, DTPA can be substituted for EDTA.
- HENS buffer – HEN buffer with 1% SDS (w/v).
- HEN/10 buffer – HEN buffer diluted 10-fold with dH2O.
- HENS/10 buffer – HEN/10 buffer with 1% SDS (w/v)
- Neutralization buffer (25 mM HEPES, 100 mM NaCl, 1mM EDTA, 0.5% Triton X-100, pH 7.5).
- Wash buffer – neutralization buffer containing 600 mM NaCl.
- 25% SDS in dH2O (w/v).
- 70% acetone in dH2O (v/v).
- Solutions to prepare immediately before assay:
- 10% MMTS in N,_N_-dimethylformamide (DMF) (v/v).
- 200 mM sodium ascorbate in HEN buffer (stored in the dark on ice).
- 2.5 mg/ml Biotin-HPDP in DMSO.
- Elution buffer – HEN/10 containing 1% beta-mercaptoethanol (v/v).
- Sample Protocol for the BST
- SAMPLE PREPARATION: Cells are lysed as desired. For example, a 6 cm plate of HEK293 cells can be lysed in 0.40 ml of 25 mM HEPES, 50 mM NaCl, 0.1 mM EDTA (DTPA may be a better metal chelator; note also that neocuproine is present in HEN buffer), 1% NP-40, 0.5 mM PMSF + protease inhibitors, pH 7.4. Repeated passage through a 28-gauge needle will increase lysis efficiency. It is critical to avoid DTT (or other reductants) in the lysis buffer because they will destabilize SNOs and interfere with the BST. Following centrifugation, protein concentrations are measured (e.g. BCA assay, Pierce). Samples are adjusted with lysis buffer to achieve equal concentrations of total protein.
- BLOCKING: For experiments employing 0.5 – 2 mg of protein, samples are diluted to 1.8 ml with HEN buffer. Next, 0.2 ml of 25% SDS is added along with 20 µl of 10% MMTS (final 2.0 ml volume with 2.5% SDS and 0.1% MMTS). Samples are incubated at 50 °C in the dark for 15 – 20 min with frequent vortexing. This step is frequently performed in 15 ml conical tubes. Alternatively, MMTS may be added directly to the lysis buffer to initiate blocking immediately following lysis, though it should be noted that MMTS interferes with the BCA protein assay. For some proteins, the blocking step can be efficiently performed at room temperature (e.g. _S_-nitrosylated caspase-3), though this should be determined empirically for each protein.
- PRECIPITATION: Three volumes of cold acetone (6 ml) are added to each sample. Proteins are precipitated for 20 min at −20 °C, and collected by centrifugation at 2,000 g for 5 min. The clear supernatant is aspirated and the protein pellet is gently washed with 70% acetone (4 × 5 ml).
- LABELING: Following resuspension in 0.24 ml HENS buffer, the material is transferred to a fresh 1.7 ml microfuge tube containing 30 µl biotin-HPDP (2.5 mg/ml). The labeling reaction is initiated by adding 30 µl of 200 mM sodium ascorbate (we tend to use a final concentration of 20 mM ascorbate, though less amounts may suffice [36] and higher amounts can be used as necessary [63]). Alternatively, an equivalent concentration of NaCl can be used as an ascorbate-free control. Samples are rotated at room temperature in the dark for 1 h. It is critical to avoid any sources of sunlight during this step.
- PRECIPITATION: Three volumes of cold acetone (0.9 ml) are added to each sample. Proteins are precipitated for 20 min at −20 °C, and collected by centrifugation at 5,000 g for 5 min. The clear supernatant is aspirated and the protein pellet is gently washed with 70% acetone (4 × 1 ml).
- PULLDOWN: Following complete resuspension in 0.25 ml HENS/10 buffer, 0.75 ml of neutralization buffer is added. A small fraction of each sample (e.g. 10 µl) is removed for analysis of protein “input”. The remaining material is transferred to a fresh 1.7 ml microfuge tube containing 25 – 50 µl of pre-washed avidin-affinity resin. It is critical that bead volumes in each sample are the same. The samples are gently rotated for 12 – 18 h at 4 °C.
- WASHING: Avidin beads are collected by centrifugation at 200 g × 10 s in a swinging-bucket rotor, followed by washing with wash buffer (4 × 1 ml). After the final wash, the beads are fully dried via gentle aspiration with a 28-gauge needle.
- ELUTION: To each sample is added 30 – 50 µl of elution buffer. Proteins are eluted at room temperature with frequent agitation, followed by centrifugation at 5,000 g × 30 s. Supernatant is collected without disturbing the pelleted resin, and mixed with 6x Laemmli loading buffer. For analysis of protein-SNOs via immunodetection of biotinylation, elution is performed by heating the beads to 95 °C in 30 – 50 µl of HENS/10 buffer containing non-reducing Laemmli loading buffer (all reductants must be avoided).
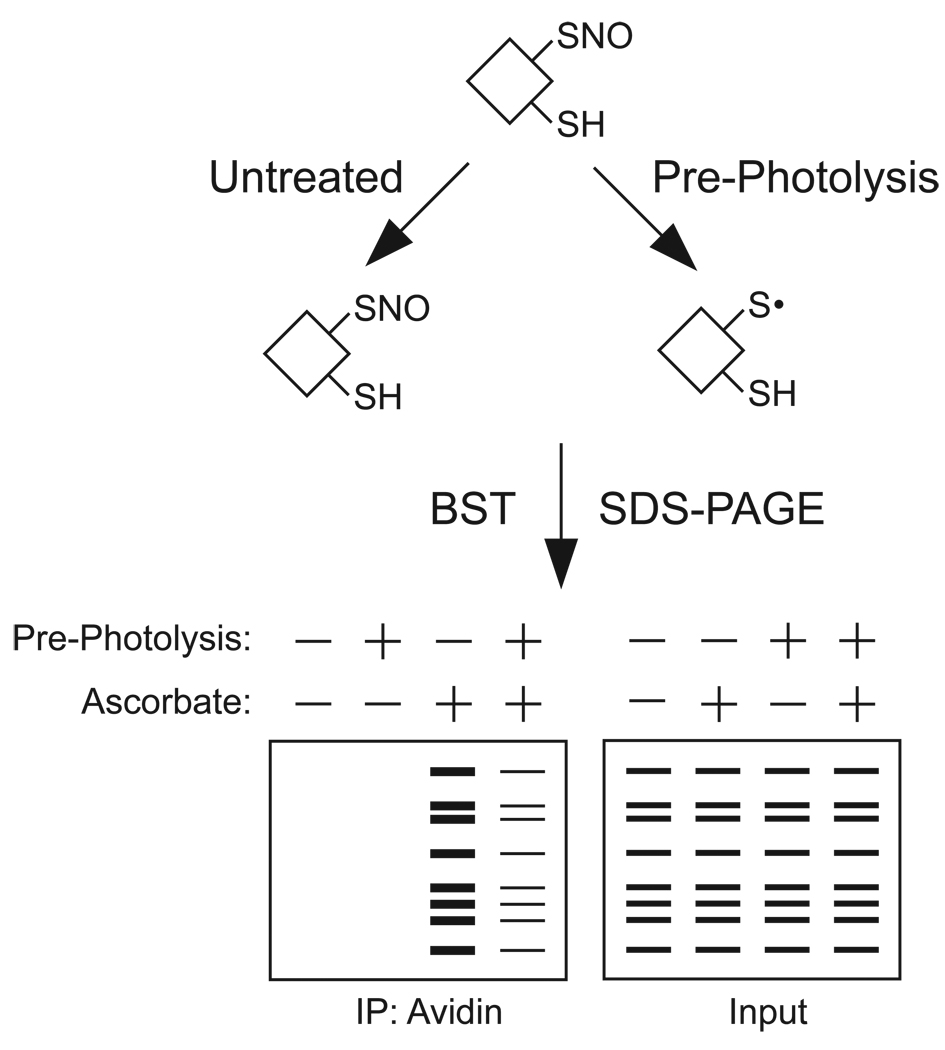
Fig. 5.
An illustration employing UV pre-photolysis and ascorbate controls within the BST for the analysis of a single lysate. One sample is split to two fractions, which are either untreated or exposed to a strong UV light source (“pre-photolysis”) prior to performing the BST. The pre-photolysis step leads to homolytic cleavage of the S-NO into NO and an unstable thiyl radical, which is either reduced to a thiol or oxidized to a higher _S_-oxide. In either case, the signal from an SNO will be attenuated by pre-photolysis, while free thiols or disulfides are unaffected. Inclusion or exclusion of ascorbate during the labeling step of the BST can also be employed to assay for protein-SNOs. This approach, which employs internal controls (e.g. pre-photolysis and ascorbate), is ideal when external controls such as NOS inhibition or activation are not feasible (e.g. human tissue samples) or ineffective (as in the case of reactions regulated by denitrosylation).
ACKNOWLEDGEMENTS
The authors would like to thank Alfred Hausladen and Abhijit Chakladar for their thoughtful comments and technical expertise. This work was supported by the National Institutes of Health grant P01-HL075443.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 2.Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 3.Kokkola T, Savinainen JR, Monkkonen KS, Retamal MD, Laitinen JT. S-nitrosothiols modulate G protein-coupled receptor signaling in a reversible and highly receptor-specific manner. BMC Cell Biol. 2005;6:21. doi: 10.1186/1471-2121-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nozik-Grayck E, Whalen EJ, Stamler JS, McMahon TJ, Chitano P, Piantadosi CA. S-nitrosoglutathione inhibits alpha1-adrenergic receptor-mediated vasoconstriction and ligand binding in pulmonary artery. Am J Physiol Lung Cell Mol Physiol. 2006;290:L136–L143. doi: 10.1152/ajplung.00230.2005. [DOI] [PubMed] [Google Scholar]
- 5.Whalen EJ, Foster MW, Matsumoto A, Ozawa K, Violin JD, Que LG, Nelson CD, Benhar M, Keys JR, Rockman HA, Koch WJ, Daaka Y, Lefkowitz RJ, Stamler JS. Regulation of beta-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell. 2007;129:511–522. doi: 10.1016/j.cell.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 6.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chanvorachote P, Nimmannit U, Wang L, Stehlik C, Lu B, Azad N, Rojanasakul Y. Nitric oxide negatively regulates Fas CD95-induced apoptosis through inhibition of ubiquitin-proteasome-mediated degradation of FLICE inhibitory protein. J Biol Chem. 2005;280:42044–42050. doi: 10.1074/jbc.M510080200. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann J, Haendeler J, Zeiher AM, Dimmeler S. TNFalpha and oxLDL reduce protein S-nitrosylation in endothelial cells. J Biol Chem. 2001;276:41383–41387. doi: 10.1074/jbc.M107566200. [DOI] [PubMed] [Google Scholar]
- 9.Kim JE, Tannenbaum SR. S-Nitrosation regulates the activation of endogenous procaspase-9 in HT-29 human colon carcinoma cells. J Biol Chem. 2004;279:9758–9764. doi: 10.1074/jbc.M312722200. [DOI] [PubMed] [Google Scholar]
- 10.Kim YM, Talanian RV, Billiar TR. Nitric oxide inhibits apoptosis by preventing increases in caspase-3-like activity via two distinct mechanisms. J Biol Chem. 1997;272:31138–31148. doi: 10.1074/jbc.272.49.31138. [DOI] [PubMed] [Google Scholar]
- 11.Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y, Man HY, Sekine-Aizawa Y, Han Y, Juluri K, Luo H, Cheah J, Lowenstein C, Huganir RL, Snyder SH. S-nitrosylation of N-ethylmaleimide sensitive factor mediates surface expression of AMPA receptors. Neuron. 2005;46:533–540. doi: 10.1016/j.neuron.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 13.Lipton SA, Choi YB, Pan ZH, Lei SZ, Chen HS, Sucher NJ, Loscalzo J, Singel DJ, Stamler JS. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- 14.Mustafa AK, Kumar M, Selvakumar B, Ho GP, Ehmsen JT, Barrow RK, Amzel LM, Snyder SH. Nitric oxide S-nitrosylates serine racemase, mediating feedback inhibition of D-serine formation. Proc Natl Acad Sci U S A. 2007;104:2950–2955. doi: 10.1073/pnas.0611620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi H, Shin Y, Cho SJ, Zago WM, Nakamura T, Gu Z, Ma Y, Furukawa H, Liddington R, Zhang D, Tong G, Chen HS, Lipton SA. Hypoxia enhances S-nitrosylation-mediated NMDA receptor inhibition via a thiol oxygen sensor motif. Neuron. 2007;53:53–64. doi: 10.1016/j.neuron.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowenstein CJ. Nitric oxide regulation of protein trafficking in the cardiovascular system. Cardiovasc Res. 2007;75:240–246. doi: 10.1016/j.cardiores.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsushita K, Morrell CN, Cambien B, Yang SX, Yamakuchi M, Bao C, Hara MR, Quick RA, Cao W, O'Rourke B, Lowenstein JM, Pevsner J, Wagner DD, Lowenstein CJ. Nitric oxide regulates exocytosis by S-nitrosylation of N-ethylmaleimide-sensitive factor. Cell. 2003;115:139–150. doi: 10.1016/s0092-8674(03)00803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrell CN, Matsushita K, Chiles K, Scharpf RB, Yamakuchi M, Mason RJ, Bergmeier W, Mankowski JL, Baldwin WM, 3rd, Faraday N, Lowenstein CJ. Regulation of platelet granule exocytosis by S-nitrosylation. Proc Natl Acad Sci U S A. 2005;102:3782–3787. doi: 10.1073/pnas.0408310102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G, Moniri NH, Ozawa K, Stamler JS, Daaka Y. Nitric oxide regulates endocytosis by S-nitrosylation of dynamin. Proc Natl Acad Sci U S A. 2006;103:1295–1300. doi: 10.1073/pnas.0508354103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SF, Huri DA, Snyder SH. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science. 2005;310:1966–1970. doi: 10.1126/science.1119407. [DOI] [PubMed] [Google Scholar]
- 21.Tian J, Kim SF, Hester L, Snyder SH. S-nitrosylation/activation of COX-2 mediates NMDA neurotoxicity. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0804852105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu L, Han C, Lim K, Wu T. Activation of cytosolic phospholipase A2alpha through nitric oxide-induced S-nitrosylation. Involvement of inducible nitric-oxide synthase and cyclooxygenase-2. J Biol Chem. 2008;283:3077–3087. doi: 10.1074/jbc.M705709200. [DOI] [PubMed] [Google Scholar]
- 23.Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 24.Azad N, Vallyathan V, Wang L, Tantishaiyakul V, Stehlik C, Leonard SS, Rojanasakul Y. S-nitrosylation of Bcl-2 inhibits its ubiquitin-proteasomal degradation. A novel antiapoptotic mechanism that suppresses apoptosis. J Biol Chem. 2006;281:34124–34134. doi: 10.1074/jbc.M602551200. [DOI] [PubMed] [Google Scholar]
- 25.Chanvorachote P, Nimmannit U, Stehlik C, Wang L, Jiang BH, Ongpipatanakul B, Rojanasakul Y. Nitric oxide regulates cell sensitivity to cisplatin-induced apoptosis through S-nitrosylation and inhibition of Bcl-2 ubiquitination. Cancer Res. 2006;66:6353–6360. doi: 10.1158/0008-5472.CAN-05-4533. [DOI] [PubMed] [Google Scholar]
- 26.Li F, Sonveaux P, Rabbani ZN, Liu S, Yan B, Huang Q, Vujaskovic Z, Dewhirst MW, Li CY. Regulation of HIF-1alpha stability through S-nitrosylation. Mol Cell. 2007;26:63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim KH, Ancrile BB, Kashatus DF, Counter CM. Tumour maintenance is mediated by eNOS. Nature. 2008;452:646–649. doi: 10.1038/nature06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomsen LL, Scott JM, Topley P, Knowles RG, Keerie AJ, Frend AJ. Selective inhibition of inducible nitric oxide synthase inhibits tumor growth in vivo: studies with 1400W, a novel inhibitor. Cancer Res. 1997;57:3300–3304. [PubMed] [Google Scholar]
- 29.Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 30.Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson's disease. Proc Natl Acad Sci U S A. 2007;104:18742–18747. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu Z, Kaul M, Yan B, Kridel SJ, Cui J, Strongin A, Smith JW, Liddington RC, Lipton SA. S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 2002;297:1186–1190. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- 32.Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E, Uehara T, Lipton SA. Nitrosative stress linked to sporadic Parkinson's disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci U S A. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durham WJ, Aracena-Parks P, Long C, Rossi AE, Goonasekera SA, Boncompagni S, Galvan DL, Gilman CP, Baker MR, Shirokova N, Protasi F, Dirksen R, Hamilton SL. RyR1 S-nitrosylation underlies environmental heat stroke and sudden death in Y522S RyR1 knockin mice. Cell. 2008;133:53–65. doi: 10.1016/j.cell.2008.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez DR, Beigi F, Treuer AV, Hare JM. Deficient ryanodine receptor S-nitrosylation increases sarcoplasmic reticulum calcium leak and arrhythmogenesis in cardiomyocytes. Proc Natl Acad Sci U S A. 2007;104:20612–20617. doi: 10.1073/pnas.0706796104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehnart SE, Mongillo M, Bellinger A, Lindegger N, Chen BX, Hsueh W, Reiken S, Wronska A, Drew LJ, Ward CW, Lederer WJ, Kass RS, Morley G, Marks AR. Leaky Ca2+ release channel/ryanodine receptor 2 causes seizures and sudden cardiac death in mice. J Clin Invest. 2008;118:2230–2245. doi: 10.1172/JCI35346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 37.Bartberger MD, Mannion JD, Powell SC, Stamler JS, Houk KN, Toone EJ. S-N dissociation energies of S-nitrosothiols: on the origins of nitrosothiol decomposition rates. J Am Chem Soc. 2001;123:8868–8869. doi: 10.1021/ja0109390. [DOI] [PubMed] [Google Scholar]
- 38.Stamler JS, Toone EJ. The decomposition of thionitrites. Curr Opin Chem Biol. 2002;6:779–785. doi: 10.1016/s1367-5931(02)00383-6. [DOI] [PubMed] [Google Scholar]
- 39.Marzinzig M, Nussler AK, Stadler J, Marzinzig E, Barthlen W, Nussler NC, Beger HG, Morris SM, Jr, Bruckner UB. Improved methods to measure end products of nitric oxide in biological fluids: nitrite, nitrate, and S-nitrosothiols. Nitric Oxide. 1997;1:177–189. doi: 10.1006/niox.1997.0116. [DOI] [PubMed] [Google Scholar]
- 40.Moore KP, Mani AR. Measurement of protein nitration and S-nitrosothiol formation in biology and medicine. Methods Enzymol. 2002;359:256–268. doi: 10.1016/s0076-6879(02)59190-4. [DOI] [PubMed] [Google Scholar]
- 41.Saville B. A Scheme for the colorimetric determination of microgram amounts of thiols. Analyst. 1958;83:670–672. [Google Scholar]
- 42.Park JK, Kostka P. Fluorometric detection of biological S-nitrosothiols. Anal Biochem. 1997;249:61–66. doi: 10.1006/abio.1997.2159. [DOI] [PubMed] [Google Scholar]
- 43.King M, Gildemeister O, Gaston B, Mannick JB. Assessment of S-nitrosothiols on diaminofluorescein gels. Anal Biochem. 2005;346:69–76. doi: 10.1016/j.ab.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 44.Kojima H, Nakatsubo N, Kikuchi K, Kawahara S, Kirino Y, Nagoshi H, Hirata Y, Nagano T. Detection and imaging of nitric oxide with novel fluorescent indicators: diaminofluoresceins. Anal Chem. 1998;70:2446–2453. doi: 10.1021/ac9801723. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez J, Specian V, Maloney R, Jourd'heuil D, Feelisch M. Performance of diamino fluorophores for the localization of sources and targets of nitric oxide. Free Radic Biol Med. 2005;38:356–368. doi: 10.1016/j.freeradbiomed.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 46.Hausladen A, Rafikov R, Angelo M, Singel DJ, Nudler E, Stamler JS. Assessment of nitric oxide signals by triiodide chemiluminescence. Proc Natl Acad Sci U S A. 2007;104:2157–2162. doi: 10.1073/pnas.0611191104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stamler JS, Jaraki O, Osborne J, Simon DI, Keaney J, Vita J, Singel D, Valeri CR, Loscalzo J. Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc Natl Acad Sci U S A. 1992;89:7674–7677. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doctor A, Platt R, Sheram ML, Eischeid A, McMahon T, Maxey T, Doherty J, Axelrod M, Kline J, Gurka M, Gow A, Gaston B. Hemoglobin conformation couples erythrocyte S-nitrosothiol content to O2 gradients. Proc Natl Acad Sci U S A. 2005;102:5709–5714. doi: 10.1073/pnas.0407490102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmer LA, Doctor A, Chhabra P, Sheram ML, Laubach VE, Karlinsey MZ, Forbes MS, Macdonald T, Gaston B. S-nitrosothiols signal hypoxia-mimetic vascular pathology. J Clin Invest. 2007;117:2592–2601. doi: 10.1172/JCI29444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmer LA, Gaston B. S-nitrosothiol assays that avoid the use of iodine. Methods Enzymol. 2008;440:157–176. doi: 10.1016/S0076-6879(07)00809-9. [DOI] [PubMed] [Google Scholar]
- 51.Jia L, Bonaventura C, Bonaventura J, Stamler JS. S-nitrosohaemoglobin: a dynamic activity of blood involved in vascular control. Nature. 1996;380:221–226. doi: 10.1038/380221a0. [DOI] [PubMed] [Google Scholar]
- 52.McMahon TJ, Moon RE, Luschinger BP, Carraway MS, Stone AE, Stolp BW, Gow AJ, Pawloski JR, Watke P, Singel DJ, Piantadosi CA, Stamler JS. Nitric oxide in the human respiratory cycle. Nat Med. 2002;8:711–717. doi: 10.1038/nm718. [DOI] [PubMed] [Google Scholar]
- 53.Stamler JS. S-nitrosothiols in the blood: roles, amounts, and methods of analysis. Circ Res. 2004;94:414–417. doi: 10.1161/01.RES.0000122071.55721.BC. [DOI] [PubMed] [Google Scholar]
- 54.Mannick JB, Schonhoff C, Papeta N, Ghafourifar P, Szibor M, Fang K, Gaston B. S-Nitrosylation of mitochondrial caspases. J Cell Biol. 2001;154:1111–1116. doi: 10.1083/jcb.200104008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haendeler J, Hoffmann J, Tischler V, Berk BC, Zeiher AM, Dimmeler S. Redox regulatory and anti-apoptotic functions of thioredoxin depend on S-nitrosylation at cysteine 69. Nat Cell Biol. 2002;4:743–749. doi: 10.1038/ncb851. [DOI] [PubMed] [Google Scholar]
- 56.Park HS, Huh SH, Kim MS, Lee SH, Choi EJ. Nitric oxide negatively regulates c-Jun N-terminal kinase/stress-activated protein kinase by means of S-nitrosylation. Proc Natl Acad Sci U S A. 2000;97:14382–14387. doi: 10.1073/pnas.97.26.14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun J, Xin C, Eu JP, Stamler JS, Meissner G. Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc Natl Acad Sci U S A. 2001;98:11158–11162. doi: 10.1073/pnas.201289098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu L, Eu JP, Meissner G, Stamler JS. Activation of the cardiac calcium release channel (ryanodine receptor) by poly-S-nitrosylation. Science. 1998;279:234–237. doi: 10.1126/science.279.5348.234. [DOI] [PubMed] [Google Scholar]
- 59.Hausladen A, Privalle CT, Keng T, DeAngelo J, Stamler JS. Nitrosative stress: activation of the transcription factor OxyR. Cell. 1996;86:719–729. doi: 10.1016/s0092-8674(00)80147-6. [DOI] [PubMed] [Google Scholar]
- 60.Holmes AJ, Williams DLH. Reaction of ascorbic acid with S-nitrosothiols: clear evidence for two distinct reaction pathways. J Chem Soc, Perkin Trans. 2000;2:1639–1644. [Google Scholar]
- 61.Leis JR, Rios A, Rodriguez-Sanchez L. Reactivity of phenolic nucleophiles towards nitroso compounds. Part 2. Reaction with alkyl nitrite (O-nitroso compounds) J Chem Soc, Perkin Trans 2. 1998:2729–2733. [Google Scholar]
- 62.Munro AP, Williams DLH. Reactivity of nitrogen nucleophiles towards S-nitrosopenicillamine. J Chem Soc, Perkin Trans 2. 1999:1989–1993. [Google Scholar]
- 63.Forrester MT, Foster MW, Stamler JS. Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J Biol Chem. 2007;282:13977–13983. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- 64.Eu JP, Liu L, Zeng M, Stamler JS. An apoptotic model for nitrosative stress. Biochemistry. 2000;39:1040–1047. doi: 10.1021/bi992046e. [DOI] [PubMed] [Google Scholar]
- 65.Nathan C. Nitric oxide as a secretory product of mammalian cells. FASEB J. 1992;6:3051–3064. [PubMed] [Google Scholar]
- 66.Stuehr DJ, Marletta MA. Mammalian nitrate biosynthesis: mouse macrophages produce nitrite and nitrate in response to Escherichia coli lipopolysaccharide. Proc Natl Acad Sci U S A. 1985;82:7738–7742. doi: 10.1073/pnas.82.22.7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodriguez J, Maloney RE, Rassaf T, Bryan NS, Feelisch M. Chemical nature of nitric oxide storage forms in rat vascular tissue. Proc Natl Acad Sci U S A. 2003;100:336–341. doi: 10.1073/pnas.0234600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhelyaskov VR, Gee KR, Godwin DW. Control of NO concentration in solutions of nitrosothiol compounds by light. Photochem Photobiol. 1998;67:282–288. [PubMed] [Google Scholar]
- 69.Landino LM, Koumas MT, Mason CE, Alston JA. Ascorbic acid reduction of microtubule protein disulfides and its relevance to protein S-nitrosylation assays. Biochem Biophys Res Commun. 2006;340:347–352. doi: 10.1016/j.bbrc.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 70.Mayer B, Schrammel A, Klatt P, Koesling D, Schmidt K. Peroxynitrite-induced accumulation of cyclic GMP in endothelial cells and stimulation of purified soluble guanylyl cyclase. Dependence on glutathione and possible role of S-nitrosation. J Biol Chem. 1995;270:17355–17360. doi: 10.1074/jbc.270.29.17355. [DOI] [PubMed] [Google Scholar]
- 71.van der Vliet A, Hoen PA, Wong PS, Bast A, Cross CE. Formation of S-nitrosothiols via direct nucleophilic nitrosation of thiols by peroxynitrite with elimination of hydrogen peroxide. J Biol Chem. 1998;273:30255–30262. doi: 10.1074/jbc.273.46.30255. [DOI] [PubMed] [Google Scholar]
- 72.Schrammel A, Gorren AC, Schmidt K, Pfeiffer S, Mayer B. S-nitrosation of glutathione by nitric oxide, peroxynitrite, and (*)NO/O(2)(*-) Free Radic Biol Med. 2003;34:1078–1088. doi: 10.1016/s0891-5849(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 73.Huang B, Chen C. An ascorbate-dependent artifact that interferes with the interpretation of the biotin switch assay. Free Radic Biol Med. 2006;41:562–567. doi: 10.1016/j.freeradbiomed.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 74.Giustarini D, Dalle-Donne I, Colombo R, Milzani A, Ranieri R. Is ascorbate able to reduce disulfide bridges? A cautionary note. Nitric Oxide. 2008;19 doi: 10.1016/j.niox.2008.07.003. (In Press) [DOI] [PubMed] [Google Scholar]
- 75.Walmsley TA, Abernethy MH, Fitzgerald HP. Effect of daylight on the reaction of thiols with Ellman's reagent, 5,5'-dithiobis(2-nitrobenzoic acid) Clin Chem. 1987;33:1928–1931. [PubMed] [Google Scholar]
- 76.Riddles PW, Blakeley RL, Zerner B. Reassessment of Ellman's reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- 77.Buettner GR. The pecking order of free radicals and antioxidants: lipid peroxidation, alpha-tocopherol, and ascorbate. Arch Biochem Biophys. 1993;300:535–543. doi: 10.1006/abbi.1993.1074. [DOI] [PubMed] [Google Scholar]
- 78.May JM, Huang J, Qu ZC. Macrophage uptake and recycling of ascorbic acid: response to activation by lipopolysaccharide. Free Radic Biol Med. 2005;39:1449–1459. doi: 10.1016/j.freeradbiomed.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 79.May JM, Li L, Qu ZC, Huang J. Ascorbate uptake and antioxidant function in peritoneal macrophages. Arch Biochem Biophys. 2005;440:165–172. doi: 10.1016/j.abb.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 80.Winkler BS. Unequivocal evidence in support of the nonenzymatic redox coupling between glutathione/glutathione disulfide and ascorbic acid/dehydroascorbic acid. Biochim Biophys Acta. 1992;1117:287–290. doi: 10.1016/0304-4165(92)90026-q. [DOI] [PubMed] [Google Scholar]
- 81.Winkler BS, Orselli SM, Rex TS. The redox couple between glutathione and ascorbic acid: a chemical and physiological perspective. Free Radic Biol Med. 1994;17:333–349. doi: 10.1016/0891-5849(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 82.Njus D, Kelley PM. Vitamins C and E donate single hydrogen atoms in vivo. FEBS Lett. 1991;284:147–151. doi: 10.1016/0014-5793(91)80672-p. [DOI] [PubMed] [Google Scholar]
- 83.Fleming JE, Bensch KG, Schreiber J, Lohmann W. Interaction of ascorbic acid with disulfides. Zeitschrift fuer Naturforschung, C: Journal of Biosciences. 1983;38C:859–861. [Google Scholar]
- 84.Nishikimi M, Ozawa T. Stabilization of ascorbate solution by chelating agents that block redox cycling of metal ions. Biochem Int. 1987;14:111–117. [PubMed] [Google Scholar]
- 85.Oikawa S, Kawanishi S. Distinct mechanisms of site-specific DNA damage induced by endogenous reductants in the presence of iron(III) and copper(II) Biochim Biophys Acta. 1998;1399:19–30. doi: 10.1016/s0167-4781(98)00092-x. [DOI] [PubMed] [Google Scholar]
- 86.Satoh K, Ida Y, Kochi M, Tajima M, Kashimata M, Sakagami H. Effect of metals and their antagonists on the radical intensity and cytotoxicity of ascorbates. Anticancer Res. 1997;17:3355–3360. [PubMed] [Google Scholar]
- 87.Saxena P, Saxena AK, Cui XL, Obrenovich M, Gudipaty K, Monnier VM. Transition metal-catalyzed oxidation of ascorbate in human cataract extracts: possible role of advanced glycation end products. Invest Ophthalmol Vis Sci. 2000;41:1473–1481. [PubMed] [Google Scholar]
- 88.Wang X, Kettenhofen NJ, Shiva S, Hogg N, Gladwin MT. Copper dependence of the biotin switch assay: modified assay for measuring cellular and blood nitrosated proteins. Free Radic Biol Med. 2008;44:1362–1372. doi: 10.1016/j.freeradbiomed.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Landgraf W, Regulla S, Meyer HE, Hofmann F. Oxidation of cysteines activates cGMP-dependent protein kinase. J Biol Chem. 1991;266:16305–16311. [PubMed] [Google Scholar]
- 90.Schoneich C. Selective Cu2+/ascorbate-dependent oxidation of alzheimer's disease beta-amyloid peptides. Ann N Y Acad Sci. 2004;1012:164–170. doi: 10.1196/annals.1306.013. [DOI] [PubMed] [Google Scholar]
- 91.Stohs SJ, Bagchi D. Oxidative mechanisms in the toxicity of metal ions. Free Radic Biol Med. 1995;18:321–336. doi: 10.1016/0891-5849(94)00159-h. [DOI] [PubMed] [Google Scholar]
- 92.Chevion M. A site-specific mechanism for free radical induced biological damage: the essential role of redox-active transition metals. Free Radic Biol Med. 1988;5:27–37. doi: 10.1016/0891-5849(88)90059-7. [DOI] [PubMed] [Google Scholar]
- 93.Gutierrez-Correa J, Stoppani AO. Inactivation of yeast glutathione reductase by Fenton systems: effect of metal chelators, catecholamines and thiol compounds. Free Radic Res. 1997;27:543–555. doi: 10.3109/10715769709097858. [DOI] [PubMed] [Google Scholar]
- 94.Shapiro MP, Setlow B, Setlow P. Killing of Bacillus subtilis spores by a modified Fenton reagent containing CuCl2 and ascorbic acid. Appl Environ Microbiol. 2004;70:2535–2539. doi: 10.1128/AEM.70.4.2535-2539.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Clarkson TW, Magos L. Studies on the binding of mercury in tissue homogenates. Biochem J. 1966;99:62–70. doi: 10.1042/bj0990062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Raftery MJ. Enrichment by organomercurial agarose and identification of cys-containing peptides from yeast cell lysates. Anal Chem. 2008;80:3334–3341. doi: 10.1021/ac702539q. [DOI] [PubMed] [Google Scholar]
- 97.Zalups RK. Molecular interactions with mercury in the kidney. Pharmacol Rev. 2000;52:113–143. [PubMed] [Google Scholar]
- 98.Zhang Y, Keszler A, Broniowska KA, Hogg N. Characterization and application of the biotin-switch assay for the identification of S-nitrosated proteins. Free Radic Biol Med. 2005;38:874–881. doi: 10.1016/j.freeradbiomed.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 99.Greco TM, Hodara R, Parastatidis I, Heijnen HF, Dennehy MK, Liebler DC, Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc Natl Acad Sci U S A. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Han P, Chen C. Detergent-free biotin switch combined with liquid chromatography/tandem mass spectrometry in the analysis of S-nitrosylated proteins. Rapid Commun Mass Spectrom. 2008;22:1137–1145. doi: 10.1002/rcm.3476. [DOI] [PubMed] [Google Scholar]
- 101.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci U S A. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Camerini S, Polci ML, Restuccia U, Usuelli V, Malgaroli A, Bachi A. A novel approach to identify proteins modified by nitric oxide: the HIS-TAG switch method. J Proteome Res. 2007;6:3224–3231. doi: 10.1021/pr0701456. [DOI] [PubMed] [Google Scholar]
- 103.Kettenhofen NJ, Wang X, Gladwin MT, Hogg N. In-gel detection of S-nitrosated proteins using fluorescence methods. Methods Enzymol. 2008;441:53–71. doi: 10.1016/S0076-6879(08)01204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kettenhofen NJ, Broniowska KA, Keszler A, Zhang Y, Hogg N. Proteomic methods for analysis of S-nitrosation. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;851:152–159. doi: 10.1016/j.jchromb.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Han P, Zhou X, Huang B, Zhang X, Chen C. On-gel fluorescent visualization and the site identification of S-nitrosylated proteins. Anal Biochem. 2008;377:150–155. doi: 10.1016/j.ab.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 106.Friedman DB, Lilley KS. Optimizing the difference gel electrophoresis (DIGE) technology. Methods Mol Biol. 2008;428:93–124. doi: 10.1007/978-1-59745-117-8_6. [DOI] [PubMed] [Google Scholar]
- 107.Wu TL. Two-dimensional difference gel electrophoresis. Methods Mol Biol. 2006;328:71–95. doi: 10.1385/1-59745-026-X:71. [DOI] [PubMed] [Google Scholar]
- 108.Hausladen A, Gow AJ, Stamler JS. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc Natl Acad Sci U S A. 1998;95:14100–14105. doi: 10.1073/pnas.95.24.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]