PAK4 kinase activity and somatic mutation promote carcinoma cell motility and influence inhibitor sensitivity (original) (raw)
. Author manuscript; available in PMC: 2013 Oct 18.
Published in final edited form as: Oncogene. 2012 Jun 11;32(16):2114–2120. doi: 10.1038/onc.2012.233
Abstract
Hepatocyte growth factor (HGF) and its receptor (c-Met) are associated with cancer cell motility and invasiveness. p21-activated kinase 4 (PAK4), a potential therapeutic target, is recruited to and activated by c-Met. In response, PAK4 phosphorylates LIM kinase 1 (LIMK1) in an HGF-dependent manner in metastatic prostate carcinoma cells. PAK4 overexpression is known to induce increased cell migration speed but the requirement for kinase activity has not been established. We have used a panel of PAK4 truncations and mutations in a combination of over-expression and RNAi rescue experiments to determine the requirement for PAK4 kinase activity during carcinoma cell motility downstream of HGF. We find that neither the kinase domain alone nor a PAK4 mutant unable to bind Cdc42 is able to fully rescue cell motility in a PAK4-deficient background. Nevertheless, we find that PAK4 kinase activity and associated LIMK1 activity are essential for carcinoma cell motility, highlighting PAK4 as a potential anti-metastatic therapeutic target. We also show here that overexpression of PAK4 harboring a somatic mutation, E329K, increased the HGF-driven motility of metastatic prostate carcinoma cells. E329 lies within the G-loop region of the kinase. Our data suggest E329K mutation leads to a modest increase in kinase activity conferring resistance to competitive ATP inhibitors in addition to promoting cell migration. The existence of such a mutation may have implications for the development of PAK4-specific competitive ATP inhibitors should PAK4 be further explored for clinical inhibition.
Introduction
The progression of cancer by oncogenic transformation from localised primary solid tumours to more widely disseminated metastasis is associated with poor patient prognosis and increased mortality. As such, regulators of metastatic cell migration represent attractive therapeutic targets. The oncogenic receptor tyrosine kinase c-Met represents such a target for cancer therapeutics since it plays a dual role promoting tumour formation in addition to stimulating cell motility and metastasis (1). Indeed a variety of c-Met-targeting agents are currently under evaluation in clinical trials (2). Activation of c-MET by hepatocyte growth factor (HGF) leads to recruitment of a plethora of proteins including Gab-1 (3). Recently, p21-activated kinase 4 (PAK4) was identified as a novel Gab-1 binding partner (4). PAK4 is a group II PAK that specifically interacts with Cdc42 (5). However, the regulation of PAK4 activity and the role of Cdc42 interaction is poorly understood. It is known that PAK4 binds and phosphorylates a number of cytoskeletal protein targets including GEF-H1 (6), paxillin (7), β5 integrin (8) and the ADF/cofilin regulators LIMK1 and slingshot homologue (SSH-1) (9,10). Cofilin activity is important for supporting lamellipodia protrusion by promoting F-actin disassembly (11) and its regulation has been implicated in promoting and directing cancer cell motility (12). PAK4 is tumorogenic in vitro and in vivo (13,14) and overexpression or genetic amplification of PAK4 occurs in numerous cancer cell lines and tumours (reviewed in 15). Further, two somatic mutations adjacent to- and within-the PAK4 kinase domain (A279T and E329K) have been identified in colon carcinoma patients (16); although the functional consequences of these mutations has not been explored. Several recent studies have also implicated PAK4 in pancreatic, breast and ovarian carcinoma cell invasion (17,18,19). There is currently much interest in targeting group II PAKs therapeutically (reviewed by 20 and 21). Indeed, a PAK4-targeting competitive ATP inhibitor, PF3758309 has been reported (22). However, whether PAK4 promotes motility via kinase-dependent or –independent mechanisms has not been addressed. In this study we used a systematic approach to study domains of PAK4 required for HGF signal transduction in a prostate carcinoma cell model of motility.
Results and Discussion
PAK4 is required for HGF-induced LIMK1-mediated PC3 cell migration
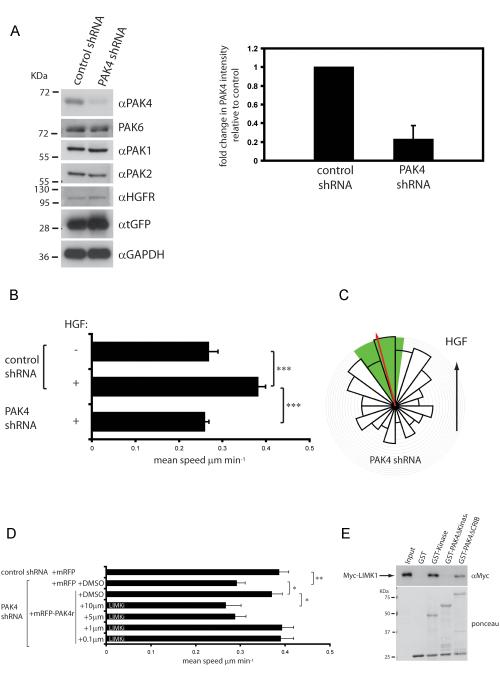
To facilitate our investigation we have used PC3 cells, which do not form cell:cell junctions or prominent actin stress fibres. We generated stable cell lines expressing control non-targeting or PAK4 specific shRNA and tGFP from a bicistronic operon. There is a ~80% reduction in PAK4 expression in cells stably expressing PAK4 shRNA, without affecting PAK1, PAK2, PAK6 or HGFR/c-Met expression (Fig 1 A and Fig S1A). We found that depletion of PAK4 significantly reduced cell motility in response to HGF (control shRNA cell mean speed ± s.e.m. 0.38 ± 0.018 μm/minute; PAK4 shRNA cell mean speed ± s.e.m. 0.26 ± 0.011 μm/minute; P<0.0001) (Fig 1B, movies 1 and 2). These results are consistent with previous reports using PAK4 null fibroblasts (23) and our previous data (24). Indeed, similar data have also implicated PAK4 in pancreatic ductal adenocarcinoma, breast, and ovarian carcinoma cell invasion, although these studies have tended to rely on transwell assays that preclude microscopic observation of motile cells and involve subjective measurement rather than directly measuring cell migration speed (17,18,19). We have previously shown that PC3 cells migrate up a linear gradient of HGF (24). Here we find that PAK4 depleted PC3 cells exhibit positive chemotaxis (Fig 1C) albeit moving at a reduced mean speed of migration (mean speed ± s.e.m. 0.17 ± 0.010 μm/minute), thus knockdown of PAK4 attenuates the mean speed of migration, but not the directionality of PC3 cell motility. As a further control, we transiently transfected PAK4 knockdown cells with shRNA-resistant mRFP-tagged PAK4 (PAK4r, (8)). Crucially, mRFP-PAK4r, was able to rescue the mean speed of cell migration of PAK4 depleted cells (Fig 1D). HGF stimulation of PC3 cells leads to PAK4-mediated phosphorylation and activation of LIMK (24), therefore, we sought to determine if LIMK activity was required for PAK4 mediated migration. We incubated mRFP-PAK4r-rescued cells with _N_-{5-[2-(2,6-Dichloro-phenyl)-5-difluoromethyl-2H-pyrazol-3-yl]-thiazol-2-yl}-isobutyramide, a LIMK inhibitor (LIMKi) (25) or DMSO as a control. LIMKi inhibited HGF-induced PAK4r-mediated cell migration in a dose dependent manner (Fig 1D). These findings confirm a specific requirement for LIMK1 in PAK4-mediated HGF-induced cell motility. Several studies advocate a role for LIMK in promoting cell migration (26), but our findings conflict with data using LIMKi on breast cancer cells where the authors suggested that LIMKs are not required for 2D cell motility, though are required for invasion (25). The seeming discrepancy between these data is likely due to experimental design. Scott et al, analysed the effect of LIMK inhibition of cells migrating in media containing serum on cell-derived matrices and as confluent monolayers in modified scratch assays, where multiple cues to stimulate cell migration exist. In our assays, the cells were serum starved prior to specific activation with HGF, compelling the cells to migrate in response to HGF using the preferential PAK4-LIMK-cofilin pathway (24).
Figure 1.
PAK4 is required for HGF-mediated cell motility, but not chemotaxis. (A) For western blot analysis cells were lysed in all experiments as follows. 10 minutes in lysis buffer (0.5% NP-40, 30 mM sodium pyrophosphate, 50 mM Tris-HCl pH 7.6, 150 mM NaCl, 0.1 mM EDTA, 50 mM NaF, 1 mM Na3VO4 and complete mini-EDTA free protease inhibitor (Roche)) then clarification by centrifugation at 14,000 g for 10 minutes. Lysates were immunoblotted according to standard procedures. Blots were developed by enhanced chemiluminescence (ECLplus, GE Healthcare). Control and PAK4 shRNA expressing cell lysates were probed for PAK4/6/1/2, c-Met, tGFP and GAPDH for details of antibodies see supplemental materials and methods. PAK4 expression from cell lysates stably expressing control and PAK4 shRNA from 4 independent experiments were quantified relative to control ± s.d. (B and D) PC3 cells stably expressing control and PAK4 shRNA were maintained in low serum for 24 h, stimulated with HGF and imaged or PC3 cells stably expressing PAK4 shRNA were transfected with plasmids encoding mRFP or mRFP-PAK4r and serum starved for 24 h. Cells were then stimulated with HGF in the presence of either DMSO or LIMKi and imaged. Cell images were collected using an axiovert 100 microscope and Sensicam (PCO Cook) CCD camera, taking a frame every 10 min for 21 h using AQM acquisition software (Kinetic Imaging Ltd, Belfast, U.K.). Subsequently cells were tracked for the whole of the time-lapse sequence using Motion Analysis software (Kinetic Imaging Ltd). Unless indicated, at least 50 cells were tracked over nine separate films from three separate experiments for each experimental condition. Mathematical analysis was then carried out using Mathematica 6.0™ workbooks 37. Mean track speeds for each condition were compared using the Student’s T-test and statistical significance was accepted for a min P≤0.05. See Supplemental Materials and Methods for details of migration analysis. C) PAK4-depleted cells were maintained in low serum for 24 h, exposed to a gradient of HGF in a Dunn chemotaxis chamber for 12 h and individual cells (n=123) from two separate experiments tracked. A chemotaxis circular histogram was generated to show the proportion of cells with a direction of migration lying within each 18° segment (source of HGF at the top). Red arrow indicates mean direction of cell migration; green segment 99% confidence interval calculated from a Rayleigh test. E) purified GST, GST-PAK4 kinase domain, GST-PAK4Δkinase and GST-PAK4ΔCRIB beads were used to pulldown Myc-LIMK1 from cell lysates. Samples were analysed by anti-Myc immunoblotting and ponceau staining. Representative of three separate experiments. GST-pull down assays were performed according to standard protocols, for details see supplemental Materials and methods.
The kinase domain of PAK4 is sufficient for substrate interaction, but not motility
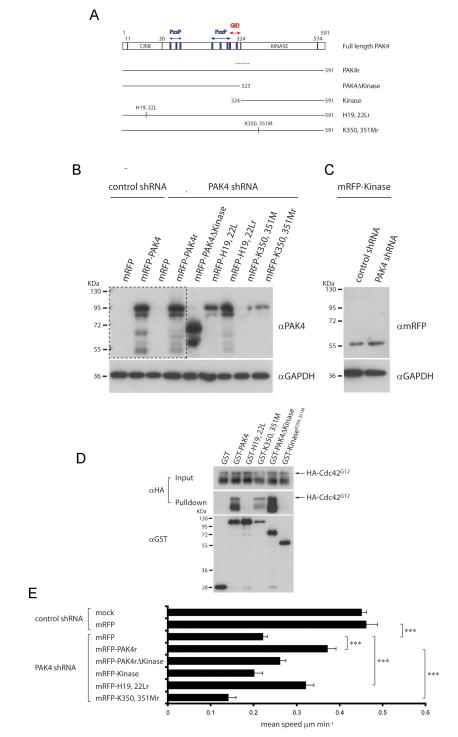
Given that inhibiting LIMK impairs PAK4-mediated PC3 cell motility (Fig 1D), we further explored the PAK4:LIMK1 interaction. GST pulldown assays revealed that the C terminal kinase domain of PAK4 is capable of binding to LIMK1 (Fig 1E). Taken together with our LIMKi studies this finding validates LIMK1 as a direct PAK4 target downstream of HGF. To test whether the kinase domain is sufficient to rescue the attenuated motility of PAK4 depleted cells, we expressed shRNA resistant (Figure 2A-C) mRFP-tagged PAK4 kinase domain (mRFP-Kinase, αα 324-591) and the N-terminus of PAK4 (mRFP-PAK4ΔKinase, αα1-323). mRFP-PAK4ΔKinase and mRFP-Kinase were not able to rescue the speed of cell migration of PAK4-depleted cells (Fig 2E). In contrast, Li et al., found that the PAK4 kinase domain (amino acids 323-591) was sufficient to promote haptotactic migration towards vitronectin 8. This difference in requirement for minimal PAK4 sequences might result from the fact that the β5 integrin binding site is also within the PAK4 kinase domain (27), whereas the PAK4 Gab-1 interaction domain (GID) required to recruit PAK4 to c-Met, is not (4). We found that PAK4 derivatives lacking the first 132 amino acid residues were mislocalised to the nucleus, even if they retained the Gab-1 interaction domain (data not shown). Our finding agrees with a recent report that identified N-terminal sequences that mediate PAK4 nuclear localisation (27) and emphasises that sequences outside the kinase domain are required to correctly localise PAK4 during HGF signal transduction.
Figure 2.
Active PAK4 kinase domain is necessary, but not sufficient for motility (A) Domain structure of PAK4 and GST- and mRFP-tagged shRNA-resistant PAK4 proteins. CRIB: Cdc42 and Rac interactive binding motif; PxxP: Pro-x-x-Pro amino acid sequence motifs. GID: GEF-H1/Gab-1 interaction domain. Dotted line indicates corresponding PAK4 shRNA target region. (B) Expression of shRNA resistant mRFP-PAK4 derivatives in PAK4 depleted cells. lysates were probed for PAK4 expression and GAPDH. A higher exposure of the area contained within the dotted line is shown below to visualise endogenous PAK4 in control shRNA cell lysates. (C) Control and PAK4 shRNA expressing PC3 cell lysates transfected with mRFP-PAK4 Kinase domain were probed for mRFP expression and GAPDH. (D) Mean migration speed ±s.e.m of cells stably expressing PAK4 or control shRNA transiently transfected with plasmids encoding mRFP or mRFP-PAK4 derivatives in response to HGF stimulation. n= ≥40 cells for each population over 3 separate experiments see figure 1 for image capture and tracking analysis details. Statistical significance was calculated using Student’s t-test, ***p<0.0001. (E) GST, or GST-tagged PAK4 derivatives were used to pulldown co-overexpressed HA-Cdc42G12V from 293 cell lysates. Blot is representative of 3 separate experiments.
GTPase interaction and kinase activity are required for motility
Because the N-terminus and the C-terminal kinase domain of PAK4 are both necessary to rescue cell migration of PAK4-depelted cells, we considered whether both a GTPase interaction and a kinase activity were similarly both required to stimulate migration. To test this hypothesis, we introduced mutations in conserved histidine residues within the N-terminal CRIB domain (H19, 22L) and mutations to inactivate the kinase domain (K350, 351M, (5,13) ) into both PAK4r (Fig 2A) and ‘wild-type’ PAK4 (Fig 4A). We confirmed that PAK4H19, 22L and the PAK4 kinase domain are not able to bind constitutively active Cdc42G12V (Fig 2D). Further, we confirmed that both PAK4H19, 22L and K350, 351M mutants were able to bind LIMK1 (data not shown). In motility assays, we found that the Cdc42 deficient binding mutant PAK4H19, 22L was able to partially rescue motility of PAK4 depleted cells (Fig 2E) suggesting that interaction with Cdc42 is required for full motility. In contrast, kinase dead PAK4K350, 351M failed to rescue PAK4 depletion and PAK4 depleted cells expressing PAK4K350, 351M did not display any significant cell motility (Fig 2E). The underlying mechanism of migration inhibition in this context is unclear but we could speculate that PAK4K350, 351M is acting to sequester substrate utilised by not only PAK4 but other kinases and thus inhibits additional signalling pathways.
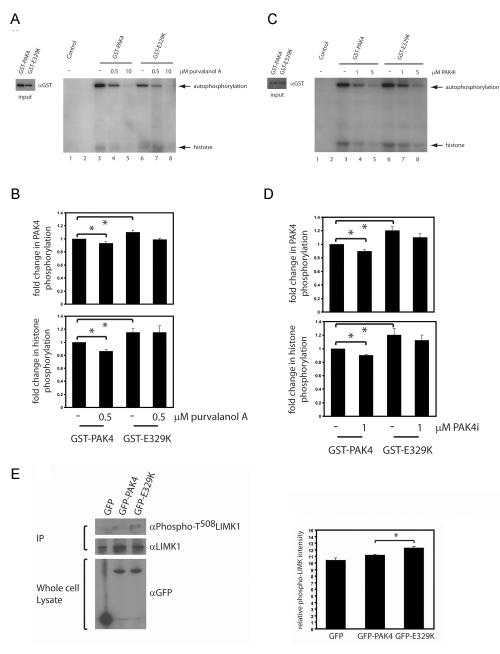
Figure 4.
E329K is an active kinase. Kinase assays were performed as previously described (43) (A -D) GST-PAK4 and GST-PAK4E329K kinase activity was assayed without inhibitor (Lanes 3 and 6) or with either Purvalanol A ((A): 0.5μM, lanes 4 and 7; 10μM, lanes 5 and 8) or PAK4i ((C): 1μM, lanes 4 and 7; 5μM, lanes 5 and 8). For inhibitor experiments, 0.5μM or 10μM Purvalanol A or 1μM or 5μM PAK4i was added to the kinase buffer. The reaction was stopped by adding SDS-page loading buffer. Autoradiographs and western blots were quantified using Andor IQ software (Belfast, UK) and the level of phosphorylation normalised to GST-protein levels. Phosphorylation of histone and autophosphorylation are indicated by arrows. Retained purified PAK4 proteins were subjected to GST immunoblotting to monitor expression levels (input). (A and C) PAK4 autophosphorylation and histone phosphorylation relative to wildtype PAK4 without Purvalanol A ±s.e.m. (B) or without PAK4i (D) ±s.e.m from 5 and 4 separate experiments, respectively. Statistical significance was calculated using student t-test, *p<0.05.
To complement our rescue experiments, we overexpressed PAK4 derivatives in cells to determine their ability to enhance HGF-mediated cell migration. Consistent with our previous findings (24), overexpression of full length (mRFP-) PAK4 significantly enhances HGF-mediated cell motility (Fig 3A-B). mRFP-PAK4H19, 22L, -PAK4ΔKinase and –Kinase domain in contrast, failed to enhance HGF-mediated cell migration. However, once again we observed the most dramatic effect on cell motility by overexpressing PAK4K350, 351M (Fig 3B), suggesting that PAK4K350, 351M overexpression has a dominant negative effect on wildtype PC3 cells expressing endogenous PAK4. Siu et al. also observed a similar inhibition of haptotactic transwell migration of breast carcinoma cells expressing kinase dead PAK4 (19). We did not observe any alteration in the directional persistence of cells expressing mRFP or any of the mRFP-tagged PAK4 derivatives in our motility assays. Kinase dead PAK4K350, 351M expressing cells exhibited a slight increase in persistent migration, though this is likely due to their diminished motility (Supp Fig 1B). Taken together with the findings that PAK4 depletion attenuated cell motility in response to HGF and that removing the PAK4 kinase domain abolishes rescue of motility, we conclude that PAK4 kinase activity is critical for cell motility. These data confirm that inhibition of PAK4 kinase activity is an attractive therapeutic target in efforts to inhibit metastatic spread.
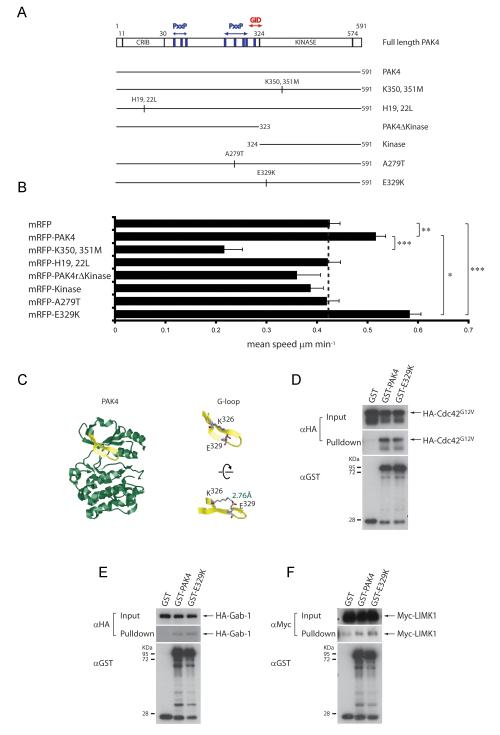
Figure 3.
PAK4 somatic mutation E329K enhances PC3 cell motility. (A) Domain structure of PAK4 and GST/mRFP-tagged PAK4 proteins used in overexpression experiments (Abbreviated as in Fig 2). (B) Mean migration speed ±s.e.m of cells transiently transfected with plasmids encoding either mRFP or mRFP-tagged PAK4 derivatives in response to HGF stimulation. n= ≥48 cells for each population (except cells expressing mRFP-kinase (n=30) and -PAK4K350, 351M (n=33)) over 3 separate experiments. Statistical significance was calculated using Student’s t-test, ***p<0.0001, **p<0.001, *p<0.05. (C) (left) Ribbon diagram of the kinase domain structure of human PAK4 (2X4Z) bound to PF-3758309 ATP analog (not shown) 22 generated using Rasmol. Glycine-rich loop region (G-loop) shown in yellow. (right top) Highlighted G-loop showing basic (Lysine 326) and acidic (Glutamic acid 329) side chains preceeding the G-loop motif (right bottom) Rotated view of G-loop showing electrostatic interactions and distance between K326 and E329 (D, E and F) GST, GST-PAK4 or GST-PAK4E329K were used to pulldown co-overexpressed HA-Cdc42G12V, HA-Gab-1 or Myc-LIMK1 from 293 cell lysates. Blots are representative of 3 separate experiments.
Somatic mutation E329K enhances PC3 cell motility
Having found clear evidence for the regulation of cell migration by the kinase domain of PAK4, we sought to determine how PAK4 somatic mutations (16) adjacent to and within the kinase domain would affect cell motility. To that end, we generated A279T and E329K mutations in PAK4 (Fig 3A). mRFP-PAK4A279T failed to enhance migration speed above the level of mRFP controls (Fig 3B). Residue A279 lies within a PxxP sequence, and it is tempting to speculate that the substitution of hydrophobic alanine to the larger and nucleophilic threonine might disrupt an SH3 domain interaction, or even engage different SH3 domain-containing proteins. From our data, it is not clear what advantage metastatic cells gain from the A279T mutation, but it might be proliferative rather than migratory. Neither expression of mRFP-PAK4, -PAK4A279T or PAK4E329K effected the persistence of random cell migration (Supp Fig 1B). However, we found that overexpression of mRFP-PAK4E329K significantly enhanced mean migration speed (Fig 3B) even beyond the level induced by overexpressing wildtype PAK4.
Mutation of E329K does not impair interaction with Cdc42, Gab-1 or LIMK1
To explore how E329K mutation influences PAK4 biology, we examined the location of this residue in the kinase domain crystal structure of PAK4 (22, 28). E329 resides on β strand 1b within the glycine-rich loop (G-loop) of the kinase, a conserved structural feature that contributes to ATP binding and orientation for catalysis (29). The E329 side chain extends upward from the G-loop away from the bound ATP and is juxtaposed to the nearby basic side chain K326 at a distance indicative of a salt bridge interaction (Fig 3C). In fact, G-loop salt bridges are a common feature in a large number of diverse protein kinases (30). To determine whether this mutation affects binding to either upstream regulators (Cdc42/Gab-1) or substrates (LIMK1), we performed interaction assays. These assays revealed that similarly to PAK4, PAK4E329K is able to interact with Cdc42, Gab-1 and LIMK1 (Fig 3D-F).
Mutation of E329K elevates kinase activity
Mutation of G-loop amino acid residues can have opposing effects on the catalytic activity of kinases; mutation can impair catalytic activity, as is the case for Lyn (30). In contrast, mutation of E255 in Abl tyrosine kinase does not diminish catalytic activity (31,32) and can modestly increase the transforming activity of Bcr-Abl fusion proteins (33). We therefore sought to determine the activity of PAKE329K using an in vitro kinase assay. We found that PAKE329K retains autophosphorylation and substrate kinase activity (Fig. 4A, lanes 1, 3 and 6), moreover, there is a modest (and significant) increase in PAKE329K activity compared to wildtype PAK4 (Fig 4 A-D) These data show that PAKE329K is catalytically active in cells, and suggest that PAK4E329K mediated increased cell migration speed is likely due to increased kinase activity, providing further evidence that there is a correlation between the level of PAK4 kinase activity and cell migration speed in PC3 cells (Fig 2E). We did not find a significant difference between the Km(ATP) of wildtype PAK4 and PAKE329K (our unpublished data) but did confirm a small but significant difference in kinase activity using a luminescence assay (supp Fig 1C). Km(ATP) measurements do not distinguish between autophosphorylation and substrate phosphorylation and we would speculate that the increased activity we detect is centred on the interaction between PAKE329K and its substrate (s). Indeed, overexpression of PAKE329K increases the level of LIMK1 phosphorylation in cells compared to overexpression of wildtype PAK4 (Fig 4E). The E329K mutation delivers an increase over wildtype level activity which translates to approximately 10%. Whilst this number seems modest, previous work (7) has already shown that high levels of PAK4 kinase activity induce a loss of cell adhesion and rounding. Therefore, a 10% increase in kinase activity translated into an increased migration speed as we have demonstrated here, is more likely to convey a metastatic advantage than high levels of kinase activity, where cellular function is likely to be significantly impaired. Indeed, this very mutation was found in a cancer patient sample (16).
Mutation of E329K confers resistance to competitive ATP inhibitors
Bcr-Abl E255K or E255V mutations (analogous to the PAK4 E329K mutation) have been strongly implicated in clinical resistance to the competitive ATP inhibitor Imatinib/Gleevec (34, 35) in chronic myeloiod leukimia. We speculated that PAK4 E329K mutation might confer resistance to competitive ATP inhibitors. To test this, we performed in vitro kinase assays in the presence of Purvalanol A 28 and PAK4i, both potent PAK4 inhibitors (22). At 10μM Purvalanol A, we observed inhibition of both wildtype PAK4 and PAK4E329K kinase activity (Fig. 4A, lanes 5 and 8). In the presence of 0.5μM Purvalanol A PAK4E329K appeared to exhibit a modest resistance to inhibition, exhibiting moderately elevated substrate phosphorylation in comparison to wildtype PAK4 (Fig. 4A and B, lanes 4 and 7). This effect was also evident in the presence of PAK4i. PAK4E329K substrate phosphorylation in the presence of 1μM PAK4i, in particular, was significantly higher than wildtype PAK4 (Fig 4C lanes 4 and 7 and 4D). Moreover, this effect was not solely due to the incorporation of a point mutation in the kinase domain, as GST-PAK4S474D exhibited no evidence of resistance to PAK4i inhibition (data not shown). Taken together, these data suggest that E329K somatic mutation of the PAK4 kinase domain confers resistance to inhibition with competitive ATP inhibitors. Indeed, IC50 values for PAK4i were calculated as ~0.45μM and ~0.65μM for wildtype PAK4 and PAK4E329K respectively (Figure S2). Whilst mutations conferring resistance to tyrosine kinase inhibitors are well established, less is known about development of resistance to inhibitors of serine/threonine kinases. Girdler et al. recently reported the induction of drug resistant mutations in Aurora kinase by treatment of cells with inhibitors (36). PAK4E329K may therefore prove an important tool in aiding drug design and development, but also highlights the importance of developing multiple and alternative therapeutic strategies (for example allosteric PAK inhibitors (21)) to offset the risks associated with clinical drug resistance.
Supplementary Material
1
2
3
movie1
movie2
Acknowledgements
A.D.W., and work in the laboratories of G. E. J. and C. M. W. is supported by a grant from Cancer Research UK. A.D is supported by a grant from Breast Cancer Campaign. We would like to thank Matthias Krause for gateway destination and pGIPZ-control shRNA vectors. We also thank Mary Holdom for helpful discussion and advice with protein structure analysis and Mike Olsen for practical advice with using LIMKi.
Footnotes
The authors declare there is no conflict of interest.
Author contribution ADW, GEJ and CMW planned the experiments. ADW, AD and CMW conducted the experiments. MH performed migration analysis. ADW, GEJ and CMW wrote the paper. CW and GEJ contributed equally to the paper.
References
- 1.Cecchi F, Rabe DC, Bottaro DP. Targeting the HGF/Met signalling pathway in cancer. Eur J Cancer. 2010;46:1260–1270. doi: 10.1016/j.ejca.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knudsen BS, Woude GV. Showering c-MET-dependent cancers with drugs. Curr opin Genetics Dev. 2008;18:87–96. doi: 10.1016/j.gde.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Lai AZ, Abella JV, Park M. Crosstalk in Met receptor oncogenesis. Trends Cell Biol. 2009;19:542–551. doi: 10.1016/j.tcb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Paliouras GN, Naujokas MA, Park M. Pak4, a novel Gab1 binding partner, modulates cell migration and invasion by the Met receptor. Mol Cell Biol. 2009;29:3018–3032. doi: 10.1128/MCB.01286-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abo A, Qu J, Cammarano MS, Dan C, Fritsch A, Baud V, et al. PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. Embo J. 1998;17:6527–6540. doi: 10.1093/emboj/17.22.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callow MG, Zozulya S, Gishizky ML, Jallal B, Smeal T. PAK4 mediates morphological changes through the regulation of GEF-H1. J Cell Sci. 2005;118:1861–1872. doi: 10.1242/jcs.02313. [DOI] [PubMed] [Google Scholar]
- 7.Wells CM, Whale AD, Parsons M, Masters JR, Jones GE. PAK4: a pluripotent kinase that regulates prostate cancer cell adhesion. J Cell Sci. 2010;123:1663–1673. doi: 10.1242/jcs.055707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Zhang H, Lundin L, Thullberg M, Liu Y, Wang Y, et al. p21-activated Kinase 4 phosphorylation of integrin β5 Ser-759 and Ser-762 regulates cell migration. J Biol Chem. 2010;285:23699–23710. doi: 10.1074/jbc.M110.123497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dan C, Kelly A, Bernard O, Minden A. Cytoskeletal changes regulated by the PAK4 serine/threonine kinase are mediated by LIM kinase 1 and cofilin. J Biol Chem. 2001;276:32115–32121. doi: 10.1074/jbc.M100871200. [DOI] [PubMed] [Google Scholar]
- 10.Soosairajah J, Maiti S, Wiggan O, Sarmiere P, Moussi N, Sarcevic B, et al. Interplay between components of a novel LIM kinase-slingshot phosphatase complex regulates cofilin. Embo J. 2005;24:473–486. doi: 10.1038/sj.emboj.7600543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai FPL, Szczodrak M, Block J, Faix J, Breitsprecher D, Mannherz HG, et al. Arp2/3 complex interactions and actin network turnover in lamellipodia. Embo J. 2008;27:982–992. doi: 10.1038/emboj.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS, et al. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 13.Callow MG, Clairvoyant F, Zhu S, Schryver B, Whyte DB, Bischoff JR, et al. Requirement for PAK4 in the anchorage-independent growth of human cancer cell lines. J Biol Chem. 2002;277:550–558. doi: 10.1074/jbc.M105732200. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Xiao H, Tian Y, Nekrasova T, Hao X, Lee HJ, et al. The pak4 protein kinase plays a key role in cell survival and tumorigenesis in athymic mice. Mol Cancer Res. 2008;6:215–224. doi: 10.1158/1541-7786.MCR-08-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whale A, Hashim FN, Fram S, Jones GE, Wells CM. Signalling to cancer cell invasion through PAK family kinases. Front Biosci. 2011;16:849–864. doi: 10.2741/3724. [DOI] [PubMed] [Google Scholar]
- 16.Parsons DW, Wang TL, Samuels Y, Bardelli A, Cummins JM, DeLong L, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 17.Kimmelman AC, Hezel AF, Aguirre AJ, Zheng H, Paik J, Ying H, et al. Genomic alterations link Rho family of GTPases to the highly invasive phenotype of pancreas cancer. Proc Natl Acad Sci U S A. 2008;105:19372–19377. doi: 10.1073/pnas.0809966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Lock JG, Olofsson H, Kowalewski JM, Teller S, Liu Y, et al. Integrin-mediated cell attachment induces a PAK4-dependent feedback loop regulating cell adhesion through modified integrin αvβ5 clustering and turnover. Mol Biol Cell. 2010;21:3317–3329. doi: 10.1091/mbc.E10-03-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siu MKY, Chan HY, Konga DSH, Wong ESY, Wong OGW, Ngan HYS, et al. p21-activated kinase 4 regulates ovarian cancer cell proliferation, migration, and invasion and contributes to poor prognosis in patients. Proc Natl Acad Sci U S A. 2010;107:18622–18627. doi: 10.1073/pnas.0907481107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eswaran J, Soundararajan M, Knapp S. Targeting group II PAKs in cancer and metastasis. Cancer Metastasis Rev. 2009;28:209–217. doi: 10.1007/s10555-008-9181-4. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Z, Manser E. Do PAKs make good drug targets? Faculty of 1000. 2010;2(70) doi: 10.3410/B2-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray BW, Guoa C, Piraino J, Westwick JK, Zhang C, Lamerdin J, et al. Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. Proc Natl Acad Sci U S A. 2010;107:9446–9451. doi: 10.1073/pnas.0911863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finkelstein E, Chang W, Chao P-HG, Gruber D, Minden A, Hung CT, et al. Roles of microtubules, cell polarity and adhesion in electric-field-mediated motility of 3T3 fibroblasts. J Cell Sci. 2004;117:1533–1545. doi: 10.1242/jcs.00986. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed T, Shea K, Masters JR, Jones GE, Wells CM. A PAK4-LIMK1 pathway drives prostate cancer cell migration downstream of HGF. Cell Signal. 2008;20:1320–1328. doi: 10.1016/j.cellsig.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 25.Scott RW, Hooper S, Crighton D, Li A, König I, Munro J, et al. LIM kinases are required for invasive path generation by tumor and tumor-associated stromal cells. J Cell Biol. 2010;191:169–185. doi: 10.1083/jcb.201002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshioka K, Foletta V, Bernard O, Itoh K. A role for LIM kinase in cancer invasion. Proc Natl Acad Sci U S A. 2003;100:7247–7252. doi: 10.1073/pnas.1232344100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baldassa S, Calogero AM, Colombo G, Zippel R, Gnesutta N. N-Terminal interaction domain implicates PAK4 in translational regulation and reveals novel cellular localization signals. J. Cell. Physiol. 2010;224:722–733. doi: 10.1002/jcp.22172. [DOI] [PubMed] [Google Scholar]
- 28.Eswaran J, Lee WH, Debreczeni JE, Filippakopoulos P, Turnbull A, Fedorov O, et al. Crystal Structures of the p21-activated kinases PAK4, PAK5, and PAK6 reveal catalytic domain plasticity of active Group II PAKs. Structure. 2007;15:201–213. doi: 10.1016/j.str.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant BD, Hemmer W, Tsigelny I, Adams JA, Taylor SS. Kinetic analyses of mutations in the glycine-rich loop of cAMP-dependent protein kinase. Biochemistry. 1998;37:7708–7715. doi: 10.1021/bi972987w. [DOI] [PubMed] [Google Scholar]
- 30.Barouch-Bentov R, Che J, Lee CC, Yang Y, Herman A, Jia Y, et al. A conserved salt bridge in the G loop of multiple protein kinases is important for catalysis and for in vivo Lyn function. Molecular Cell. 2009;33:43–52. doi: 10.1016/j.molcel.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griswold IJ, MacPartlin M, Bumm T, Goss VL, O’Hare T, Lee KA, et al. Kinase domain mutants of Bcr-Abl exhibit altered transformation potency, kinase activity, and substrate utilization, irrespective of sensitivity to Imatinib. Mol Cell Biol. 2006;26:6082–6093. doi: 10.1128/MCB.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto M, Kurosu T, Kakihana K, Mizuchi D, Miura O. The two major imatinib resistance mutations E255K and T315I enhance the activity of BCR/ABL fusion kinase. Biochem Biophys Res Commun. 2004;319:1272–1275. doi: 10.1016/j.bbrc.2004.05.113. [DOI] [PubMed] [Google Scholar]
- 33.Skaggs BJ, Gorre ME, Ryvkin A, Burgess MR, Xie Y, Han Y, et al. Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR-ABL mutants. Proc Natl Acad Sci U S A. 2006;103:19466–19471. doi: 10.1073/pnas.0609239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 35.Pavlovsky C, Kantarjian H, Cortes JE. First-line therapy for chronic myeloid leukemia: Past, present, and future. Am J Hematol. 2009;84:287–293. doi: 10.1002/ajh.21380. [DOI] [PubMed] [Google Scholar]
- 36.Girdler F, Sessa F, Patercoli S, Villa F, Musacchio A, Taylor S, et al. Molecular basis of drug resistance in aurora kinases. Chem. Biol. 2008;15:552–562. doi: 10.1016/j.chembiol.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 37.Zicha D, Dunn G, Jones G. Analyzing chemotaxis using the Dunn direct-viewing chamber. Methods Mol Biol. 1997;75:449–57. doi: 10.1385/0-89603-441-0:449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
movie1
movie2