Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation (original) (raw)
. Author manuscript; available in PMC: 2013 Jul 3.
Published in final edited form as: Nature. 2010 Sep 30;467(7315):591–595. doi: 10.1038/nature09385
Abstract
During immune responses, antibodies are selected for their ability to bind to foreign antigens with high affinity, in part by their ability to undergo homotypic bivalent binding. However, this type of binding is not always possible. For example, the small number of gp140 glycoprotein spikes displayed on the surface of the human immunodeficiency virus (HIV disfavours homotypic bivalent antibody binding1–3. Here we show that during the human antibody response to HIV, somatic mutations that increase antibody affinity also increase breadth and neutralizing potency. Surprisingly, the responding naive and memory B cells produce polyreactive antibodies, which are capable of bivalent heteroligation between one high-affinity anti-HIV-gp140 combining site and a second low-affinity site on another molecular structure on HIV. Although cross-reactivity to self-antigens or polyreactivity is strongly selected against during B-cell development4, it is a common serologic feature of certain infections in humans, including HIV, Epstein-Barr virus and hepatitis C virus. Seventy-five per cent of the 134 monoclonal anti-HIV-gp140 antibodies cloned from six patients5 with high titres of neutralizing antibodies are polyreactive. Despite the low affinity of the polyreactive combining site, heteroligation demonstrably increases the apparent affinity of polyreactive antibodies to HIV.
Although most B cells in the nascent repertoire are poly- or self-reactive, only 5% of the B cells in the mature naive B cell compartment retain this unusual form of reactivity4,6.
Surprisingly, however, B cells can re-acquire poly- and self-reactivity during the germinal centre reaction7, and self-reactivity is also associated with serum antibody responses in several persistent infections, including vaccinia virus in mice8 and HIV, Epstein-Barr virus and hepatitis C virus in humans. This strange though relatively common feature of antibodies was first documented in studies of anti-hapten-specific myeloma proteins over three decades ago9; however, its role (if any) in the antibody response to a specific antigen has yet to be explored.
One possible function for polyreactivity would be to increase antibody affinity for a pathogen where simple homotypic bivalent ligation is not feasible. To test this idea we studied antibody response to HIV because its gp140 surface spike is present at a very low density of only 10–15 viral spikes per virion2,3,10,11. Homotypic bivalent binding is therefore unlikely, but an anti-HIV antibody might increase its overall apparent affinity to the virus by heterogenous ligand binding or heteroligation. In this model, one combining site would bind with high affinity to gp140, whereas the other would bind with low affinity to any of several other ligands on the viral surface.
We studied 134 unique anti-gp140 and 52 control non-gp140 reactive antibodies5 (Supplementary Table 1). All of the anti-HIV antibodies were compared with previously reported IgG antibodies cloned from the memory B cells of uninfected controls7, non-gp140 binding antibodies from the same patients5, and previously characterized broadly neutralizing anti-HIV antibodies 2F5, 4E10, 2G12 and b12.
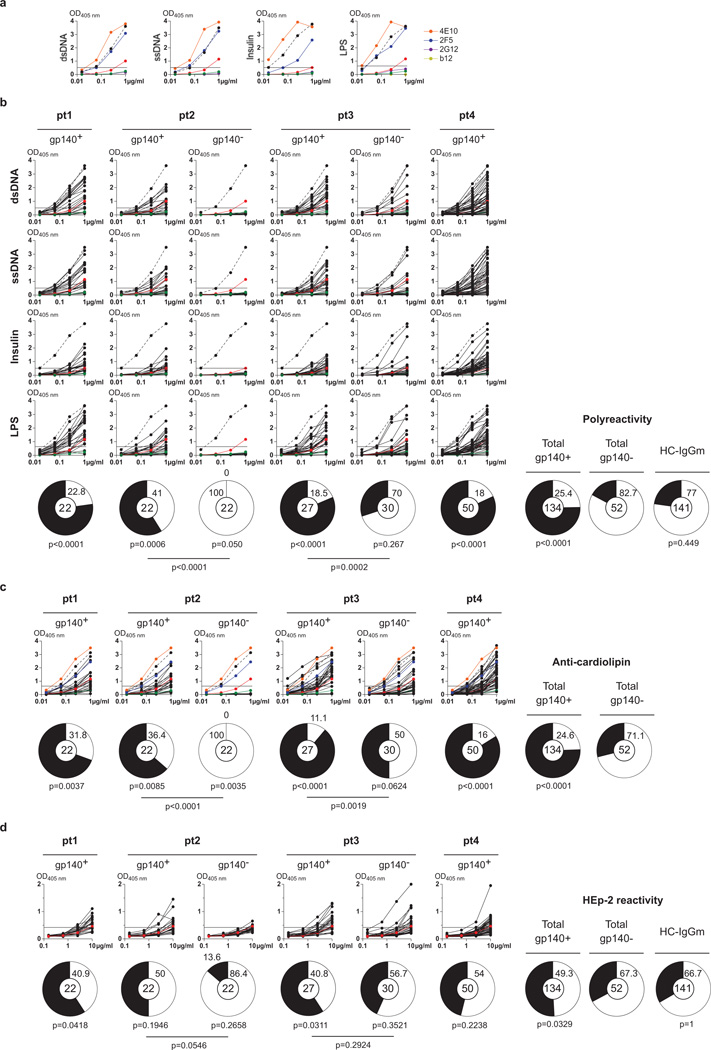
To determine whether anti-HIV antibodies are polyreactive, we measured binding to single-stranded DNA, double-stranded DNA, lipopolysaccharide, insulin and keyhole limpet haemocyanin (KLH) by enzyme-linked immunosorbent assay (ELISA)4 (Fig. 1a, b, Supplementary Figs 1a and 2 and Supplementary Table 1). Seventy-five per cent of all anti-gp140 memory antibodies were polyreactive (P < 0.0001 compared with uninfected controls, Fig. 1b). Although the frequency of polyreactive antibodies varied between patients (from 59 to 82%), the difference between gp140 binding and non-binding antibodies was highly significant in all cases, whether compared with all antibodies or with those within an individual (Fig. 1b and Supplementary Fig. 1a). Cardiolipin binding was highly correlated with polyreactivity (Fig. 1c and Supplementary Figs 1b and 3, P < 0.0001 compared with control Fig. 1c and Supplementary Fig. 4). Similarly, anti-gp140 antibodies were more reactive in the HEp-2 ELISA than matched or historical controls (51% anti-gp140+ versus 33% gp140−, P = 0.033, Fig. 1d and Supplementary Fig. 1c). We conclude that the majority of all anti-gp140 antibodies studied are polyreactive and more likely to bind to cardiolipin and self-antigens in HEp-2 cell lysates compared with control antibodies.
Figure 1. Polyreactivity, anti-cardiolipin and HEp-2 ELISAs.
a, ELISAs measuring the reactivity of 4E10 (ref. 23), b12 (ref. 24), 2F5 (ref. 25) and 2G12 (ref. 26) against double-stranded DNA, single-stranded DNA, insulin and lipopolysaccharide. Dotted lines represent the positive control antibody ED38 (ref. 27). Horizontal lines show cut-off OD405 nm for positive reactivity. Green and red lines show the negative and low positive control antibodies, respectively4,28. b, As in a but for IgG antibodies cloned from gp140+ cells from patients 1–4 and for gp140− cells from patients 2 and 3. Pie charts summarize the frequency of polyreactive (black) and non-polyreactive (white) IgG+ clones, and for all pooled gp140+, gp140− and previously published control IgG antibodies (HC-IgGm)7 (right). P values are in comparison with pooled gp140− IgGs or between gp140+ and gp140− for patients 2 and 3 as indicated. c, Graphs show anti-cardiolipin ELISAs for the same antibodies as in b. Orange and blue lines show the positive control antibodies 4E10 and 2F5 (refs 23, 25), respectively. Pie charts summarize the frequency of cardiolipin reactive (black) and non-reactive (white) IgGs. P values are in comparison with pooled gp140− IgGs (right) or between gp140+ and gp140− for patients 2 and 3 as indicated. d, Same as b except for HEp-2 ELISAs.
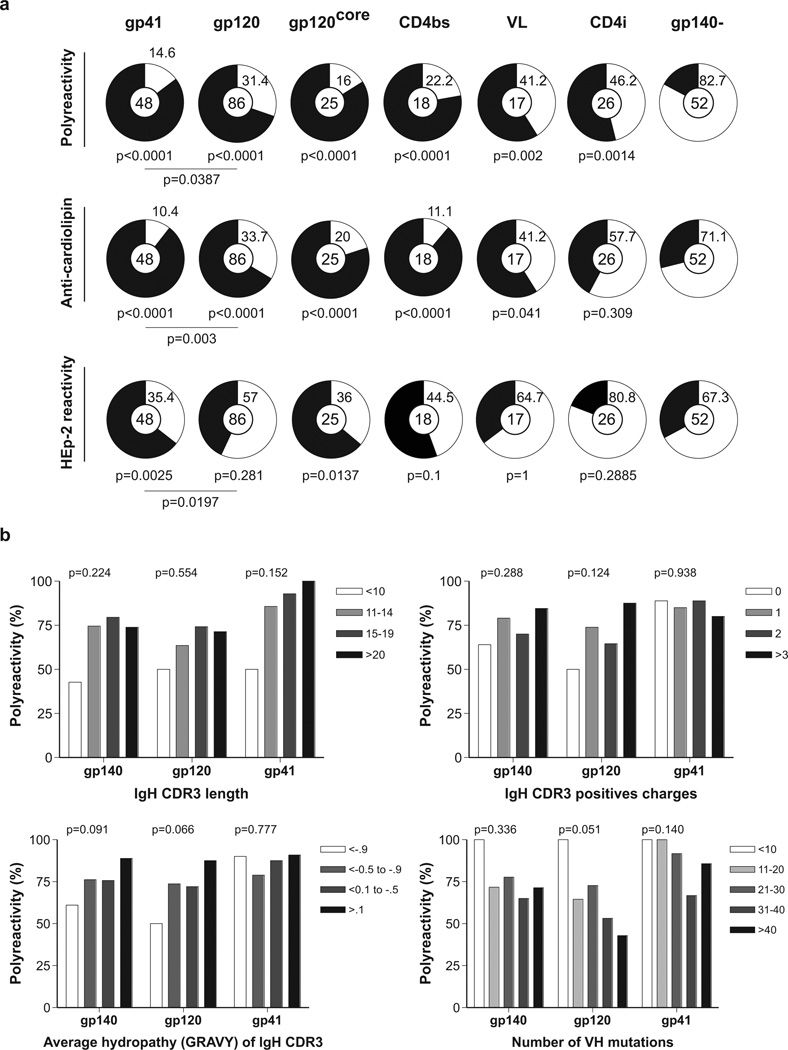
To determine whether polyreactivity, cardiolipin or HEp-2 reactivity is associated with binding to specific epitopes on gp140, we segregated the ELISA results according to reactivity with gp120, gp41, the CD4-binding site (CD4bs), the CD4-induced site (CD4i), the variable loops and the gp120 core antigen (gp120core5). Anti-gp41 antibodies were more reactive than anti-gp120 in all assays (Fig. 2a). Among the anti-gp120s, antibodies to gp120core, CD4bs and the variable loops were more likely to be poly-, cardiolipin- or HEp-2 reactive than antibodies to CD4i (Fig. 2a).
Figure 2. IgH chain gene features and reactivity.
a, Pie charts summarize polyreactivity, anti-cardiolipin and HEp-2 reactivity of gp140+ antibodies grouped by antibody specificity for gp41, gp120, gp120core, CD4bs, variable loops and CD4i. Reactive (black) and non-reactive (white) IgGs are shown; numbers in the centre indicate number of antibodies tested. P values are in comparison with gp140− IgGs unless otherwise indicated. b, Polyreactivity of all gp140-, gp120- and gp41-specific IgG antibodies grouped according to IgH CDR3 length, IgH CDR3-positive charges, IgH CDR3 hydropathy (GRAVY) or number of VH mutations.
Several features of antibodies have been associated with polyreactivity, including the length, charge and hydrophobicity of the IgH complementarity-determining region 3 (CDR3). However, these features have never been examined in the context of polyreactive antibodies that specifically recognize a particular antigen12,13. We found that none of these features were highly correlated with polyreactivity in the anti-gp140 antibodies (Fig. 2b).
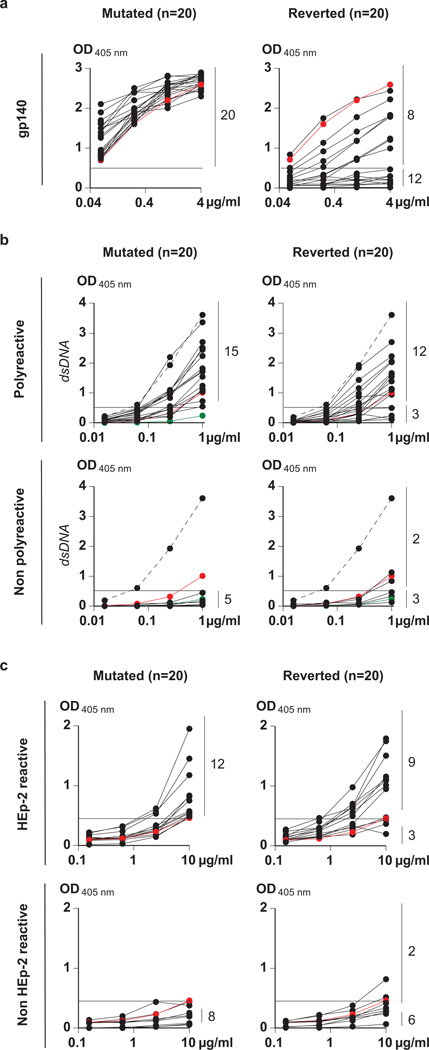
Anti-gp140 antibodies are highly somatically mutated5. To examine the relationship between somatic mutation and antibody specificity, we reverted the somatic mutations in 20 randomly selected anti-gp140 antibodies and tested them for specific reactivity and polyreactivity. Gp140 binding was undetectable in 12 out of 20 reverted antibodies (Fig. 3a). The eight antibodies that retained gp140 binding showed lower binding activity in ELISA, consistent with a loss in binding affinity (Fig. 3a, Supplementary Fig. 5 and Supplementary Table 2). When tested for neutralizing activity, five of the eight reverted antibodies that retained residual gp140 binding showed complete loss of activity (Supplementary Tables 2 and 3). The other three showed loss of breadth and potency (Supplementary Table 3). We conclude that somatic hypermutation enhances the affinity and neutralizing activity of the anti-gp140 antibodies cloned from memory B cells of the selected patients with high serum titres of broadly neutralizing antibodies.
Figure 3. Reactivity of reverted antibodies.
a, ELISAs against gp140 for 20 selected polyreactive and non-polyreactive antibodies (Supplementary Table 2), before (mutated) and after (reverted) reversion. The red line indicates the reactivity of the control b12 antibody24. b, Anti-double-stranded DNA ELISAs (as example for polyreactivity) for the mutated and reverted antibodies that were initially polyreactive (top) or non-polyreactive (bottom). Numbers on the right indicate antibodies in reactive or non-reactive categories before and after reversion. c, As in b except for HEp-2 reactivity. Positive and negative control antibodies are as in Fig. 1.
To examine the relationship between polyreactivity and somatic mutation, we measured the polyreactivity of the reverted antibodies (Supplementary Table 2). Fifteen of the 20 anti-gp140 somatically mutated antibodies were polyreactive before reversion (Fig. 3b, c). After reversion, 12 were still polyreactive (Fig. 3b, 3c and Supplementary Figs 2d and 5). Of the five that were not polyreactive before reversion, two were polyreactive after reversion. Thus 70% of the antibodies tested were polyreactive before entering the germinal centre as opposed to 5% in the overall naive repertoire4,6. We conclude that polyreactive antibodies are positively selected during the anti-HIV antibody response even before the germinal centre reaction and before somatic mutation; this reactivity is preferentially retained throughout affinity maturation despite hypermutation.
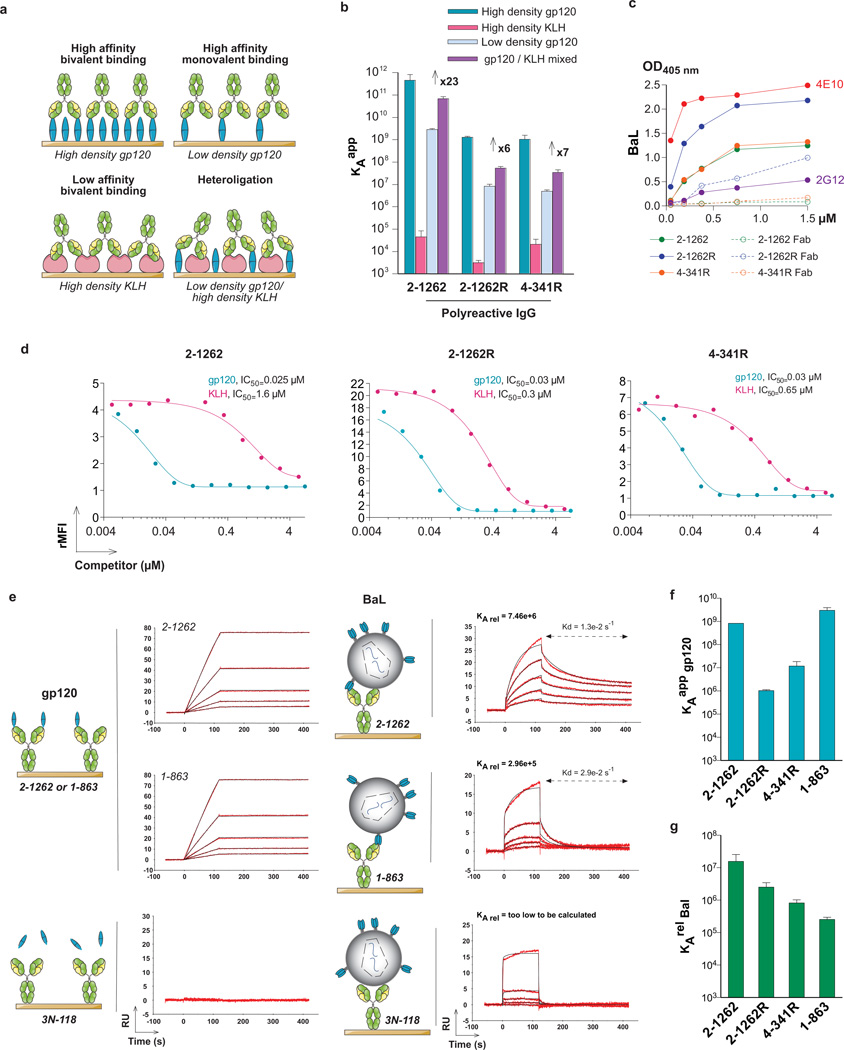
Polyreactive anti-gp140 antibodies would be positively selected if they bind more strongly to HIV than non-polyreactive antibodies (Fig. 4a and Supplementary Data 1). To determine whether heteroligation increases the affinity of anti-HIV antibodies we performed surface plasmon resonance (SPR) experiments (Fig. 4, Supplementary Figs 6 and 7 and Supplementary Table 4). We selected KLH to use as a representative low-affinity ligand because its degree and spectrum of reactivity in ELISA was intermediate in the polyreactivity ELISAs and because it is readily coupled to the SPR chips with gp120 (Supplementary Figs 2 and 6a). Chips bearing high densities of gp120 were designed to allow for bivalent antibody binding. In contrast, chips bearing low densities of gp120 should only allow for monovalent antibody binding. Finally, mixed chips derivatized with a combination of a small amount of gp120 and high-density KLH were designed to evaluate heteroligation by high-affinity monovalent binding to gp120 and simultaneous low-affinity binding to KLH (Fig. 4a and Supplementary Fig. 8).
Figure 4. Heteroligation.
a, Representation of bivalent and monovalent homotypic binding versus heteroligation on high- and low-density gp120, high-density KLH and mixed chips. b, Apparent _K_a (_K_aapp) as measured by SPR on chips derivatized with high or low concentrations of gp120 or high concentrations of KLH, or a combination of low-gp120 and high-KLH. Numbers indicate the fold enhancement that can be attributed to heteroligation. See also Supplementary Table 4. Error bars, s.e.m. c, ELISA binding studies for IgG and corresponding Fabs on HIV-BaL22. d, Solution competition-binding assay for fluorescent BaL virus22 using gp120 or KLH as competitors. Coloured lines show fitted curves. The y axis indicates the relative mean fluorescence index (rMFI), the x axis the competitor concentration in micromoles. Gp120 and KLH concentrations required to achieve 50% binding inhibition (IC50) are indicated. e, SPR sensorgrams for binding to BaL virus22 or gp120 protein to IgG antibody-immobilized sensor chips as illustrated by the schematic. RU, response units. f, Apparent _K_a (_K_aapp) for gp120 protein as measured by SPR. Error bars, s.e.m. g, As f but for the relative _K_a to BaL virus (_K_arelBaL).
The three polyreactive and two non-polyreactive antibodies tested bound to the high-density gp120 chips, with values of high-affinity _K_aappranging from 5.6 × 108 to 4.5 × 1011 M−1 and, as expected, the reverted antibody 2–1262R bound with lower affinity to gp120 at higher density than its mutated counterpart (Fig. 4b and Supplementary Table 4, _K_aapp 1.3 × 109 versus 4.5 × 1011 M−1, respectively). Also as expected, the values of _K_aapp measured on the chips derivatized with the low-density gp120 were significantly lower than on the high-density gp120 chips (Fig. 4b, Supplementary Fig. 7a and Supplementary Table 4) and there was no binding by the polyreactive anti-gp41 controls to the low-density gp120-derivatized surface (Supplementary Fig. 6d). The three polyreactive antibodies bound to KLH with low affinities, with values of _K_aapp ranging from 3.1 × 103 to 4.6 × 104 M−1, whereas binding by the non-polyreactive antibodies was too low to calculate. Interestingly, the combination of low amounts of gp120 with high-density KLH on a mixed surface resulted in an increase in polyreactive antibody affinity (_K_aapp) of up to nearly 25-fold over the gp120 alone at low density (Fig. 4b). In contrast there was no such effect for the non-polyreactive anti-gp120 and polyreactive but non-gp120-reactive controls (Supplementary Fig. 7a and Supplementary Table 4). The net values of _K_aapp produced by heteroligation were intermediate between monovalent and divalent homotypic binding to gp120 (Supplementary Table 4). Fabs did not show enhanced binding on the mixed chips (Supplementary Table 5 and Supplementary Fig. 7b, c). In addition, polyreactive but not non-polyreactive antibodies were inhibited by KLH when the antigen was present at low concentrations (Supplementary Fig. 9). Finally, the intact antibodies bound strongly to the virus in ELISA, whereas binding by Fab fragments of the same antibodies in ELISA was difficult to detect (Fig. 4c and Supplementary Fig. 10a). As expected, binding to the virus was inhibited by KLH at concentrations ranging from one to two orders of magnitude higher than gp120, and higher concentrations of KLH were required to inhibit binding of anti-HIV antibodies that showed higher levels of polyreactivity (Fig. 4d and Supplementary Figs 6a and 10b, c).
To determine whether polyreactivity enhances antibody binding to HIV, we performed SPR experiments comparing soluble gp120 and the virus. We included 1–863, which is not polyreactive, but like 2–1262 also binds to the CD4-binding site5, and 3N-1185, which does not bind to gp120 but is polyreactive (Supplementary Table 1 and Supplementary Figs 6a and 7a). The highest affinity for soluble gp120 was shown by 1–863, followed in order by 2–1262, 4–341R and 2–1262R (Fig. 4f, Supplementary Fig. 10d and Supplementary Table 5). In striking contrast, 2–1262 had the highest relative affinity for the virus, followed by 4–341R, 2–1262R and finally the non-polyreactive 1–863 antibody (Fig. 4e, g, Supplementary Fig. 10d and Supplementary Table 5). Also consistent with heteroligation, competition SPR experiments using the virion and HIV-mimic liposomes showed enhanced antibody dissociation from the virus in the presence of the liposomes (Supplementary Fig. 11). We conclude that polyreactivity enhances the relative affinity of anti-gp120 antibodies to the HIV virion.
Antibodies specific for conserved regions of the HIV spike protein have the ability to neutralize the virus and prevent infection in non-human primates14,15, and it is generally agreed that such antibodies would be an important component of any successful anti-HIV vaccine16–18. Our data show that somatic hypermutation is essential to increased antibody affinity, neutralizing potency and breadth.
Antibodies are traditionally considered to engage antigen in a monospecific bivalent manner, which enhances apparent affinity by decreasing the rate of dissociation from the ligand. Our experiments elucidate an unanticipated mechanism for increasing antibody affinity in cases when homotypic bivalent binding is not possible. Heteroligation appears to be a conserved feature of immune recognition, because a related mechanism is used by the T-cell receptor to facilitate signal transduction19. Although heteroligation increases apparent affinity and is positively selected during the immune response to HIV, this feature is not required for neutralization; for example b12 and 2G12 show little if any polyreactivity20 (Fig. 1a and Supplementary Fig. 6a).
Heteroligation improves antibody affinity when homotypic bivalent binding is disfavoured, but it may be particularly difficult to achieve because only a small fraction of the B cells in the naive repertoire are polyreactive4. This paucity of polyreactive B cells in the naive repertoire may in part explain the delayed and only occasional development of high-affinity broadly neutralizing antibodies during HIV infection. Finally, the existence of heteroligation and its contribution to anti-HIV antibody affinity suggests that mimicking the low-density viral antigens encountered during natural infection should be considered as a means to enhance anti-HIV immunization.
METHODS SUMMARY
Antibody reactivity
Anti-gp140, reverted anti-gp140 and control antibodies cloned from the peripheral blood IgG memory B cells from six HIV-infected patients5 were tested for polyreactivity, HEp-2 cells and other antigens by ELISA as previously described4,21.
Antibody heteroligation analyses
Antibody affinities to gp120, KLH or both were measured by SPR using a Biacore T100 (Biacore). Antibody binding to HIV virus was tested by ELISA using BaL virus as antigen and by bead-based flow cytometry assay using a green fluorescent protein (GFP)–Gag BaL virus22. Competition experiments were performed with different concentrations of purified YU2-gp120 and KLH proteins. The relative affinity of the selected IgG antibodies to BaL virus22 was measured by SPR using IgG-immobilized surfaces.
METHODS
Antibodies
Anti-gp140 and control antibodies were produced from peripheral blood IgG memory B cells isolated from six HIV-infected patients as described5,29. Twenty randomly selected anti-gp140 antibodies were reverted to their germline sequence by PCR and expressed as previously described7,28.
Fabs
Fab fragments were produced from mutated and reverted anti-gp140 IgG antibodies by papain digestion using Fab preparation kit (Pierce). Their purity was checked on R-250 blue-stained 4–12% NuPAGE gel (Invitrogen).
Serum IgGs
IgGs were isolated from serum from HIV, and from patients with systemic lupus erythematosus and healthy humans5,21 using Protein G sepharose beads (GE Healthcare) according to the manufacturer’s instructions.
ELISAs
Antibodies and serum IgGs were tested for polyreactivity, cardiolipin, HEp-2 cell, KLH (Sigma), gp140 and gp120 reactivity by ELISA as previously described4,21. Antibodies were considered polyreactive when they recognized at least two structurally different antigens out of the four tested: single-stranded DNA, double-stranded DNA, insulin and lipopolysaccharide4. Threshold values for reactivity were determined by using control antibodies mGO53 (negative), eiJB40 (low positive) and ED38 (high positive) for polyreactivity ELISAs. The polyreactive ELISA used to detect antibody binding to various antigens is a sensible assay that cannot allow an absolute measurement of antibody affinities to the substances tested. Additional positive and negative control sera for HEp-2 reactivity were used as per manufacturer’s suggestion (INOVA Diagnostics)4,27.
Competition ELISA
High-binding ELISA plates (Costar) were coated with low- and high-density gp120 at a concentration of 0.5 µg ml−1 and 5 µg ml−1, respectively. Plates were blocked with 2% BSA-0.05% Tween 20-1 mM EDTA-PBS and incubated for 2 h with IgG antibodies at 1 µg ml−1 in 1:2 serially diluted KLH-containing PBS (with a KLH concentration from 0.09 to 5.55 µM). Plates were developed as previously described4,21. Data shown are representative for at least three independent experiments.
Neutralization assay
Virus neutralization was measured using a TZM-bl cell assay as previously described5.
Liposomes
HIV-mimic liposomes were produced by mixing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS), brain sphingomyelin and cholesterol dissolved in chloroform (Avanti Polar Lipids) at a molar ratio of 9.35:19.25:8.25:18.15:45.00 (ref. 29). The mixture was dried in the fume hood under a gentle stream of nitrogen and then by a high vacuum overnight. The lipid pellet was suspended in PBS above the transition temperature for 1 h by vigorous mixing and five repeated freeze/thaw cycles. The suspension was sonicated in a sonicator bath (two cycles of 15 min with a 10-s pulse and a 15-s pulse-off) and passed through a 100-nm polycarbonate membrane (15 times) using a mini-extruder (Avanti Polar Lipids). Before SPR experiments, the liposome suspension was sonicated again (one cycle as described above) above the transition temperature.
Surface plasmon resonance
All experiments were performed with a Biacore T100 (Biacore) in HBS-EP+ running buffer (Biacore) at 25 °C. Samples were analysed in kinetic experiments performed at least in duplicate. KLH, YU-2 gp120 and IgGs were immobilized on CM5 chips (Biacore) by amine coupling at pH 4.5. The concentration of gp120 was 0.0125 mg ml−1 or 0.125 mg ml−1 for low and high immobilization levels resulting in 100 RU and 10,000 RU, respectively. KLH was immobilized at 0.25 mg ml−1, resulting in 15,000 RU for high-density KLH surface. For the mixed surface, gp120 was at 0.0125 mg ml−1 and KLH at 0.25 mg ml−1 KLH, resulting in a total immobilization level of 15,000 RU. IgGs were immobilized at 0.1 mg ml−1, resulting in 5,000 RU. For kinetic measurements on the gp120- and KLH-derivatized chips, IgGs or Fabs were injected through flow cells in at least five different concentrations (1,379 nM, 689.7 nM, 344.8 nM, 172.4 nM, 86.21 nM, 43.1 nM, 21.55 nM, 10.78 nM for IgGs, and 1000 nM, 500 nM, 250 nM, 125 nM, 62.5 nM for Fabs) in HBS-EP+ running buffer (Biacore) at flow rates of 100 µl min−1 with 2-min association and 5-min dissociation. For the HIV-binding analyses on the IgG-immobilized surfaces, purified BaL virus22 was injected through flow cells at a p24 concentration of 78.1 nM (with four successive 1:2 dilutions) in HBS-EP+ running buffer at a flow rate of 40 µl min−1 with 2 min association and 5 min dissociation. The purified gp120 at 43.10 nM used as control and liposomes at 1,333 µg min−1 (with four successive 1:2 dilutions) were injected through flow cells under the same conditions. Competition experiments for BaL virus binding to the IgG-immobilized surfaces using liposomes were performed using the same experimental settings as for the BaL virus binding analyses except that increasing concentrations of liposomes (from 20.8 to 1333 µg ml−1) were mixed with the purified virus (p24 at 78.1 nM). The sensor surface was regenerated between each experiment with a 30-s injection of 10 mM glycine-HCl pH 2.5 at a flow rate of 50 µl min−1. Off rate (Kd (s−1)), on rate (Ka (M−1 s−1 and RU−1 s−1 for bivalent binding Ka2)) and binding constants (_K_D (M) or _K_A (M−1 and RU−1 for bivalent binding _K_A2)) were calculated after subtraction of backgrounds (binding to a control flow cell and signal of the HBS-EP+ running buffer) using Biacore T100 Evaluation software using the kinetic analysis and the bivalent model (IgGs on high-density gp120 and KLH surfaces), 1:1 binding model (IgGs and Fabs on low-density gp120 surface) or heterogeneous model (IgGs on mixed surface). The _K_A1 value was used as an estimation of apparent _K_A (_K_Aapp) because _K_A1 represents most of the binding31. _K_D of the Fab binding to KLH and gp120/KLH mixed surfaces were determined by Scatchard analysis using nonlinear regression (one-site binding model). The relative immobilization level of gp120 on the mixed versus low-density gp120 surfaces was estimated by the specific binding of the non-polyreactive anti-gp120 1–863 antibody to gp120 ligand on both chips. Maximal binding capacity (_B_max) of 1–863 to each surface was determined by Scatchard linear plotting, and the relative immobilization level of gp120 protein on the mixed surface was given by the _B_max ratio ((_B_max low gp120/high KLH / _B_max low gp120) × 100) (Supplementary Fig. 8). Fold increase attributed to heteroligation was calculated using the following formula: _K_Aapp mixed low gp120/high KLH / (_K_Aapp KLH + _K_Aapp low gp120). To compare the antibody affinities to BaL virus, we calculated a relative _K_A (_K_Arel BaL) using an arbitrary concentration of 100 nm of virus for 43.1 mM of p24 protein. For the analysis of the gp120 binding (secondary binding) to the IgG antibodies bound to gp120 immobilized on the low- and high-density chips (100 and 10,000 RU, respectively) (primary binding), the antibodies were injected through flow cells (IgG concentrations indicated above) in HBS-EP+ running buffer at flow rates of 30 µl min−1 with 2-min association and 160-s dissociation. Purified YU-2 gp120 (at a final concentration of 450 nM) was then injected in HBS-EP+ running buffer at a flow rate of 30 µl min−1 with 2-min association. The sensor surface was regenerated as described above.
HIV-binding antibody assays
BaL and GFP–Gag-labelled BaL viruses were produced, purified and concentrated at 2–2.5 µg ml−1 of p24 protein as previously described22. BaL-strain-coated ELISA plates were incubated with IgG and Fab at 1.5 µM and four consecutive 1:2 dilutions. IgG and Fab bindings were detected with peroxydase-conjugated goat anti-human IgG F(ab′)2 antibodies (Jackson ImmunoResearch).
Using an HIV bead-based flow cytometry assay, varying concentrations of YU2-gp120 and KLH were used to compete for the antibody binding (IgG concentration of 25 nm) to GFP–Gag BaL virus. GFP–HIV-antibody complexes bound to human IgG1 capture beads (BD Pharmingen) were detected by GFP fluorescence emission using a FACS Calibur flow cytometer (Becton Dickinson). Relative mean fluorescence index was calculated by dividing the sample mean fluorescence index by the mean fluorescence index given by negative control mGO53. Data shown are representative for at least two independent experiments.
Statistics
P values for Ig gene repertoire analyses, analysis of lengths, positive charges, hydropathy of IgH CDR3 and antibody reactivity were calculated by 2 × 2 or 2 × 5 Fisher’s exact test. Polyreactivity and anti-cardiolipin ELISA OD405 nm values and number of virus strains neutralized and off-target reactivity were compared using Spearman’s correlation test.
Supplementary Material
01
Acknowledgements
We thank J. R. Mascola and R. T. Wyatt for discussion and supplying gp140 and gp120 proteins. This research was supported by the Rockefeller University, the National Institutes of Health (NIH 1 PO1 AI081677), the International AIDS Vaccine Initiative and the Bill and Melinda Gates Foundation. T.J.H. was supported by the National Institutes of Health (R01 AI047770). M.J.Z. and H.W. were supported the German Research Foundation (GRK1121). B.D.W. and M.C.N. are Howard Hughes Medical Institute investigators.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
The authors declare no competing financial interests.
References
- 1.Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein JS, et al. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc. Natl Acad. Sci. USA. 2009;106:7385–7390. doi: 10.1073/pnas.0811427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wardemann H, et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 5.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 6.Tsuiji M, et al. A checkpoint for autoreactivity in human IgM+ memory B cell development. J. Exp. Med. 2006;203:393–400. doi: 10.1084/jem.20052033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiller T, et al. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26:205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zinkernagel RM, et al. Virus-induced autoantibody response to a transgenic viral antigen. Nature. 1990;345:68–71. doi: 10.1038/345068a0. [DOI] [PubMed] [Google Scholar]
- 9.Michaelides MC, Eisen HN. The strange cross-reaction of menadione (vitamin K3) and 2,4-dinitrophenyl ligands with a myeloma protein and some conventional antibodies. J. Exp. Med. 1974;140:687–702. doi: 10.1084/jem.140.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chertova E, et al. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 2002;76:5315–5325. doi: 10.1128/JVI.76.11.5315-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlsson GB, Gao F, Robinson J, Hahn B, Sodroski J. Increased envelope spike density and stability are not required for the neutralization resistance of primary human immunodeficiency viruses. J. Virol. 1996;70:6136–6142. doi: 10.1128/jvi.70.9.6136-6142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguilera I, Melero J, Nunez-Roldan A, Sanchez B. Molecular structure of eight human autoreactive monoclonal antibodies. Immunology. 2001;102:273–280. doi: 10.1046/j.1365-2567.2001.01159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ichiyoshi Y, Casali P. Analysis of the structural correlates for antibody polyreactivity by multiple reassortments of chimeric human immunoglobulin heavy and light chain V segments. J. Exp. Med. 1994;180:885–895. doi: 10.1084/jem.180.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibata R, et al. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nature Med. 1999;5:204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 15.Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Natl. Rev. 2004;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlsson Hedestam GB, et al. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nature Rev. Microbiol. 2008;6:143–155. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- 17.Mascola JR. HIV/AIDS: allied responses. Nature. 2007;449:29–30. doi: 10.1038/449029a. [DOI] [PubMed] [Google Scholar]
- 18.Sather DN, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 2009;83:757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogsgaard M, et al. Agonist/endogenous peptide-MHC heterodimers drive T cell activation and sensitivity. Nature. 2005;434:238–243. doi: 10.1038/nature03391. [DOI] [PubMed] [Google Scholar]
- 20.Haynes BF, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 21.Yurasov S, et al. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J. Exp. Med. 2005;201:703–711. doi: 10.1084/jem.20042251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Julie IJ, et al. Modulation of viscoelasticity and HIV transport as a function of pH in a reversibly crosslinked hydrogel. Adv. Funct. Mater. 2009;19:2969–2977. doi: 10.1002/adfm.200900757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchacher A, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 24.Burton DR, et al. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl Acad. Sci. USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muster T, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trkola A, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meffre E, et al. Surrogate light chain expressing human peripheral B cells produce self-reactive antibodies. J. Exp. Med. 2004;199:145–150. doi: 10.1084/jem.20031550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiller T, et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheid JF, et al. A method for identification of HIV gp140 binding memory B cells in human blood. J. Immunol. Methods. 2009;343:65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alam SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc. Natl Acad. Sci. USA. 2010;107:5972–5977. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheskis B, Freedman LP. Modulation of nuclear receptor interactions by ligands: kinetic analysis using surface plasmon resonance. Biochemistry. 1996;35:3309–3318. doi: 10.1021/bi952283r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01