Glycan-receptor specificity as a useful tool for characterization and surveillance of influenza A virus (original) (raw)
. Author manuscript; available in PMC: 2015 Nov 1.
Published in final edited form as: Trends Microbiol. 2014 Aug 6;22(11):632–641. doi: 10.1016/j.tim.2014.07.002
Abstract
Influenza A viruses are rapidly evolving pathogens with the potential for novel strains to emerge and result in pandemic outbreaks in humans. Some avian-adapted subtypes have acquired the ability to bind to human glycan receptors and cause severe infections in humans but have yet to adapt to and transmit between humans. The emergence of new avian strains and their ability to infect humans has confounded their distinction from circulating human virus strains through linking receptor specificity to human adaptation. Herein we review the various structural and biochemical analyses of influenza hemagglutinin–glycan receptor interactions. We provide our perspectives on how receptor specificity can be used to monitor evolution of the virus to adapt to human hosts so as to facilitate improved surveillance and pandemic preparedness.
Keywords: Influenza, Hemagglutinin, Glycan, Specificity, Topology, Network
Glycans as host receptors for influenza A viruses
Influenza A, a zoonotic disease, represents a substantial public health burden, especially in the case of epidemic or pandemic outbreaks [1, 2]. Influenza A virus subtypes, found naturally in aquatic birds, are identified according to their surface antigens: hemagglutinin (HA) and neuraminidase (NA). Novel strains of influenza emerge due to mutations (antigenic drift) and reassortment among subtypes (antigenic shift). In random and unpredictable instances, predominantly through the process of antigenic shift, altered influenza viruses can emerge that efficiently infect humans, are highly transmissible via aerosol between humans, and potentially pathogenic, resulting in a pandemic outbreak [3–6]. Such pandemics have occurred several times in the 20th century, including in 1918 (H1N1), 1958 (H2N2), and 1967 (H3N2). Among these subtypes, H1N1 and H3N2 have established sustained circulation in the human population (and hence will be referred to as human viruses) by underdoing antigenic drift, which results in seasonal flu outbreaks each year. More recently even among these human virus subtypes, a novel H1N1 strain in 2009 emerged from reassortment of viral gene segments among avian, swine, and human viral strains and were able to successfully establish circulation via efficient human-to-human transmission [7–9].
The previous rapid introduction and spread of novel influenza strains and subtypes in the population has increased the surveillance and study of avian-adapted strains that have been documented to infect (but not spread in) humans. Of particular interest are the H5N1, H7(N2,N7 and N9), and H9(N1 and N2) subtypes. Quite recently in fact, a novel avian-adapted H7N9 strain emerged in China that caused severe infection and whose fatality was around 25% (May 2013 statistics see http://www.who.int/csr/don/2013_05_29/en/) that is much higher than the 0.1% observed with seasonal influenza A viruses. Although this subtype has not completely adapted yet to humans, it already possesses partial phenotypic features characteristic of human adapted-viruses [10–15]. Therefore, the adaptation of avian-adapted subtypes to the human host poses a constant threat of pandemic outbreak (due to the poor preexisting immunity for novel subtypes). Significant effort has been focused on determining the genetic determinants for human host adaptation, virulence, and aerosol transmissibility [16–18]. The binding specificity of the viral surface HA to sialylated glycan receptors (glycans terminated by α-D-N-acetyl neuraminic acid; Neu5Ac) on the host cell surface is one of many factors that critically govern adaptation of influenza to the human host. Avian virus HA binds with high specificity and affinity to glycans terminated by α2→3-linked sialic acid which are found in abundance in the avian gut and lower respiratory tract of humans (these glycans will henceforth be referred to as α2→3 glycans or avian receptors) [19–22]. Human virus HAs possess characteristic glycan receptor binding properties; their HA predominantly binds with high affinity (or avidity) to glycan receptors terminated by α2→6-linked sialic acid, which are predominantly expressed in the upper respiratory epithelia of humans (these glycans will henceforth be referred to as α2→6 glycans or human receptors) [21, 23, 24]. The human upper respiratory epithelium is the primary target site for infection of human-adapted viruses and is thought to be a prerequisite for efficient human-to-human transmission via respiratory droplets. Thus, it appears that human adaptation of an HA is associated with a switch in its binding preference from avian to human receptors. Notably, this switch is a necessary but not sufficient change required for human adaptation, which ultimately involves other genetic modifications within the viral genome and emergence of phenotypic characteristics such as efficient respiratory droplet transmission in ferret animal models [18].
To address in greater detail the binding of HA to its glycan receptors, advances in the synthesis of complex glycan structures have been coupled with technologies to display these structures on various glycan array platforms and interrogate HA receptor specificity [25–27]. Using such technologies, the glycan receptor binding properties has been defined in many ways in different studies. For example, some studies using glycan arrays characterize glycan receptor binding properties based on the ratio of the number of α2→6 to α2→3 sialylated glycans that bind to a specific HA or virus analyzed at a high titer or concentration [23, 28]. Other studies have defined binding specificity based on the ratio of binding affinity (or avidity) of HA (or virus) to α2→6 vs. α2→3 glycans [29, 30]. A meaningful comparison of binding signals from glycan arrays will be to compare across glycans with similar or common substructures and linkers that link them to the array surface. The varied descriptions of glycans and their linker structures within and across array platforms make this process more tedious at the present time.
In parallel with these advances, efforts have been ongoing to routinely solve co-crystal structures of HA–glycan complexes for a variety of HA subtypes, including H1, H2, H3, H5, H7, and H9 [28, 31–39]. Detailed structural information has provided a wealth of information on key interactions within the glycan-receptor binding site (RBS) of HA with surrogates of either avian or human receptors or both, leading to the identification of hallmark residues that distinguish binding of HAs to both avian and human receptors.
Despite the valuable information offered by these studies, there still remain key unanswered questions to our understanding of HA–glycan specificity. For example, it is difficult to assess the effect of hallmark residues on avian and human-receptor binding in the context of natural sequence evolution of HA. Introducing amino acid changes in different natural strains of H5 HA based on prototypic amino acids that contribute to human receptor-binding human virus HAs results in drastically different glycan binding properties unrelated to the human receptor binding preference [40, 41]. It has been difficult as well to link prototypic glycan-array based receptor binding properties of HA with the physiological tissue tropism; for example, as is seen in the recently emerged H7N9 HA [14]. Finally, the varying definitions of glycan receptor binding have complicated the distinction between binding of human and avian virus HA to human receptors. The human virus HA binding to human receptors is one of the factors that distinguish the efficient aerosol transmission of human virus from the lack of such transmission of avian virus [18].
In the context of the above questions, herein we review the current tools to structurally and biochemically characterize HA–glycan interactions. We also offer our perspective how human receptor specificity can be benchmarked as a tool for monitoring the evolution of avian virus HAs. The eventual goal enabled by this understanding is to improve surveillance methods to advance preparedness in the event of emergence of novel influenza strains, enabling implementation of countermeasures that can avoid or mute future epidemics or pandemics.
HA–glycan receptor interactions: structural and biochemical aspects of receptor specificity
Glycan receptor conformation and overall topology in RBS of HA
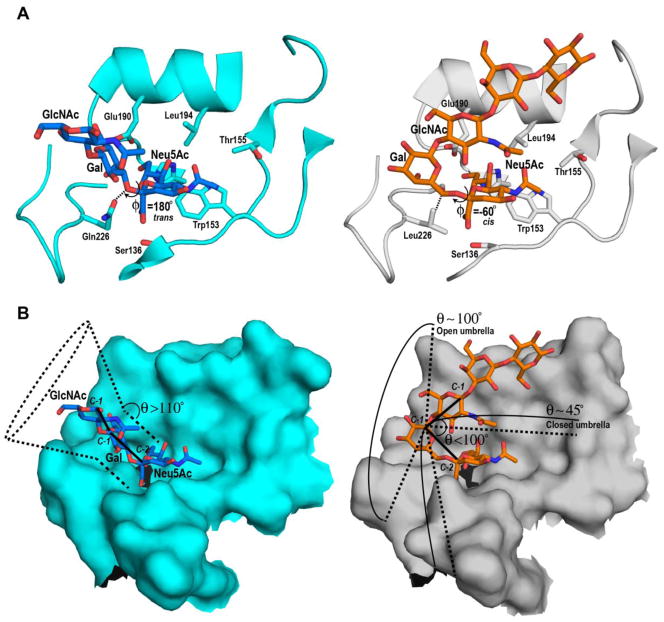
Several X-ray crystallographic structures of HA–glycan receptor complexes have been solved [28, 31–39]. Notably, the most commonly used glycans to represent avian and human receptors, respectively are LS-tetrasaccharide a (or LSTa; Neu5Acα2-3Galβ1-3GlcNAcβ1-3Galβ1-4Glc) and LS-tetrasaccharide c (or LSTc; Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4Glc) where Gal, GlcNAc and Glc are abbreviations for the hexopyranose sugars D-galactose, N-acetyl-D-glucosamine and D-glucose respectively. Based on some of the earliest X-ray co-crystal structures, the conformations of LSTa and LSTc have been characterized primarily by the glycosidic torsion angles of the terminal sialic acid linkage [20] (Figure 1A). In the case of the Neu5Acα2-3Gal-linkage in LSTa complexed with avian-adapted HA, the torsion angle φ (C1-C2-O-C3) is ~180° and is described as the trans conformation. In the trans conformation the glycosidic oxygen is pointed towards the base of the RBS. In contrast, the Neu5Acα2-6Gal linkage in LSTc complexed with human-adapted HA, φ (C1-C2-O-C6), is ~ −60°, or a cis conformation. In this conformation the glycosidic oxygen points away from the base of the RBS and the C6 atom of the penultimate Gal sugar points towards the base of the RBS.
Figure 1. Glycan receptor conformation and topology in HA RBS.
A, Left panel shows the trans conformation adopted by LSTa in the RBS of avian-adapted H3 HA (PDB ID: 1MQM). Right panel shows the cis conformation adopted by LSTc in the RBS of pandemic H3 HA (PDB ID:2YPG). The RBS is shown in cartoon with side chains of key residues labelled and shown. The glycans are shown in the stick representation where LSTa and LSTc are respectively colored by atoms (C:blue; O:red; N:dark blue) and (C:orange; O:red; N:dark blue). The distinguishing interactions involving the residue at the 226 position are indicated using dotted lines. B, Left panel shows the topological description of avian HA-LSTa glycan complex with where θ is the angle between C-2 atom of Neu5Ac, C-1 atom of Gal and C-1 atom of GlcNAc. For θ > 110°, the possible conformations sampled by the avian receptor spans a surface on the RBS that resembles a cone (shown in dotted lines). Right panel shows the topological description of the pandemic H3 HA-LSTc glycan complex where for θ < 100°, LSTc spans a much larger surface on the RBS that resembles an umbrella in an open state (θ ~ 100°) to a closed state (θ ~ 45°).
The cis and trans definition of glycan receptor conformation enabled distinguishing key contacts of residues within the RBS to either LSTa or LSTc, leading to the concept of ‘hallmark’ residues within the RBS of avian- and human-adapted HAs. In the case of avian-adapted HAs, residue Glu-190 and Gln-226 (residue numbering based on H3 HA throughout the text) are hallmark residues wherein Glu-190 is positioned to interact with sialic acid and Gln-226 in the base of the RBS is positioned to make ionic contacts with the glycosidic oxygen of Neu5Acα2-3Gal in the trans conformation [20]. In the case of human-adapted H2N2 and H3N2 HAs, the 226-position typically has a hydrophobic residue such as Leu, Ile or Val, which facilitates hydrophobic interactions with the C6 atom of Neu5Acα2-6Gal in the cis conformation [20]. Additionally, the presence of a hydrophobic residue in the 226-position does not enable favorable contacts with the glycosidic oxygen of Neu5Acα2-3Gal and hence is detrimental for avian-receptor binding. Therefore, the Gln226→Leu amino acid change has been considered a hallmark mutation for switching the receptor preference (leading to human adaptation) for H2 and H3 HAs.
On the other hand, both avian- and human-adapted H1 HAs have Gln-226, which based on the RBS structure of this subtype is not favorable for hydrophobic contacts with the C6 atom of Neu5Acα2-6Gal. Instead, an Asp in the 225-position (which is typically a Gly in avian-adapted H1 HAs) in human-adapted H1 HA provides additional contacts with the penultimate Gal sugar in the trans conformation [20, 33, 39]. In H1 HA, the differences in contacts between avian- and human-receptors go beyond the distinct contacts with Neu5Acα2-6Gal and Neu5Acα2-3Gal in the base of the RBS. Another amino acid typically observed in human-adapted H1 HA is Asp-190, which favors specific contacts with the third GlcNAc sugar (from the non-reducing end) which is not the case with Glu-190, which is typically present within avian-adapted HAs [20, 33]. Therefore the Glu vs. Asp in the 190-position in H1 HA likely plays a key role in distinguishing avian and human receptor specificity [42]. Consequently, in H1 HA, Glu190→Asp and Gly225→Asp have been considered as hallmark amino acid changes to switch receptor specificity leading to human adaptation [42, 43].
While, the cis and trans definition of glycan conformation has been useful to characterize the distinct interactions with the terminal Neu5Ac α2→3Gal or Neu5Ac α2→6Gal motif, this definition does not fully describe HA binding to a range of structurally diverse glycans, either present on glycan array platforms or present in glycomic analysis of human respiratory cells and tissues [24]. This limitation motivated studies that revisited the definition of glycan conformation, extending the conformational analysis beyond the terminal sialic acid linkage to describe overall topology and dynamics of the glycan receptor upon binding to the RBS of avian and human-adapted HAs [24, 44]. To capture this topology, a parameter, θ, has been defined to measure the angle between the Neu5Ac, the penultimate Gal and the third GlcNAc sugar (measured using anomeric carbon atoms as shown in Figure 1B). The θ parameter permits classification of the ensemble of conformations sampled by the avian and human receptors within the HA binding site.
In the case of avian receptors, the conformations sampled by the Neu5Acα2→3Gal linkage (keeping the Neu5Ac anchored) and the sugars beyond this linkage (at the reducing end) span a region on the binding surface of HA that resemble a cone. Therefore the term cone-like topology has been used to capture the ensemble of these conformations - characterized by a θ angle > 110°. The different conformations sampled by Neu5Acα2→6Gal linkage (keeping the Neu5Ac anchored) and the sugars beyond this linkage (at the reducing end) span a wider area on the HA binding surface [24]. A portion of these RBS-receptor contacts can be described as a cone-like surface, whereas the other portion is more correctly described as umbrella-like, and is characterized by θ angle <100°. Given the conformationally more flexible form, depending on the attributes of the HA RBS and the receptor sequence, the umbrella-like conformation ensemble can be fully folded (θ ~ 45°) to fully open (θ ~90°) [24]. Regardless, the stem of the umbrella is defined by the Neu5Acα2-6Gal- motif and the spokes of the umbrella are occupied by monosaccharides at the reducing end of Gal.
Examination of a range of avian-adapted HAs indicate that the defining characteristic of a cone-like topology is that the majority of contacts with the HA RBS are through a three-sugar (or trisaccharide) (Neu5Acα2→3/6Galβ1→3/4GlcNAc-) motif as well as monosaccharide substitutions such as O-sulfation (of Gal or GlcNAc) or fucosylation (at GlcNAc). On the other hand, the umbrella-like topology was such that monosaccharides beyond a trisaccharide make substantial contacts with the HA RBS. Using these shape-based definitions of the flexible glycan conformation, it was shown that the umbrella-like topology is predominantly adopted by human receptors which possessed at least 4 sugars including Neu5Ac, for example, poly-lactosamine branches terminated by α2→6-linked Neu5Ac (referred to as long α2→6). The cone-like topology was shown to be adopted by both avian and human receptors [24].
Taken together, the shape- or topology-based definitions of glycan receptor conformation have been able to provide additional structural perspectives on HA–glycan interactions in the context of glycan diversity going beyond terminal sialic acid linkage. Furthermore, this framework has led to the identification of additional key residue positions within the RBS of different HA subtypes that are involved in binding to avian and human receptors [24, 44]. Finally, in conjunction with experimental information (detailed below), this structural framework has enabled robust classification of avian and human receptors.
Measuring and characterizing HA–glycan interactions
As is the case with many virus-receptor interactions, binding between HA receptor is multivalent. A variety of biochemical methods have been used to characterize the specificity in the context of multivalent HA–glycan interactions (see review by Shriver et al. [45] for an overview of these methods). Among the various tools, glycan array platforms are rapidly emerging as a popular tool to probe finer nuances of glycan structures that are recognized by various HAs [25–27].
Glycan platforms consist of hundreds of synthetic glycan motifs (typically present on N- and O-linked glycoproteins and glycolipids) displayed on the surface of the array. Multiple types of arrays have been developed that utilize different strategies including the formation of neoglycolipids [46–48], neoglycoproteins [49], or the direct application of glycans to various surfaces [26, 50–55]. Studies have also begun to adapt these technologies towards the presentation of natural glycans by harvesting glycans from the cells or tissues and imprinting these on a glycan array format [56, 57], thus allowing one to probe the glycan repertoire of a biological system.
Both whole viruses and recombinantly expressed trimeric HA units have been analyzed on glycan array platforms. While analysis of viruses on glycan arrays permits obtaining qualitative binding characteristics and offers a better chance for identifying binding to low-affinity glycan ligands, it is exigent, for two major reasons, to quantify these interactions so as to compare binding properties across different viruses. First, unless directly labeled, virus concentration is commonly expressed as hemagglutination units (HAU) based on the virus’s ability to agglutinate red blood cells. This is not a true measure of concentration; depending on agglutination potential of a virus, an HAU value could correspond to a very different concentration of HA. Emphasizing this fact, glycomic analysis of red blood cells has shown that the glycans on red blood cells do not necessarily recapitulate those observed in human respiratory cells [58]. Thus, viruses that bind well to human respiratory tissue may nonetheless fail to agglutinate red blood cells [58]. Second, depending on the morphology of the virus, the distribution of the trimeric HAs vary between different viruses. In turn, the differences in distribution of the trimeric HA units impinge on the avidity of glycan binding.
In contrast, analyzing recombinantly expressed turmeric HA units offers a means to circumvent these challenges. First, HA concentration can be precisely measured. Also, an approach to precomplex the trimeric HA unit with primary and secondary antibodies (in HA:primary:secondary ratio of 4:2:1) prior to analysis on glycan array enables one to address issues associated with multivalency [25, 59]. Due to the stoichiometry of the precomplex, this approach ensures that the predominant species in the analyte corresponds to four HA trimeric units that are spatially constrained relative to each other (due to the antibody interactions). The apparent affinity constant defined by such an assay can be used to compare quantitative binding of different HAs analyzed. Notably, this parameter does not have any independent significance from the standpoint of physical chemistry.
A survey of studies completed to date indicates that the scope of array-binding assays varies across different studies. In many studies, glycan arrays are used as a primary screen for qualitatively analyzing the frequency and type of α2→3 and α2→6 glycans that demonstrate binding to recombinant HA (or whole virus). In these studies, glycan-binding specificity is defined on the basis of the ratio of α2→6 to α2→3 glycans that show binding signals. This type of a screening approach, while informative, does not offer quantitative information, particularly relating to the relative human and avian receptor-binding affinity of an HA. Quantitative studies focus on selection of a small sampling of glycans representative of avian and human receptor and performing a dose response curve by varying either HA (virus) concentration or glycan concentration [30, 47, 59, 60]. The trade-off of using this approach is, of course, the loss of information regarding the diversity of glycan structures that are recognized by a given HA.
Although glycan array platforms display hundreds of diverse glycan structures, they still likely do not capture the physiological context or diversity of glycan receptors encountered by influenza A viruses in human or animal models. This issue has been addressed in part by examining binding of HA or whole virus to tissue sections of the respiratory tract of humans (upper respiratory tract:pharynx and trachea; middle respiratory tract: bronchus; and lower respiratory tract:alveolar) and/or animal models, such as mice, ferrets, or pigs [21, 22, 24, 61, 62]. Plant lectins such as SNA-I (specifically binds to α2→6 glycans), MAL-II (shows binding to α2→3 glycans), Jacalin (marker for O-linked mucin glycans), and Con A (marked for N-linked glycans) can be used to characterize the glycans present in these sections and hence provide structural information on HA recognition. Additionally, analytical tools such as mass spectrometry have been used to perform detailed structural characterization of sialylated glycans isolated from the respiratory tissues and cell lines [24, 63, 64].
Avian-adapted viruses and HA extensively stain the alveolar sections in the human lower respiratory tract that predominantly express avian receptors [22]. On the other hand, human-adapted viruses and HA show characteristic binding to apical surface of human tracheal sections that predominantly express human receptors [21, 24, 62]. Also within the human respiratory epithelium, avian-adapted viruses have been shown to primarily infect ciliated cells while human-adapted viruses have been shown to infect non-ciliated cells [65]. This cell tropism may be important to the physiology of influenza transmission, especially for human-adapted viruses. Binding to non-ciliated, goblet cells, especially those with heavily glycosylated mucins on their surface, may play a role in droplet formation and transmission.
It is clear that these tools provide diverse yet related information on HA–glycan interactions. Therefore, integration of information from analysis of HA or virus binding to glycan arrays and physiological tissues can provide additional information beyond that available in arrays, including tropism of HA. In our view, integrated information from both tissue staining and array analysis provides the most detailed biochemical information on HA–glycan interactions that is best suited to phenotypic characterization of HA. To ensure accurate assessment of results, an important step is to benchmark the measurements made to prototypic human-adapted viruses such as the pandemic strains.
Characteristic human receptor binding of pandemic viruses and their relationship to aerosol transmissibility of pandemic viruses
The 1918 H1N1 subtype is among the most studied viruses. The ability to reconstruct the pandemic 1918 H1N1 virus through reverse genetics and test its virulence in ferrets permitted a systematic exploration of the roles for various viral genes in its virulence and transmissibility [16–18, 66]. Based on the notion of hallmark Asp-190 and Asp-225 residues playing a key role in human-receptor binding of H1N1, single amino acid changes at these positions were made on a prototypic pandemic HA (A/South Carolina/1/1918 or SC18). This resulted in two variants, NY18 (Asp225→Gly mutant of SC18) and AV18 (Asp-190→Glu mutant of NY18). Analysis of the aerosol transmissibility of these viruses in ferrets demonstrated that SC18 transmitted efficiently via respiratory droplet, NY18 showed transmission but was inefficient and AV18 did not transmit at all [43]. Given that all the other genes were identical between SC18, NY18 and AV18, these results showed a clear and direct link between altering the RBS of HA and transmissibility of the virus.
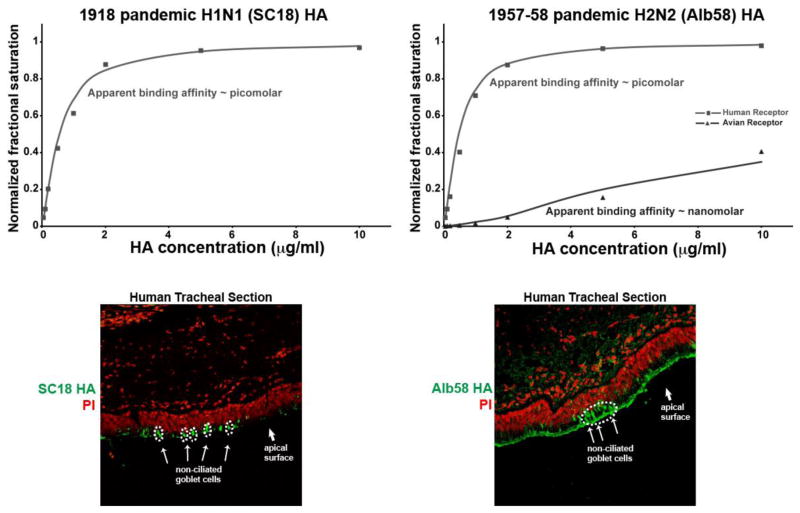
The glycan receptor-binding properties of SC18, NY18 and AV18 HA have been subsequently characterized in several other studies [23, 59] including recent crystallographic analysis of SC18 and NY18 HA binding to LSTa and LSTc [39]. Dose-dependent direct binding of SC18, NY18 and AV18 analyzed by precomplexing the HA on a glycan array with representative avian and human receptors showed distinct quantitative binding properties [59]. While SC18 showed exclusive binding to human receptors with an apparent affinity (Kd′) in the picomolar range and minimal binding to avian receptors (Figure 2), AV18 showed exclusive binding to avian receptors. NY18 demonstrated intermediate binding, with two orders of magnitude lower apparent affinity to human receptors and binding to avian receptors in the sub-nanomolar range. That NY18, having Asp-190, demonstrated similar binding affinity to both avian and human receptors was surprising given that Asp-190 should have enabled NY18 to distinguish between LSTa and LSTc [39, 42].
Figure 2. Glycan receptor binding properties of representative pandemic HAs.
Top panels show dose-dependent binding of HA to representative human (Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ) and avian (Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ) receptors. Bottom panels show binding of HA to human tracheal tissue sections (HA in green against propidium iodide or PI in red). The non-ciliated goblet cell regions on the apical surface of the human tracheal section are highlighted in dotted white circles (see references [59, 68] for details).
SC18, NY18 and AV18 were also analyzed on human tracheal and alveolar sections. Both SC18 and NY18 showed apical surface staining of tracheal section in a manner that was characteristic of human-adapted HAs. However, SC18 showed a characteristic predominant staining of non-ciliated goblet cells when compared to NY18, which stained ciliated cells to a greater extent than goblet cells [59]. SC18 showed minimal to no staining of an alveolar tissue section, while, consistent with their avian receptor binding properties, NY18 and AV18 showed detectable staining of alveolar tissue.
Similar to 1918 H1N1, ferret transmission and glycan-binding studies have been completed for the a prototypic strain (A/Albany/6/58 or Alb58) of the 1958 H2N2 pandemic. This virus transmitted efficiently via respiratory droplets in ferrets [67]. Dose-dependent glycan array binding of Alb58 HA on the glycan array showed high affinity binding to human receptors (Kd′ ~ picomolar) [68]. Interestingly, unlike SC18, Alb58 also showed observable binding to avian receptors, albeit at a binding affinity that was orders of magnitude lower than that to human receptors (Kd′ ~ nanomolar) (Figure 2). Consistent with what was observed for SC18, Alb58 HA extensively stained the goblet cells. Additionally, Alb58 stained ciliated cells on the apical surface of human tracheal tissue sections as well as alveolar sections [68]. Although cross comparison of ferret transmission and glycan binding properties have not been performed on the 1967–68 pandemic H3N2 strain, the glycan binding properties of a prototypic strain (A/Aichi/1/68 or Aichi68) have been analyzed. Aichi68 shows comparable binding to both avian and human receptors with high binding affinity [37].
More recently, the aerosol transmission in ferrets and glycan-binding properties of the 2009 H1N1 pandemic strain (A/California/04/09 or Ca0409) were studied [47, 69–72]. The dose-dependent glycan binding property of Ca0409 was very similar to that of SC18 wherein binding was observed exclusively to human receptors. Human tissue of Ca0409, confirmed this analysis, and binding of Ca0409 HA was restricted to the goblet cell region on the apical surface of the tracheal section similar to SC18 [69]. Notably, however, the binding affinity of Ca0409 to human receptors (Kd′ in subnanomolar range) was substantially lower than that of SC18 [73]. Consequently, while the Ca0409 virus showed respiratory droplet transmission in ferrets, the efficiency of transmission was lower than that of SC18 [69].
Based on the studies summarized above, the HA of pandemic viruses (those that are able to achieve respiratory droplet transmission) has distinct glycan binding properties, which include high affinity binding to human receptors and a characteristic extensive staining of the goblet cells in the apical surface of human tracheal section (Figure 2). On a comparative basis, the relative binding affinity of a given HA to human receptors correlates with the efficiency of transmission in the ferret model. Indeed, this analysis has demonstrated predictive power in the case of H1 [59], H2 [67, 68], H5 [74] and H7 [14, 75–77]. Based on these observations, we postulate that the key HA determinant enabling human-to-human transmission via the respiratory droplet is the ability of the HA to bind distinct goblet cell derived glycans in the upper airways. This is an attribute that is independently measurable (through experiments in the ferret) and is a necessary determinant of a virus capable of initiating an epidemic or pandemic. Conversely, infection can occur via multiple mechanisms, independent of HA–glycan specificity or even that of HA itself, and is also a function of the immune status of the individual [78, 79].
Analyzing amino acid changes in the RBS in the context of natural sequence evolution of HA
The Ca0409 HA was shown to share high sequence identity, antigenic similarity and hallmark residues associated with human receptor binding with SC18 HA [80, 81]. Despite these similarities, this HA showed substantially lower affinity binding to human receptors relative to that of SC18 HA. A detailed structural analysis of the key residues in the RBS revealed key differences in the inter-residue interactions involving positions 219, 227, 222, 225 and 186 between CA0409, SC18 and seasonal H1N1 HAs [73]. While the interactions at these positions were of a hydrophobic nature in SC18 HA, they were ionic in the case of seasonal HAs. In contrast to either case, in Ca0409, they were neither hydrophobic nor ionic, affecting the positioning of the key Asp-190 residue. Introducing a single amino acid change Ile-219→Lys in Ca0409, made these interactions ionic in character, which in turn substantially increased human receptor-binding affinity of Ca0409 [73].
Examination of the role of glycosylation on HA in mediating the receptor binding properties of HA has extended this analysis [40, 82–84]. Molecular dynamics simulation studies predicted that HA–glycans may form interactions near the binding pocket to influence receptor binding [82]. Site-directed mutagenesis to knockout glycosylation sites on HA [40, 85] or modifying structure of N-linked glycans on the virus by enzymatic treatment or transgenic cell lines [83] have shown distinct changes in glycan-receptor binding specificity. Loss of glycosylation at a highly conserved sequon was detrimental to receptor binding by SC18 and NY18 HA, whereas it did not affect the binding of AV18 HA to avian receptors [85]. This observation was explained by analyzing the network of inter-residue interactions in the RBS of SC18, NY18, and AV18 and describing the relationship of this network to the conserved glycosylation sequon. Removal of HA glycosylation at this sequon had minimal impact on the network involving the 220-loop in the RBS of AV18, whereas loss of glycosylation disrupted the network within NY18 and SC18 HA [85] (Figure 3).
Figure 3. Network and molecular features of HA RBS.
RBS of SC18 complexed with LSTc is shown along with N-linked trimannosyl core glycan structure added at the Asn-91 position using the GlyProt tool (http://www.glycosciences.de/modeling/glyprot/php/main.php) where the SC18–LSTc cocrystal structure (PDB ID: 2WR7) was submitted for in silico glycosylation. The side chains of the key residues are shown and labeled. The RBSN of representative positions are shown as interconnected circular nodes. The nodes are colored with varying shades of red where light pink corresponds to residues with lowest RBSN score and bright red corresponds to residues with highest RBSN score. The network of interactions involving the glycosylation is indicated as a yellow box in the RBSN diagram. The key residue positions along with their RBSN provide a more robust approach to investigate amino acid changes for human adaptation of HA.
The aforementioned observations suggest that the presence of hallmark residues cannot be directly linked to conferring a specific glycan-binding property to any HA, particularly when extrapolating between subtypes. This is evidenced from attempts to introduce hallmark amino acids - observed in pandemic H1N1 or H2N2 or H3N2 HA - into H5 HA so as to confer so-called gain of function (i.e. aerosol transmissibility in ferrets). None of these H5 HA mutants showed comparable glycan receptor binding properties of pandemic HAs [40, 41] or gain of function [74]. It is interesting to view this notion in the context of other studies that have approached influenza protein evolution from the perspective of whether stabilizing mutations are constrained by epistasis (the notion that stability-affecting mutations are tolerated only after occurrence of other compensatory mutations) [86,87]. The effects of amino acid changes on glycan receptor binding is more nuanced than those affecting overall protein stability and therefore understanding these effects requires a more detailed structural analysis of HA–glycan contacts. These structural analyses would need to go beyond identifying key residues and account for interactions between key residues (and any proximal glycosylation) in the RBS.
These aforementioned key aspects are relevant to recent studies that have demonstrated mutations in 2004 (A/Vietnam/1203/04 or Viet04) and 2005 (A/Indonesia/5/05 or Ind05) strains of H5N1 HA that have conferred respiratory droplet transmission upon the virus strains [29, 86]. Based on these results, several follow-on studies were inclined to fix these mutations as so-called hallmark changes for any H5 HA [87]. However, from surveillance data, we know that the sequences of HA from current circulating strains of H5 have diverged significantly from Viet04 and Indo05 HA. Given this divergence, introducing the same set of mutations that resulted in gain of function for Viet04 or Ind05 to currently circulating H5 HAs do result in a switch in receptor preference and a gain of function [41]. Given the established framework, a detailed structural analysis of RBS of H5 HA was completed.
On the basis of phylogenetic ‘closeness’ of H5 to H2 HA, Alb58 HA was chosen as the reference human-adapted HA to identify key RBS properties within H5 HA. Four key differences were observed. First, the composition of the 130 loop of H2 HA is different from H5 HA in that the loop length is shorter by an amino acid. This deletion in the 130-loop in H5 HA relative to H2 HA was shown to critically govern the 130-loop. Second, amino acids in the ‘base’ of the RBS (such as those in 130-loop at positions 136-138, and 220-loop at positions 219-228) are different in H2 than in H5. Third, the ‘top’ of the RBS primarily comprising the ‘190-helix’ (residues 188-196) that interacts with the sugars beyond terminal Neu5Acα2→6Gal motif in the human receptor are different in H2 than in H5 (specifically at positions 188, 189, 192 and 193). Fourth, position 158 is glycosylated in H5 HA but not in H2 HA. Glycosylation at this site has been shown to influence glycan receptor binding property of H5 HA [40].
Given these differences, and to understand amino acid changes that would enable the RBS of H5 HA match with that of H2, it was important to analyze the network of inter-residue interactions within the RBS. This analysis was performed by defining a map known as RBS network or RBSN, which showed interactions between RBS residues using a 2-D graph (Figure 3). The extent of connectivity was quantified using a network score such that the higher the score of an amino acid within the RBS, the more structurally constrained it is to mutation. Finally, the differences between the RBS of H2 and H5 HA were captured using a new definition termed molecular features (one feature for each difference) which incorporated topological definition of glycan receptor in the RBS, RBS residues involved in the binding, and their RBSN maps. Four distinct features were identified that together constituted a complete description of the H5 RBS.
The molecular feature definition was then mapped onto the phylogenetic sequence analyses of H5 HA which showed that many of the clades, including currently circulating clade 1, clade 2.2, clade 2.2.1, and clade 7 had already acquired amino acid changes characteristic of one or two of Features 1, 3 and 4. However, only a subset of the rapidly evolving and currently circulating clades 2.2.1 and 7 had acquired amino acid changes to match Feature 1 and/or part of Feature 2, which are critical features of the RBS base. From this subset, it was demonstrated that select strains required as few as one or two amino acid changes to match the requisite features, switch in binding preference and demonstrate human receptor binding affinity in the same range as that of pandemic HAs [41].
Concluding remarks
Influenza A viruses have always been viewed as ‘unpredictable’ pathogens where a novel subtype could cross species into humans and potentially lead to a widespread pandemic outbreak. The concept of switch in glycan-receptor specificity needs to be defined and interpreted carefully to enable it to be a useful tool for surveillance and strain characterization (Box 1).
Box 1. Outstanding questions.
- Is the ratio of α2→6 to α2→3 binding sufficient to characterize glycan receptor binding properties of human viruses?
- How do we analyze and interpret HA–glycan receptor binding in the context of amino acid changes arising from natural sequence evolution due to host selection pressure and epistasis and other changes such as those that affect HA:NA balance so as to improve surveillance?
- How can we establish a mechanistic link between HA–glycan interactions in a physiological context such as goblet cell binding and the ability of the virus to efficiently infect and replicate versus to transmit via aerosol in the human host?
First, amino acid mutations that confer specific receptor binding properties to a particular strain and subtype of HA do not always (or even often) confer the same properties to a HA of a different virus. For example, the extent of binding to different receptors appears to vary between different subtypes. While human-adapted viruses of group 1 viruses, such as H1N1 and H2N2, show minimal or low binding affinity to avian receptors, viruses within group 2, including H3N2, show comparable binding to both human and avian receptors. These studies question the simple definition of switch as ratio of binding to α2→6 vs. α2→3 glycan receptors and emphasize the importance of identifying and understanding the structural and biochemical aspects of HA-glycan receptor interactions. This is particularly relevant when looking at subtypes that are rapidly evolving, such as H5N1. We believe that the recently developed inter-residue interaction network can potentially be evolved into a metric that can assist such surveillance efforts and provide a method to identify strains which are evolving towards human receptor specificity. Towards such a goal, additional studies need to be undertaken to systematically define network scores and network maps of RBS residues of avian-, swine- and human-adapted HAs and identifying a scoring system that discriminates the network properties of these HAs.
Second, the analyses of receptor binding properties of HA from multiple subtypes, including pandemic strains of H1 and H2, point to characteristic properties such as high affinity binding to human receptors and also distinct goblet cell staining patterns, which are at least useful descriptors and may also provide insight into a mechanistic link between receptor binding preference and aerosol transmissibility. Goblet cells secrete mucins, which can potentially assist in the aerosolization of the virus, which in turn might facilitate efficient respiratory droplet transmission. Additionally, goblet cell tropism might also shed light on why many influenza strains, including H7N9, do not demonstrate efficient respiratory droplet transmission even though they have been shown to replicate efficiently in the human respiratory tract. Therefore, it is critical to distinguish the roles of glycan specificity in virus infection with that of virus transmission.
Third, adaptive changes that affect glycan receptor binding property of HA are often accompanied by changes in influenza A neuraminidase enzyme (NA) [88, 89]. The relationship between HA binding and NA activity is thought to play a key role in balancing the human receptor engagement and viral release from the infected cell to achieve efficient respiratory droplet transmission [90, 91]. Glycan-array based methods to probe specificity and activity of NA have been developed [90] and can be employed in conjunction with measurements of receptor binding properties of HA to improve surveillance.
In summary, through a better understanding of HA–glycan interactions, we can significantly improve surveillance methods to advance preparedness and potential countermeasures in the event of emergence of novel influenza strains.
HIGHLIGHTS.
- Glycan topology beyond terminal linkage clearly demarcates hemagglutinin (HA)–glycan receptor interactions.
- Pandemic virus HAs share characteristic biochemical and physiological receptor binding.
- Hallmark mutations in naturally evolving avian HAs led to very different receptor binding.
Acknowledgments
This work was funded in part by National Institutes of Health Merit Award (R37 GM057073-13), National Research Foundation supported Interdisciplinary Research group in Infectious Diseases of SMART (Singapore MIT alliance for Research and Technology) and the Skolkovo Foundation supported Infectious Diseases Center at MIT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ahmed R, et al. Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nature immunology. 2007;8:1188–1193. doi: 10.1038/ni1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez Velasco R, et al. Systematic review of economic evaluations of preparedness strategies and interventions against influenza pandemics. PloS one. 2012;7:e30333. doi: 10.1371/journal.pone.0030333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russell CJ, Webster RG. The genesis of a pandemic influenza virus. Cell. 2005;123:368–371. doi: 10.1016/j.cell.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu Rev Pathol. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Layne SP, et al. Pandemic influenza: an inconvenient mutation. Science. 2009;323:1560–1561. doi: 10.1126/science.323.5921.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neumann G, et al. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser C, et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science. 2009;324:1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh Y, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearce MB, et al. Pathogenesis and transmission of swine origin A(H3N2)v influenza viruses in ferrets. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3944–3949. doi: 10.1073/pnas.1119945109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belser JA, et al. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature. 2013;501:556–559. doi: 10.1038/nature12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013;381:1916–1925. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao R, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. The New England journal of medicine. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 13.Kageyama T, et al. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 2013;18:20453. [PMC free article] [PubMed] [Google Scholar]
- 14.Tharakaraman K, et al. Glycan receptor binding of the influenza A virus H7N9 hemagglutinin. Cell. 2013;153:1486–1493. doi: 10.1016/j.cell.2013.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe T, et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature. 2013;501:551–555. doi: 10.1038/nature12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palese P. Influenza: old and new threats. Nat Med. 2004;10:S82–87. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- 17.Pappas C, et al. Single gene reassortants identify a critical role for PB1, HA, and NA in the high virulence of the 1918 pandemic influenza virus. Proc Natl Acad Sci U S A. 2008;105:3064–3069. doi: 10.1073/pnas.0711815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Hoeven N, et al. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc Natl Acad Sci U S A. 2009;106:3366–3371. doi: 10.1073/pnas.0813172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gambaryan AS, et al. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6′-sialyl(N-acetyllactosamine) Virology. 1997;232:345–350. doi: 10.1006/viro.1997.8572. [DOI] [PubMed] [Google Scholar]
- 20.Russell RJ, et al. Avian and human receptor binding by hemagglutinins of influenza A viruses. Glycoconj J. 2006;23:85–92. doi: 10.1007/s10719-006-5440-1. [DOI] [PubMed] [Google Scholar]
- 21.Shinya K, et al. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 22.van Riel D, et al. H5N1 virus attachment to lower respiratory tract. Science. 2006;312:399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- 23.Stevens J, et al. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol. 2006;355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Chandrasekaran A, et al. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat Biotechnol. 2008;26:107–113. doi: 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- 25.Stevens J, et al. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat Rev Microbiol. 2006;4:857–864. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang PH, et al. Glycan arrays: biological and medical applications. Curr Opin Chem Biol. 2008;12:86–92. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song X, et al. A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. The Journal of biological chemistry. 2011;286:31610–31622. doi: 10.1074/jbc.M111.274217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevens J, et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 29.Imai M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong X, et al. Receptor binding by an H7N9 influenza virus from humans. Nature. 2013;499:496–499. doi: 10.1038/nature12372. [DOI] [PubMed] [Google Scholar]
- 31.Ha Y, et al. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc Natl Acad Sci U S A. 2001;98:11181–11186. doi: 10.1073/pnas.201401198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha Y, et al. X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology. 2003;309:209–218. doi: 10.1016/s0042-6822(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 33.Gamblin SJ, et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, et al. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc Natl Acad Sci U S A. 2009;106:17175–17180. doi: 10.1073/pnas.0906849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu R, et al. Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic. J Virol. 2010;84:1715–1721. doi: 10.1128/JVI.02162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H, et al. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS pathogens. 2010;6:e1001081. doi: 10.1371/journal.ppat.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin YP, et al. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21474–21479. doi: 10.1073/pnas.1218841110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang H, et al. Structure and receptor complexes of the hemagglutinin from a highly pathogenic H7N7 influenza virus. Journal of virology. 2012;86:8645–8652. doi: 10.1128/JVI.00281-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, et al. Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution. Journal of virology. 2013;87:5949–5958. doi: 10.1128/JVI.00545-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevens J, et al. Recent avian H5N1 viruses exhibit increased propensity for acquiring human receptor specificity. J Mol Biol. 2008;381:1382–1394. doi: 10.1016/j.jmb.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tharakaraman K, et al. Structural determinants for naturally evolving H5N1 hemagglutinin to switch its receptor specificity. Cell. 2013;153:1475–1485. doi: 10.1016/j.cell.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glaser L, et al. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J Virol. 2005;79:11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tumpey TM, et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315:655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 44.Xu D, et al. Distinct glycan topology for avian and human sialopentasaccharide receptor analogues upon binding different hemagglutinins: a molecular dynamics perspective. J Mol Biol. 2009;387:465–491. doi: 10.1016/j.jmb.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shriver Z, et al. Context-specific target definition in influenza a virus hemagglutinin-glycan receptor interactions. Chem Biol. 2009;16:803–814. doi: 10.1016/j.chembiol.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukui S, et al. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat Biotechnol. 2002;20:1011–1017. doi: 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]
- 47.Childs RA, et al. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat Biotechnol. 2009;27:797–799. doi: 10.1038/nbt0909-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, et al. Altered receptor specificity and cell tropism of D222G hemagglutinin mutants isolated from fatal cases of pandemic A(H1N1) 2009 influenza virus. J Virol. 2010;84:12069–12074. doi: 10.1128/JVI.01639-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oyelaran O, Gildersleeve JC. Glycan arrays: recent advances and future challenges. Curr Opin Chem Biol. 2009;13:406–413. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blixt O, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia B, et al. Versatile fluorescent derivatization of glycans for glycomic analysis. Nat Methods. 2005;2:845–850. doi: 10.1038/nmeth808. [DOI] [PubMed] [Google Scholar]
- 52.Grun CH, et al. One-step biotinylation procedure for carbohydrates to study carbohydrate-protein interactions. Anal Biochem. 2006;354:54–63. doi: 10.1016/j.ab.2006.03.055. [DOI] [PubMed] [Google Scholar]
- 53.Mercey E, et al. Polypyrrole oligosaccharide array and surface plasmon resonance imaging for the measurement of glycosaminoglycan binding interactions. Anal Chem. 2008;80:3476–3482. doi: 10.1021/ac800226k. [DOI] [PubMed] [Google Scholar]
- 54.Karamanska R, et al. Surface plasmon resonance imaging for real-time, label-free analysis of protein interactions with carbohydrate microarrays. Glycoconj J. 2008;25:69–74. doi: 10.1007/s10719-007-9047-y. [DOI] [PubMed] [Google Scholar]
- 55.Song X, et al. Glycan microarrays. Methods Mol Biol. 2012;800:163–171. doi: 10.1007/978-1-61779-349-3_11. [DOI] [PubMed] [Google Scholar]
- 56.Byrd-Leotis L, et al. Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses. Proc Natl Acad Sci U S A. 2014;111:E2241–2250. doi: 10.1073/pnas.1323162111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song X, et al. Chemistry of natural glycan microarrays. Curr Opin Chem Biol. 2014;18:70–77. doi: 10.1016/j.cbpa.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aich U, et al. Glycomics-based analysis of chicken red blood cells provides insight into the selectivity of the viral agglutination assay. The FEBS journal. 2011;278:1699–1712. doi: 10.1111/j.1742-4658.2011.08096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srinivasan A, et al. Quantitative biochemical rationale for differences in transmissibility of 1918 pandemic influenza A viruses. Proc Natl Acad Sci U S A. 2008;105:2800–2805. doi: 10.1073/pnas.0711963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao HY, et al. Differential receptor binding affinities of influenza hemagglutinins on glycan arrays. Journal of the American Chemical Society. 2010;132:14849–14856. doi: 10.1021/ja104657b. [DOI] [PubMed] [Google Scholar]
- 61.Nicholls JM, et al. Sialic acid receptor detection in the human respiratory tract: evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir Res. 2007;8:73. doi: 10.1186/1465-9921-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jayaraman A, et al. Decoding the distribution of glycan receptors for human-adapted influenza A viruses in ferret respiratory tract. PLoS ONE. 2012;7:e27517. doi: 10.1371/journal.pone.0027517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bateman AC, et al. Glycan analysis and influenza A virus infection of primary swine respiratory epithelial cells: the importance of NeuAc{alpha}2-6 glycans. The Journal of biological chemistry. 2010;285:34016–34026. doi: 10.1074/jbc.M110.115998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walther T, et al. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS pathogens. 2013;9:e1003223. doi: 10.1371/journal.ppat.1003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matrosovich MN, et al. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci U S A. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tumpey TM, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 67.Pappas C, et al. Receptor specificity and transmission of H2N2 subtype viruses isolated from the pandemic of 1957. PLoS One. 2010;5:e11158. doi: 10.1371/journal.pone.0011158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Viswanathan K, et al. Determinants of glycan receptor specificity of H2N2 influenza A virus hemagglutinin. PLoS One. 2010;5:e13768. doi: 10.1371/journal.pone.0013768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maines TR, et al. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science. 2009;325:484–487. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang H, et al. Structure and Receptor binding properties of a pandemic H1N1 virus hemagglutinin. PLoS Curr. 2010;2:RRN1152. doi: 10.1371/currents.RRN1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bradley KC, et al. Comparison of the receptor binding properties of contemporary swine isolates and early human pandemic H1N1 isolates (Novel 2009 H1N1) Virology. 2011;413:169–182. doi: 10.1016/j.virol.2011.01.027. [DOI] [PubMed] [Google Scholar]
- 72.Yen HL, et al. Hemagglutinin-neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14264–14269. doi: 10.1073/pnas.1111000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jayaraman A, et al. A single base-pair change in 2009 H1N1 hemagglutinin increases human receptor affinity and leads to efficient airborne viral transmission in ferrets. PloS one. 2011;6:e17616. doi: 10.1371/journal.pone.0017616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maines TR, et al. Effect of receptor binding domain mutations on receptor binding and transmissibility of avian influenza H5N1 viruses. Virology. 2011;413:139–147. doi: 10.1016/j.virol.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fouchier RA, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Srinivasan K, et al. Quantitative description of glycan-receptor binding of influenza A virus H7 hemagglutinin. PloS one. 2013;8:e49597. doi: 10.1371/journal.pone.0049597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu H, et al. Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science. 2013;341:183–186. doi: 10.1126/science.1239844. [DOI] [PubMed] [Google Scholar]
- 78.Subbarao K, Katz J. Avian influenza viruses infecting humans. Cellular and molecular life sciences: CMLS. 2000;57:1770–1784. doi: 10.1007/PL00000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fukuyama S, Kawaoka Y. The pathogenesis of influenza virus infections: the contributions of virus and host factors. Curr Opin Immunol. 2011;23:481–486. doi: 10.1016/j.coi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garten RJ, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soundararajan V, et al. Extrapolating from sequence--the 2009 H1N1 ‘swine’ influenza virus. Nat Biotechnol. 2009;27:510–513. doi: 10.1038/nbt0609-510. [DOI] [PubMed] [Google Scholar]
- 82.Kasson PM, Pande VS. Structural basis for influence of viral glycans on ligand binding by influenza hemagglutinin. Biophysical journal. 2008;95:L48–50. doi: 10.1529/biophysj.108.141507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang CC, et al. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc Natl Acad Sci U S A. 2009;106:18137–18142. doi: 10.1073/pnas.0909696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang W, et al. Glycosylation at 158N of the hemagglutinin protein and receptor binding specificity synergistically affect the antigenicity and immunogenicity of a live attenuated H5N1 A/Vietnam/1203/2004 vaccine virus in ferrets. Journal of virology. 2010;84:6570–6577. doi: 10.1128/JVI.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jayaraman A, et al. Glycosylation at Asn91 of H1N1 haemagglutinin affects binding to glycan receptors. The Biochemical journal. 2012;444:429–435. doi: 10.1042/BJ20112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Herfst S, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Russell CA, et al. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science. 2012;336:1541–1547. doi: 10.1126/science.1222526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Handel A, et al. How sticky should a virus be? The impact of virus binding and release on transmission fitness using influenza as an example. Journal of the Royal Society, Interface / the Royal Society. 2014;11:20131083. doi: 10.1098/rsif.2013.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ward MJ, et al. Evolutionary interactions between haemagglutinin and neuraminidase in avian influenza. BMC evolutionary biology. 2013;13:222. doi: 10.1186/1471-2148-13-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu R, et al. Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic. J Virol. 2012;86:9221–9232. doi: 10.1128/JVI.00697-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yen HL, et al. Hemagglutinin-neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc Natl Acad Sci U S A. 2011;108:14264–14269. doi: 10.1073/pnas.1111000108. [DOI] [PMC free article] [PubMed] [Google Scholar]