Emerging role of epigenetic mechanisms in alcohol addiction (original) (raw)
. Author manuscript; available in PMC: 2018 Apr 1.
Published in final edited form as: Alcohol Clin Exp Res. 2017 Feb 18;41(4):666–680. doi: 10.1111/acer.13338
Abstract
Alcohol use disorder (AUD) is a complex brain disorder with an array of persistent behavioral and neurochemical manifestations. Both genetic and environmental factors are known to contribute to the development of AUD, and recent studies on alcohol exposure and subsequent changes in gene expression suggest the importance of epigenetic mechanisms. In particular, histone modifications and DNA methylation have emerged as important regulators of gene expression and associated phenotypes of AUD. Given the therapeutic potential of epigenetic targets, this review aims to summarize the role of epigenetic regulation in our current understanding of AUD by evaluating known epigenetic signatures of brain regions critical to addictive behaviors in both animal and human studies throughout various stages of AUD. More specifically, the effects of acute and chronic alcohol exposure, tolerance, and post-exposure withdrawal on epigenetically-induced changes to gene expression and synaptic plasticity within key brain regions and the associated behavioral phenotypes have been discussed. Understanding the contribution of epigenetic regulation to crucial signaling pathways may prove vital for future development of novel biomarkers and treatment agents in ameliorating or preventing AUD.
Keywords: Alcoholism, Amygdala, Epigenetics, Histone acetylation, Histone methylation, DNA methylation, Anxiety
Introduction
Alcohol Use Disorder (AUD) is a multi-faceted psychiatric disorder afflicting over 16 million Americans (Center for Behavioral Health Statistics and Quality, 2015) and costing the United States over $200 billion annually (Sacks et al., 2015). Those suffering from AUD present with the inability to control their drinking, the tendency to prioritize drinking over responsibilities and activities, tolerance, withdrawal upon cessation, and continued proclivity to drink despite negative consequences (American Psychiatric Association, 2013). Recent characterization of AUD astutely defines critical disorder phases as pre-exposure “craving” (preoccupation/anticipation), acute exposure (binge/intoxication), and post-exposure (withdrawal/negative affect) (Koob, 2008;Koob and Volkow, 2010). The development and regulation of AUD is believed to be a product of both genetic and environmental influences on brain function, as these factors may account for the predisposition to alcoholism (Koob, 2003; Pickens et al., 1991; Starkman et al., 2012).
Recent research on alcohol exposure and subsequent changes in gene expression suggest that chemical modifications of the genome (epigenetic mechanisms) are important emerging molecular mechanisms in better understanding the pathophysiology of AUD and addictive behaviors (Feng and Nestler, 2013; Pandey et al., 2008a; Renthal and Nestler, 2008, 2009; Starkman et al., 2012). Epigenetic mechanisms can transiently or stably manipulate gene expression (Holliday, 2006) via DNA methylation, histone acetylation and methylation, and other less understood mechanisms (Kouzarides, 2007; Krishnan et al., 2014). These modifications can occur concurrently to varying degrees at different loci within the epigenome, allowing for an extremely diverse array of modification “signatures” that regulate gene expression in unique ways, depending on the specific arrangement (Henkels and Khorasanizadeh, 2007; Jenuwein and Allis, 2001; Wang et al., 2008). These mechanisms have recently been implicated in psychiatric disorders, including AUD (Kyzar et al., 2016a; Moonat et al., 2013; Nestler et al., 2016), and warrant detailed discussion in this review article.
Studies of alcohol’s effect on epigenetic signatures and gene expression within specific brain regions have revealed numerous modifications altering cell signaling and downstream regulation of AUD phase phenotypes (Koob and Volkow, 2010; Starkman et al., 2012). The impact of epigenetic mechanisms beyond the adolescent and adult nervous system in AUD is widespread and continuing to be explored. The role of alcohol-induced epigenetic changes in fetal alcohol spectrum disorder (FASD) and peripheral organ damage, such as the liver, has been extensively reviewed elsewhere (Bekdash et al., 2014; Lo and Zhou, 2014; Mandrekar, 2011; Shukla and Lim, 2013). Given the therapeutic potential of epigenetic manipulation in AUD, this review will focus on the available epigenetic studies in AUD phenotypes focusing on histone acetylation, histone methylation, and DNA methylation mechanisms in specific brain regions crucial to addiction, such as the amygdala, frontal cortex (FC), hippocampus, and nucleus accumbens (NAc).
Histone Acetylation Mechanism
Chromatin consists of repeated units of DNA wound around an octamer of histone proteins that organize and package nuclear genetic material (Bednar et al., 1998). Histone proteins possess amino-terminal tail domains that can be post-translationally altered via acetylation, methylation, and a variety of other modifications (Bannister and Kouzarides, 2011; Smolle and Workman, 2013). Depending on the modification location and type, associated chromatin is remodeled to become more or less condensed. This configuration alters access of transcription factors (TFs) and other DNA-binding proteins, thereby regulating transcription (Guccione et al., 2006; Guertin and Lis, 2010; Kan et al., 2007; Li et al., 2011).
Histone acetylation is a permissive mark established by histone acetyltransferase (HAT) activity, which adds an acetyl group to lysine residues on the N-terminal tails of histone proteins and generates open chromatin in a transcriptionally competent state (Bannister and Kouzarides, 2011; Marmorstein and Roth, 2001). Conversely, histone deacetylases (HDACs) remove acetyl groups from histone tails. HATs and HDACs have been linked to neuronal differentiation, synaptic plasticity, cognitive function, and psychiatric disorders (Cho and Cavalli, 2014; Nestler et al., 2016; Pirooznia and Elefant, 2013; Yeh et al., 2004). Recently, histone acetylation has been implicated in addiction mechanisms, including alcoholism (Krishnan et al., 2014; Kyzar et al., 2016a; Renthal and Nestler, 2009).
Alcohol-Induced Histone Acetylation and the Regulation of CREB
There is consistent evidence that acute and chronic alcohol exposure modulate histone acetylation in the amygdaloid circuitry leading to alcohol tolerance and dependence (Pandey et al., 2008a; Sakharkar et al., 2012). Oftentimes, changes in acetylation occur at histone 3 lysine 9 (H3K9) and are regulated in specific addiction-related brain circuitry (Bardag-Gorce et al., 2007; Finegersh and Homanics, 2014; Finegersh et al., 2015;Pandey et al., 2008a; Sakharkar et al., 2014b). Furthermore, investigations into AUD have implicated altered epigenetic regulation of synaptic plasticity and gene expression in specific brain regions, specifically via cAMP response element-binding protein (CREB) and its regulated co-factor CREB binding protein (CBP) (Correa et al., 2016; W. Guo et al., 2011; Kyzar and Pandey, 2015; Pandey, 2003).
Studies of ethanol exposure have revealed significant CREB and CBP pathway activation or attenuation in various brain regions. In the central and medial nucleus of the amygdala (CeA and MeA), acute ethanol exposure induces CREB activation and CBP expression while inhibiting HDAC activity (Pandey et al., 2008a,b; Sakharkar et al., 2012). Accordingly, acute ethanol exposure increases H3K9ac and H4K8ac within the amygdala and upregulates notable CREB target genes, including the synaptic plasticity-associated brain-derived neurotrophic factor (BDNF) and activity-regulated cytoskeleton-associated protein (Arc), neuropeptide Y (NPY), and prodynorphin (PDYN) (Fig. 1) (D’Addario et al., 2013; McCarthy et al., 2012; Pandey et al., 2008a,b). These findings correlate with increased dendritic spine density within the CeA and MeA and a reduction in anxiety-like behaviors in rats (Moonat et al., 2011; Pandey, 2003; Pandey et al., 2008b; Sakharkar et al., 2012). In the cerebellum, a brain region responsible for motor coordination that is negatively affected by alcohol, expression of the CBP-HAT and associated H3ac and H4ac is reduced after chronic ethanol exposure, unlike the aforementioned brain regions (W. Guo et al., 2011; Kalkhoven, 2004).
Fig. 1.
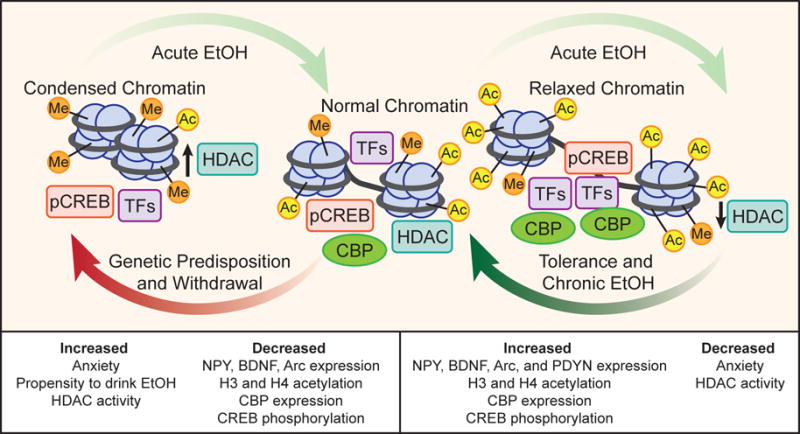
Summary of alcohol-induced histone modifications (acetylation/methylation) in the amygdala of animal models. Within the amygdala, acute ethanol (EtOH) induces histone H3 and H4 acetylation secondary to inhibited histone deacetylase (HDAC) activity and increased expression of the histone acetyltransferase [CREB-binding protein (CBP)]. These changes remodel chromatin to a less condensed form that allows for transcription factors (TFs) to access genes, such as activated CREB and CBP binding, resulting in increased expression of CREB targets, such as neuropeptide Y (NPY), prodynorphin (PDYN), brain-derived neurotrophic factor (BDNF), and activity-regulated cytoskeleton-associated protein (Arc). These modifications and related gene products in the amygdala are known to reduce anxiety-like and drinking behaviors. Tolerance and chronic exposure to EtOH normalizes molecular signatures of the epigenome. Upon withdrawal or in the case of genetic predisposition to alcoholism, HDAC activity is increased, and the subsequent histone deacetylation condenses the chromatin and downregulates the CREB pathway. This result in increased anxiety-like and drinking behaviors, decreased H3 and H4 acetylation, and decreased CBP, NPY, BDNF, and Arc expression. Ethanol will then recover normal chromatin from these states.
Similarly, in a genetic model of AUD with comorbid anxiety, alcohol-preferring (P) rats possess inherently increased HDAC2 and decreased CREB and NPY levels in the amygdala compared to non-alcohol preferring (NP) rats. Elevated HDAC2 expression in the CeA and MeA was correlated with decreased H3K9ac both globally and at promoters of Bdnf and Arc, which were innately less expressed in P rats as compared with NP rats. Upon ethanol exposure, P rats exhibited reduced anxiety-like behaviors and increased CREB activation, BDNF and Arc expression, and dendritic spine density in the CeA and MeA (Moonat et al., 2011; Pandey et al., 2005). Similarly, amygdaloid HDAC2 siRNA infusion or systemic HDAC inhibitor, Trichostatin A (TSA) treatment in P rats reduced anxiety and ethanol intake and normalized deficits in H3K9ac levels and NPY, BDNF, and Arc expression in the amygdala (Moonat et al., 2013; Sakharkar et al., 2014b). Furthermore, infusion of NPY either intracerebroventricularly (ICV) or directly into the CeA of P rats was able to attenuate anxiety and alcohol drinking behaviors and increase CREB activation and NPY levels (Gilpin et al., 2003, 2011; Zhang et al., 2010). Notably, regions of the extended amygdala, such as the CeA, MeA, and bed nucleus of the stria terminalis (BNST), have been implicated in regulating the post-exposure negative affective AUD phase (Koob and Volkow, 2010). Together, these studies strongly support a critical role of CREB pathway activation and its interaction with HAT/HDAC-associated histone acetylation mechanisms, particularly within the amygdala, in regulating AUD (Fig. 1).
Histone Acetylation Mechanisms of Alcohol Tolerance and Dependence
As discussed above, acute ethanol exposure in animal models reduces anxiety and HDAC activity and induces histone acetylation and expression of NPY in the CeA and MeA (Pandey et al., 2008a; Sakharkar et al., 2012). Likewise, HDAC2 is downregulated in the mouse cortex after acute alcohol exposure, and reduced H3K9ac at the HDAC2 promoter possibly accounts for this downregulation (Finegersh and Homanics, 2014). Other studies have also indicated that acute ethanol differentially alters various isoforms of HDAC in the periphery and brain in rats, further suggesting the role of chromatin remodeling in the action of alcohol (López-Moreno et al., 2015).
Interestingly, an identical acute dose of ethanol administered 24 hours after the first dose generates rapid ethanol tolerance and fails to produce anxiolytic effects or inhibit amygdaloid HDAC activity (Fig. 1), despite equivalent blood-alcohol concentrations to non-tolerant rats (Sakharkar et al., 2012). Treatment with TSA attenuated tolerance to the anxiolytic effect of EtOH and increased levels of NPY, H3K9ac, and H4K8ac in the CeA and MeA of rats (Sakharkar et al., 2012). These findings highlight the role of swift and dynamic epigenetic changes in modulating early molecular processes due to alcohol exposure, but these early changes can also serve as a useful molecular index of chronic tolerance that develops after long-term exposure to ethanol (Khanna et al., 1991;1992; 1996). Our laboratory has corroborated this understanding by investigating epigenetic mechanisms and gene expression similarities between rapid tolerance and chronic tolerance, finding that both are characterized by the development of tolerance to histone acetylation and anxiolytic effects of exposure (Fig. 1) (Pandey et al., 2008a; Sakharkar et al., 2012; You et al., 2014). Similarly, another lab reported that HDAC expression that was significantly altered by acute exposure returned to baseline levels after 8 repeated binge cycles in rats (López-Moreno et al., 2015). This tolerance to the anxiolytic and other behavioral effects of alcohol in animals and humans (Debatin and Barbosa, 2006; Koob et al., 1987; Koob, 2008) can encourage increased alcohol consumption and subsequent development of dependence and post-exposure withdrawal susceptibility (Tabakoff et al., 1986).
Converse to acute and chronic alcohol exposure, post-exposure withdrawal results in reduced H3K9ac, CREB activation, CBP expression, NPY levels, and expression of synaptic plasticity-associated genes in the extended amygdala (Fig. 1) (Pandey et al., 2003; 2008a,b; Sakharkar et al., 2012). Furthermore, withdrawal from both acute and chronic ethanol exposure is characterized in part by the development of anxiety (Driessen et al., 2001; Koob, 2003; Pandey et al., 2003). TSA, when administered to rats experiencing alcohol withdrawal, prevents anxiety-like behaviors and rescues the deficits in H3K9Ac, BDNF, Arc, and NPY expression within the amygdaloid brain regions (Pandey et al., 2008a; You et al., 2014). _In vitro_studies have shown that TSA induces cortical expression upregulation of the glutamate NMDA receptor, NR2B. Interestingly, the NR2B gene possesses a functional binding site for CREB that was found to be responsive to ethanol and is upregulated via H3K9ac in cases of chronic ethanol exposure and post-exposure withdrawal, and has been extensively linked to neuroplasticity, long-term potentiation, and addiction regulation (Kalivas and Volkow, 2011; Qiang et al., 2011; Rani et al., 2005). These findings cumulatively suggest the importance of histone acetylation and HDACs in regulation of multiple phases of AUD and accentuate the potential role of HDAC inhibition as treatment (Fig. 3).
Fig. 3.
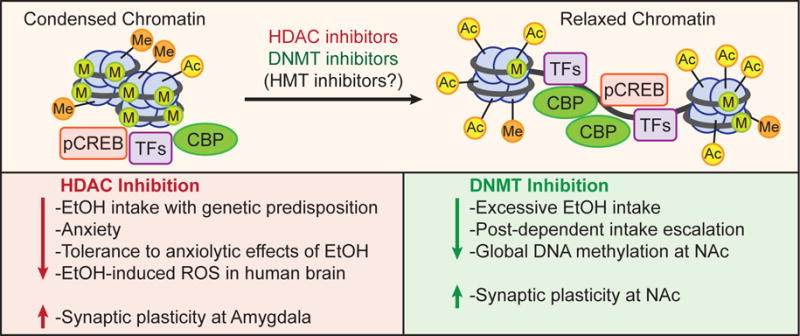
Promising pharmacotherapy treatment of alcoholism and withdrawal include DNA methyltransferase (DNMT) inhibitors, histone deacetylase (HDAC) inhibitors, and possibly histone methyltransferase (HMT) inhibitors. Current studies suggest these interventions often mimic the effects of ethanol (EtOH) by relaxing chromatin and modulating anxiolysis, synaptic plasticity, and drinking behaviors. They have been shown to alter epigenetic mechanisms, prevent alcohol-induced reactive oxygen species (ROS) in neurons, and increase synaptic plasticity within critical brain regions that regulate addiction and alcoholism, such as the nucleus accumbens (NAc) and the amygdala. Presumably as a result, numerous studies have shown that DNMT and HDAC inhibitors reduce alcohol-intake, anxiety-like behaviors, and binge-like drinking behaviors. HMT inhibitors are a promising yet currently unexplored treatment consideration in AUD.
Several studies from our and other laboratories have consistently demonstrated the molecular and behavioral impact of treatment with HDAC inhibitors. In addition to aforementioned studies, TSA and vorinostat (SAHA) were found to attenuate voluntary alcohol drinking in various animal models of alcohol intake (Jeanblanc et al., 2015; Pandey et al., 2015; Sakharkar et al. 2014b; Warnault et al., 2013), and TSA treatment also inhibits alcohol-induced reactive oxygen species (ROS) in vitro (Agudelo et al., 2011). Somewhat contrary to these findings, Qiang et al. (2014) investigated chronic EtOH exposure in mice and reported that systemic TSA treatment accelerated drinking. However, the authors note that they initiated treatments in EtOH-naive rodents and continued treatment before each EtOH exposure, while other studies provide HDAC inhibitors in conditions where chromatin architecture is innately condensed or perturbed by HDACs secondary to ethanol exposure.
The effects of other HDAC inhibitors have also been explored. Sodium butyrate (NaB) is a class I and class II HDAC inhibitor, and MS-275 is a class I HDAC inhibitor. Intraperitoneal (IP) and ICV administration of NaB and MS-275, respectively, were found to independently decrease operant self-administration of alcohol, ethanol preference, and relapse-like behaviors in alcohol dependent rats. Additionally, both treatments variably altered histone acetylation in brain regions associated with addiction and reward pathways (Jeanblanc et al., 2015; Simon-O’Brien et al., 2015). Another study indicated that_in vitro_ exposure to various HDAC inhibitors prevented the gamma-aminobutyric acid (GABA) hyposensitivity of dopaminergic neuronal firing in the ventral tegmental area (VTA) that is typically induced by chronic ethanol exposure in mice. Furthermore, this appears to be regulated by HDAC2-mediated deficits in H3K9ac in the VTA (Arora et al., 2013). Finally, valproic acid has been shown to selectively induce the degradation of HDAC2 (Krämer et al., 2003) and to dose-dependently decrease ethanol intake and preference in rats (Al Ameri et al., 2014). Together, these findings highlight the therapeutic potential of targeting epigenetic mechanisms in alcohol dependence, acute exposure tolerance, and post-exposure withdrawal when treating AUD.
Histone Acetylation in Adolescent Exposure to Alcohol
Adolescent alcohol consumption is an expanding subject in AUD and epigenetic research since adolescent exposure increases intake, risk of dependence, and addictive behaviors in adults (Kyzar et al., 2016a; Nixon and McClain, 2010). While adolescents are more resistant to locomotor disturbances, anxiolysis, and withdrawal-induced seizures, they are increasingly sensitive to withdrawal-induced anxiety, thus encouraging escalating alcohol intake and binge drinking (Chung et al., 2008; Pandey et al., 2015; Sakharkar et al., 2014a; Spear and Varlinskaya, 2005). Though the field of study is recently burgeoning, already there is a plethora of findings that suggest repeated adolescent EtOH exposure results in persistent epigenetic modifications within the specific neurocircuitry of addiction that modulates AUD phase regulation, such as that of alcohol intake and anxiety later in life (Kyzar et al., 2016a; Pandey et al., 2015).
Studies have begun revealing a complex network of histone acetylation changes in various brain regions secondary to acute and chronic alcohol exposure in adolescents. For instance, acute EtOH exposure in adolescent rats causes attenuated HDAC activity in the amygdala and BNST (Sakharkar et al., 2014a). Similarly, a study also showed increased H3K9ac, H4K5ac, and H4K12ac within the prefrontal cortex (PFC) in an adolescent binge-drinking model (Montesinos et al., 2016). Additionally, binge-like exposure in adolescent rats induced HAT activity in the PFC, resulting in increased H3Kac and H4Kac both globally and at promoters of genes such as_cfos_, which are believed to regulate addiction in other parts of the brain by modulating synaptic plasticity or transcription mechanisms (Pascual et al., 2012). In the NAc, global histone H3 and H4 acetylation is also increased (Pascual et al., 2009). These regions possibly experience transcription activation at important, specific genomic locations relevant to AUD.
In the amygdala, however, binge-like drinking via the adolescent intermittent ethanol (AIE) model induces persistent increases in global HDAC activity and HDAC2 expression (Pandey et al., 2015) alongside decreased H3K9ac both globally and at the promoter regions of Bdnf and Arc in adulthood. Predictably, these findings correlate with significant decreases in Bdnf and_Arc_ gene expression and dendritic spine density, particularly in the CeA and MeA of rats (Pandey et al., 2015). This is especially interesting as adult rats who underwent AIE also exhibit increased anxiety and alcohol intake that is abated by TSA treatment (Pandey et al., 2015).
In the hippocampus, both adolescent and adult tissues do not exhibit global histone H3 or H4 acetylation changes after binge-like ethanol exposure (Pascual et al., 2009). Interestingly, Sakharkar et al. (2016) reported that adult rats after AIE showed significant increases in hippocampal HDAC activity. Furthermore, they present with decreased hippocampal global CBP and H3K9ac levels as well as H3K9/14ac levels at the Bdnf exon IV promoter (Sakharkar et al., 2016). Importantly, adult TSA treatment reversed the changes in hippocampal histone acetylation of Bdnf gene and normalized expression of dysregulated hippocampal neurogenesis markers (Sakharkar et al., 2016). Once again, these findings implicate neuroplasticity of certain brain regions in regulating alcoholism (Kyzar et al., 2016a; Pascual et al., 2009) and highlight the therapeutic potential of HDAC inhibitors (Falkenberg and Johnstone, 2014; Kyzar and Pandey, 2015). Considering the imperative role that environment and epigenetics play in adolescent development and the ample evidence regarding epigenetic regulation of addiction neurocircuitry in adolescent alcohol exposure, it is clear that more investigation is warranted regarding this at-risk population.
Histone Methylation Mechanisms
Recently, histone methylation has gained traction in epigenetic research and so far has been linked to cancer, development, neuroplasticity, learning, addiction, and anxiety (Covington et al., 2011; Kim et al., 2013; Pattaroni and Jacob, 2013; Subbanna and Basavarajappa, 2014; Tachibana et al., 2002). Histone methyltransferases (HMTs) transfer one or more methyl groups from the methyl donor, s-adenosyl methionine (SAM), onto histone N-terminal tail lysine or arginine residues, and histone demethylators (HDMs) remove methyl groups from histones. Arginine residues can maintain one or two methyl groups, and lysine residues can be mono-, di-, or tri-methylated (Krishnan et al., 2014; Pattaroni and Jacob, 2013). Unlike histone acetylation, histone methylation has variable effects on transcription depending on the modified residues, interactions with other epigenetic factors, and the valence of methylation.
Histone Methylation as an Epigenetic Regulator in Alcoholism
Histone methylation is largely understudied relative to histone acetylation and DNA methylation, but recent findings have revealed the critical role it plays in regulating psychiatric disorders, including AUD. For example, it has been shown that acute EtOH exposure in mice significantly increased levels of H3K4me3, an activating mark often found near transcription start sites, in the cortex (Finegersh and Homanics, 2014). Similarly, another study found the activating H3K4me2 mark to be increased alongside histone acetylation at the promoters of TFs linked to synaptic plasticity and learning within the PFC of adolescent rats after binge-like alcohol exposure (Pascual et al., 2012).
Repressive histone methylation marks in the cortical structures have also emerged as important regulators of AUD. For instance, Qiang et al. (2011) investigated cell cultures of mouse cortical neurons and showed that binge-like ethanol exposure decreased the suppressive H3K9me2 and H3K9me3 marks at the aforementioned _NR2B_NMDA receptor gene. Furthermore, the decrease in repressive H3K9 methylation markers correlated with a likely causative decline in HMT expression, including that of G9a, Setdb1, and Suv39h1 (Qiang et al., 2011). Of these, G9a is largely responsible for H3K9me2 production (Shinkai and Tachibana, 2011; Tachibana et al., 2002; 2008) and has recently been investigated in addiction and alcohol exposure (Covington et al., 2011; Maze et al., 2010; Subbanna et al., 2013; Sun et al., 2012). More specifically, G9a has emerged as a key contributor to alcohol-induced gene expression regulation in FASD (Basavarajappa and Subbanna, 2016). Recently, the HMT known as PR domain-containing 2, with ZNF domain (PRDM2) was found to regulate post-dependent AUD phenotypes, including alcohol intake and stress-induced relapse. Interestingly, knockdown of Prdm2 in non-dependent rat dorsomedial PFC prompted post-dependent behaviors, revealing the future treatment potential of HMT manipulation (Barbier et al., 2016).
In the amygdala of adult rats, acute ethanol exposure significantly reduces H3K27me3 at the promoters of Pdyn and Pnoc(prepronociceptin) (D’Addario et al., 2013), which have been implicated in alcohol dependence (Karpyak et al., 2013; Xuei et al., 2006). Our laboratory found that adolescent alcohol exposure altered histone methylation in the amygdala in adulthood as well as a propensity to drink and exhibit more anxiety than their control counterparts (Kyzar et al., 2016b; Pandey et al., 2015). Specifically, the lysine demethylase, LSD1, and its neuron-specific isoform,Lsd1+8a, which demethylate H3K4me2/3 and H3K9me2/3, were both persistently downregulated in the amygdala. Furthermore, an associated increase in global and Bdnf exon IV-specific H3K9me2 but not H3K4me2 was observed in rats exposed to EtOH in adolescence compared to controls. Acute ethanol challenge to adult rats after AIE attenuated anxiety-like behaviors, deficits in_Lsd1+8a_ expression, and increase in H3K9me2 levels at_Bdnf_ exon IV in the amygdala (Kyzar et al., 2016b). Much like previously described histone acetylation reports, this data suggests adolescent exposure to alcohol results in a decrease in gene activation within the amygdala and increased activity within the PFC secondary to epigenetic reprogramming, specifically regulating the expression of genes that modulate synaptic plasticity and possibly contributing to adult psychopathology. Overall, these findings encourage future exploration, as current studies focus on the PFC and the amygdala.
Histone Methylation in Human Alcoholics
Few human studies of histone methylation currently exist, but the advancements in RNA and ChIP-sequencing have supported recent interest in the field. Specifically, the Harris lab spearheaded an exploration of genome-wide H3K4me3 activation marks in post-mortem hippocampus samples of alcohol abusers, revealing a distinct pattern of H3K4me3 at gene networks known to be functionally interconnected and either potentially or definitively linked to addiction regulation (Farris et al., 2015). Similarly, a study of the amygdala and FC of postmortem tissue in alcoholics found multiple H3K4 HMTs to be upregulated in addition to a global increase in H3K4me3 (Ponomarev et al., 2012). Another study specifically reporting on genome-wide associations of alcohol withdrawal symptoms described multiple significant polymorphisms in the gene for a HDM known as KDM4C (Wang, et al. 2012), implicating histone methylation as a regulator of AUD phenotypes that warrants further investigation. Overall, data regarding histone methylation in human alcoholics is intriguing but limited.
DNA Methylation Mechanisms
DNA methylation is a repressive mark that is characterized by methylation of the cytosine pyrimidine ring at carbon-5. Dense regions of CpG dinucleotides (CpG islands) are often found near promoter transcription start sites and their methylation is generally known to block TFs and silence associated genes (Comb and Goodman, 1990; Li and Zhang, 2014). Furthermore, some repressor complexes and TFs gravitate toward methyl-binding-proteins like MeCP2 and subsequently modify histones, thus further regulating gene expression (Chahrour et al., 2008; Jones et al., 1998; Nan et al., 1998). In order to establish DNA methylation, DNA methyltransferases (DNMT1, 3a, and 3b) relocate a methyl group from SAM to the target cytosine. Importantly, these DNMTs are abundant in fully differentiated adult neurons and are believed to play a critical role in gene regulation (Feng et al., 2005; Szulwach et al., 2011). Methylation removal from DNA also occurs, presumably via removal of the methyl group or secondary to inhibition or loss of DNMT activity (Wu and Zhang, 2010). Notably, multiple base-excision and nucleotide-excision repair mechanisms result in demethylation (Ma et al., 2009; Wu and Zhang, 2010). Furthermore, recent investigations into the ten-eleven translocation (TET) family of enzymes that hydrolyze 5-methylcytosine to 5-hydroxymethylcytosine (5hmC) suggest 5hmC may alter transcription regulation by inhibiting identification by DNA-binding enzymes such as DNMT1 or by activating the base-excision repair pathway (J.U. Guo et al., 2011; Jin et al., 2010; Valinluck and Sowers, 2007).
DNA Methylation as an Epigenetic Regulator in Animal Models
Dysregulated DNA methylation mechanisms have been implicated in psychiatric disorders including AUD (Nestler et al., 2016; Starkman et al., 2012). Studies on the effects of alcohol exposure on DNA methylation remain conflicting after decades of research. An early study of rats chronically exposed to ethanol reported more relaxed chromatin specifically in neurons and implicated non-histone protein-DNA interactions (Mahadev and Vemuri, 1998). To explain this phenomenon, multiple debatable theories and contrary findings have emerged. Firstly, chronic ethanol consumption commonly leads to folate and vitamin B deficiencies, resulting in increased homocysteine levels and downregulation of SAM (Niculescu and Zeisel, 2002). However, increased homocysteine has been elsewhere linked to global hypermethylation and presumably more condensed and transcriptionally silenced chromatin in alcoholics (Bönsch et al., 2004a). Secondly, genomic insults secondary to alcoholism may contribute to DNA hypomethylation upon base-excision repair (Chen et al., 2011). And thirdly, DNMT3b expression was found to be downregulated in adult alcoholics, but it was notably associated with genetic hypermethylation, not hypomethylation as one might expect (Bönsch et al., 2006). Ultimately, there are no conclusive theories to explain global neuronal hypomethylation. In fact, recent evidence in animal models counters the previously detailed dogma of extensive global DNA hypomethylation in neurons secondary to chronic alcohol exposure. Rather, it is possible that acute ethanol exposure may reduce DNA methylation, while chronic exposure results in DNA hypermethylation in various brain regions (Fig. 2).
Fig. 2.
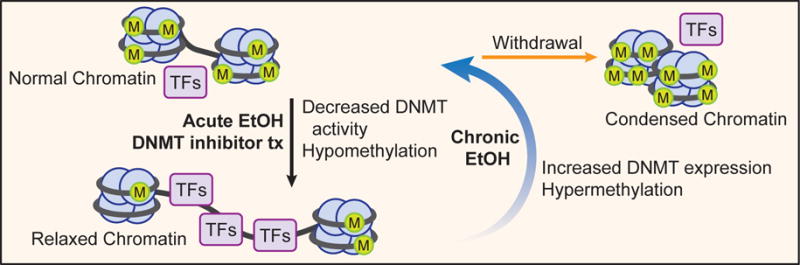
Hypothetical model indicating possible changes in DNA methylation after acute ethanol (EtOH), chronic ethanol exposure, and withdrawal upon cessation. Animal studies consistently report that acute ethanol exposure induces global DNA demethylation and presumably relaxed chromatin within various brain nuclei, altering transcription factors (TFs) access and associated gene expression. Chronic exposure in animal and human samples reportedly increases global DNA methylation in several brain regions and peripheral blood samples. Based on these findings and the role chromatin remodeling plays in anxiety and drinking behavior, we hypothesize that alcohol withdrawal results in especially hypermethylated DNA and presumably condensed chromatin as a consequence of dysregulated epigenetic mechanisms. It is possible that DNMT inhibitor treatment (Tx) or acute ethanol exposure may lead to relaxed chromatin architecture due to DNMT inhibition.
There are few studies investigating acute ethanol exposure on global DNA methylation; nonetheless, in vitro and _in vivo_evidence suggests that acute alcohol exposure and alcohol metabolites inhibit DNMT expression and activity, including that of brain tissues (Garro et al., 1991; Zhang et al., 2014). More specifically, DNMT3a expression was found to be downregulated in astrocytes secondary to acute alcohol exposure in animals (Zhang et al., 2014). Furthermore, a study of adolescent rats showed that DNMT activity was attenuated in both the BNST and the amygdala after acute alcohol exposure (Sakharkar et al., 2014a). While these studies are preliminary and limited in number, their contrast to chronic alcohol exposure-induced DNA methylation signatures encourages exploration of DNA methylation in different phases of AUD.
Chronic exposure, contrary to acute exposure, appears to induce global hypermethylation of DNA in multiple brain regions. For instance, in chronically exposed adult mice, there is reportedly an increase in DNMT1 expression within the NAc (Warnault et al., 2013). Similarly, another study which examined the NAc and medial prefrontal cortex (mPFC) of post-dependent rats after weeks of abstinence showed a persistent increase in global DNA methylation as well as a persistent increase in DNMT1 expression that was associated with the downregulation of a cluster of synaptic genes specifically within mPFC neurons (Barbier et al., 2015). These studies cumulatively suggest that chronic ethanol exposure induces global DNA hypermethylation in various brain regions of animal models associated with addiction and AUD. Furthermore, the few studies available suggest that acute but not chronic ethanol exposure may attenuate DNMT activity (Fig. 2).
The effect of acute and chronic EtOH exposure on DNA methylation of specific promoter sequences in animal models has also been investigated. For instance, in a study examining the effect of acute ethanol exposure on tissue plasminogen activator (tPA) in rat astrocytes—due to astrocytes’ essential role in neuroplasticity—the results demonstrated that acute in vitro alcohol exposure inhibited DNMT activity and DNMT3a expression, decreased DNA methylation at the tPA promoter, and increased tPA expression, suggesting a potential indirect role of DNA methylation in regulating neuroplasticity upon alcohol exposure (Zhang et al., 2014). A study of chronic ethanol exposure in cultured mouse cortical neurons showed a reduction in DNA methylation at the _NR2B_promoter, resulting in increased expression of NR2B (Marutha Ravindran and Ticku, 2004). Similarly, chronic EtOH exposure in fetal rats reportedly induced hypermethylation of Bdnf and subsequent reduction in BDNF protein expression within the olfactory bulb, suggesting yet again a role of alcohol-induced epigenetic mechanisms in regulation of synaptic plasticity (Maier et al., 1999).
Similar to HDAC inhibition, treatments of alcohol dependence models suggest that DNMT inhibition provides a promising therapeutic option in AUD by altering both global and promoter-specific DNA methylation in brain regions central to addiction neurocircuitry (Fig. 3). Mice and rats given DNMT inhibitors, such as systemic 5-azacytidine and ICV RG108, have exhibited significantly decreased alcohol intake and binge-like behaviors accompanied by decreased global DNA methylation within the NAc and increased synaptic plasticity-associated transcription (Barbier et al., 2015; Warnault et al., 2013). Furthermore, DNMT inhibition via ICV administration of RG108 also reversed alcohol-induced hypermethylation and changes in synaptotagmin 2 (Syt2) mPFC expression, which has been confirmed to regulate alcohol-drinking behaviors (Barbier et al., 2015). We also recently found that decreased expression of GADD45b increased Bdnf DNA methylation in the NAc and promoted alcohol drinking behaviors in mice (Gavin et al., 2016). These combined studies emphasize the importance of DNA methylation intervention in regulating chromatin remodeling and gene expression in alcoholism and show promise for DNMT inhibition as a treatment of AUD.
DNA Methylation in Human Alcoholics
Genome-wide and promoter-specific DNA methylation study of peripheral blood cells is a newly developing approach to isolating genes of interest and potential biomarkers in humans. At a time when biomarkers for acute and chronic substance abuse are a necessity (Andersen et al., 2015;Volkow et al., 2015), DNA methylation has emerged as a potentially reliable signature of environmental exposures for future clinical testing (Ladd-Acosta, 2015). Additionally, many gene networks and epigenetic signatures have emerged in human studies related to AUD phase phenotypes.
Firstly, there are several studies revealing changes in DNA methylation signatures within addiction related pathways. For example, two studies of alcoholic patients in the post-exposure withdrawal phase of AUD either found significant hypermethylation or no change at the dopamine transporter gene (DAT), though both reported a significant negative correlation or clear trend for negative correlation between DAT methylation and craving (Hillemacher et al., 2009a, Nieratschker et al., 2014). A separate study conversely reported decreased DNA methylation at a single CpG island site in alcoholic patients relative to controls (Jasiewicz et al., 2015). A relevant study that investigated DNA methylation of the monoamine oxidase A gene (MAOA) reported a significant association between the degree of alcohol dependence in female patients and the level of_MAOA_ methylation (Philibert et al., 2008). A very recent study detected methylation changes of extremely heavy daily drinkers at GABA receptor genes (GABRD and_GABBR1_), providing a potential future biomarker of current or recent heavy drinking (Liu et al., 2016). Furthermore, the previously discussed chronic ethanol-induced hypomethylation of_NR2B_ in alcohol-dependent rodents is also seen in humans, and the gene is increasingly hypomethylated depending on AUD severity (Biermann et al., 2009a). While not currently clear, these studies suggest potential for future biomarkers and further investigations into the DNA methylation regulation of synaptic neurotransmission in AUD.
Other gene-specific investigations have reported significant changes in pathways relevant to alcohol craving, such as pro-opiomelanocortin (POMC) and alpha-synuclein (SNCA) (Bönsch et al., 2004a,b; 2005a; Foroud et al., 2007). Specifically, a cluster of DNA methylation site alterations within the_POMC_ promoter was found to be correlated to alcohol craving in both post-exposure and pre-exposure craving phases of AUD (Muschler et al., 2010). Additionally, certain_POMC_ CpG sites were reportedly hypermethylated in particular populations of dependent patients (Zhang et al., 2013). At the SNCA promoter, hypermethylation was reported in alcoholics in the acute exposure and post-exposure withdrawal phases (Bönsch et al., 2005b).
Genome-wide studies have also emphasized a variety of genes known to be involved in alcohol metabolism and related reward circuitry. For instance, one study compared alcoholics to their nondependent siblings and revealed several genes of interest with altered methylation signatures, including that of an aldehyde dehydrogenase (ALDH1L2) involved in eliminating alcohol metabolites, a GABA receptor (GABRP) suggestive of decreased GABA binding in alcoholics, a glutamate decarboxylase (GAD1) that has been well-studied in alcohol-related GABA production, and a dopamine beta-hydroxylase (DBH) that has been strongly linked to alcohol tolerance (Zhao et al., 2013). Patient AUD phase, however, is unclear. A separate preliminary genome-wide investigation also reported epigenetically altered alcohol metabolism in several similar gene clusters and pathways previously associated with alcoholism, such as the aldehyde dehydrogenase family, stress pathways, and inflammatory responses (Zhang et al., 2013).
Recent studies have begun to combine genome-wide analysis of DNA methylation with that of intriguing phenotypic metrics. For instance, a recent study confirmed a genome-wide report of hypomethylation at the ganglioside-induced differentiation associated protein 1 (GDAP1) gene and further found that_GDAP1_ DNA methylation was significantly associated with severity of patient AUD (Brückmann et al., 2016). Another recent study examined DNA methylation in monozygotic twins with discordant AUD status and complemented this work with personality evaluation and MRI imaging during an impulsiveness task (Ruggeri et al., 2015). They found a significant association of hypermethylation at the 3′-protein-phosphatase-1G gene (PPM1G) with AUD as well as two established risk factors of AUD—adolescent escalation of alcohol intake and impulsivity (Ruggeri et al., 2015). These multi-faceted approaches garner invaluable information considering the complexity of AUD and support the notion that stratification of patient phenotypes assists in elucidating specific underlying mechanisms.
The genes noted here are believed to play a significant role in previously recognized AUD regulation mechanisms, but the number of other gene specific and genome-wide studies has greatly increased in recent years, resulting in a plethora of gene candidates available for further investigation (Zhang and Gelernter, 2016). As genome-wide epigenetic studies continue to be produced in different populations, it is likely that more unrecognized mechanisms will begin to unfold and the role of DNA methylation in AUD can further be elucidated.
DNA Methylation in Human Post-mortem Brain Tissue
Post-mortem tissue studies of humans diagnosed with AUD provide an important opportunity to investigate brain region-specific changes in DNA methylation. Unfortunately, such studies are significantly limited in breadth of tissue and sample size due to obvious restrictions in tissue availability, and so far they have elicited a variety of results. Minimal investigation of post-mortem precuneus and putamen brain regions of alcoholics has been completed, though gene networks involved in the immune and inflammatory response, lipid metabolism, and gastrointestinal disease have been identified as having significantly altered DNA methylation status in alcoholics (Hagerty et al., 2016).
In the amygdala and FC of alcoholics, studies are more extensive, though results vary greatly. In one study, global DNA hypomethylation and decreased transcription of DNMT1 was found, contrary to several animal studies (Ponomarev et al., 2012). Similar studies in human FC detected extensive hypermethylation in males but not females (Wang et al., 2016) or no such change (Manzardo et al., 2012). Interestingly, the Ponomarev study also suggested that GC content altered the dysregulation pattern of genes. Specifically, GC-rich and GC-poor gene networks were respectively upregulated and downregulated in alcoholics (Ponomarev et al., 2012).
These studies have also revealed epigenetic signature shifts and dysregulation in a myriad of gene networks, including synaptic transmission genes in FC neurons of alcoholics, which is consistent with epigenetic animal studies (Krishnan et al., 2014; Ponomarev et al., 2012). Similarly, multiple genes involved in chromatin remodeling, histone deacetylation, and transcription repression were significantly upregulated in the amygdala and FC of alcoholics (Ponomarev et al., 2012), and several histone gene promoters were found to be heavily methylated in the FC (Manzardo et al., 2012), echoing the growing relevance of histone regulation in AUD.
Upon investigation of specific genetic loci, Taqi et al. (2011) investigated known_PDYN_ CpG polymorphisms associated with increased risk of alcoholism and discovered differential DNA methylation in the post-mortem PFC of alcoholics. This study has specifically emphasized the value of combining genome-wide population studies with epigenomic studies in order to effectively isolate risk-associated epigenomic shifts in certain populations. Other studies have begun investigating comorbidities and revealed altered TET1 expression in PFC of comorbid psychotic and alcoholic post-mortem brains, broadening the scope of alcohol-induced chromatin remodeling (Guidotti et al., 2013). Though the study of post-mortem epigenomic signatures in alcoholism is in its infancy, investigations of various brain regions will likely continue to increase, hopefully elucidating the significance of these findings and guiding animal model investigations as well.
Conclusions
We have provided an evaluation of well-characterized epigenetic regulation of AUD phenotypes via modulation of gene expression within brain circuitry pivotal to anxiety, tolerance, dependence, and withdrawal regulation. Across diverse epigenetic pathways in preclinical and human studies, some consistent notions have emerged.
- A distinct epigenetic signature is present in unique phases of AUD and potentially varies based on disease phase and severity. Koob and Volkow (2010) elegantly portrayed phases of AUD as pre-exposure “craving” (preoccupation/anticipation), acute exposure (binge/intoxication), and post-exposure (withdrawal/negative affect). The reviewed studies consistently identified unique epigenetic regulation in post-dependence, acute exposure/binge-drinking, and withdrawal states. Similarly, a very recent review concluded that three domains of addictive disorders which correlate with the aforementioned phases of AUD, could be utilized to guide genetic, clinical, and therapeutic studies of the complex and multi-faceted addiction disorders (Kwako et al., 2016).
Alcoholics present with extremely varied levels of dependence, exposure, tolerance, and withdrawal symptoms. Perhaps this complexity contributes to the rare correspondence of results among studies and the ever-increasing list of genes of interest. Notably, preclinical studies that specifically study the withdrawal phase have had promising success in epigenetic investigation (Biermann et al., 2009a; Hillemacher et al., 2009b; Pandey et al., 2003; 2008a; Wang et al., 2012). Furthermore, previous attempts to stratify patients in manners such as Lesch’s typology have provided intriguing insights, such as revealing a potential relationship between DNA methylation of homocysteine-induced endoplasmic reticulum protein (HERP) and drinking by type (Biermann et al., 2009b; Bleich et al., 2006). It will be important going forward to critically stratify subject populations at least by disease severity and AUD phase upon sample retrieval in order to further understand the phases of alcohol abuse and severity of disease, as they are likely uniquely epigenetically regulated. - The synaptic plasticity and interconnectivity of brain regions involved in drinking, craving, reward, or negative affect consistently prove instrumental to the AUD and associated phenotypes across various epigenetic mechanisms reviewed here. The CREB pathway, which modulates connectivity and synaptic plasticity via BDNF and Arc expression, has repeatedly emerged an important molecular mechanism in alcoholism and addiction (Carlezon Jr et al., 2005; W. Guo et al., 2011; McCarthy et al., 2012; Pandey, 2003; Pandey et al., 2004). Upstream and downstream regulation of epigenetic processes in the amygdala, cerebellum, PFC, NAc, and other brain regions has contributed to acute exposure effects, tolerance, adolescent exposure-induced phenotypes, and withdrawal effects (Krishnan et al., 2014; Maze and Nestler, 2011; Moonat et al., 2013; Pandey et al., 2008a; 2015; Sakharkar et al., 2016). Recently, epigenetic transgenerational effects of drug exposure have been implicated in addiction as well (Rachdaoui and Sarkar, 2014; Vassoler et al., 2013).
Limitations
There are certainly limitations to be considered in AUD epigenetic research. First, chronic alcohol exposure impacts many non-neuronal tissues, resulting in substantial and perhaps confounding or mediating sequelae. For example, blood sample cell composition shifts in alcoholics versus controls may alter findings. Notably, there are correction methods available for the composition shifts for DNA methylation, and future studies should be diligent in accounting for such changes (Houseman et al., 2012; Jaffe and Irizarry, 2014).
Second, there is reasonable concern as to whether or not epigenetic signatures from blood can serve as reliable proxies for the brain because of the tissue-specific nature of epigenetic mechanisms. While one study investigated DNA methylation signatures in brain regions relative to blood samples and concluded that blood-based studies offered limited value beyond the cortex (Hannon et al., 2015), several other studies have reported significant similarity in inter-individual variation between blood samples and distinct brain regions (Davies et al., 2012;Ewald et al., 2014; Tylee et al., 2013). Notably, the value of blood biomarkers remains a promising opportunity for study and clinical application, regardless.
A third limitation resides in the sheer complexity of AUD, which is characterized by widely varied severity in multiple phases, each affected by environmental and genetic influences. This complexity and the emerging understanding that individual phases of AUD are uniquely regulated makes disease phase and severity stratifications important. It’s possible that discrepancies between studies occur due to a lack of appropriate patient stratification. For instance, some studies report on “healthy” undiagnosed individuals based on their self-reported drinking, stratify the population as drinker versus non-drinker, or do not provide sufficient information regarding the current phase of AUD patients (Philibert et al., 2012; Zhang et al., 2011; Zhu et al., 2012).
The importance of accurate disease characterization and severity stratification in epigenetic analyses is further highlighted in a recent pair of studies. In the first study, self-reported alcohol intake of patients with unknown AUD status was used for DNA methylation analysis (Philibert et al., 2012), while the second study specifically included patients recruited from alcohol treatment centers who consumed significantly more alcohol than the previous study (Philibert et al., 2014). The latter study detected low-grade changes and effectively determined a widespread shift in DNA methylation associated with acute sustained intake of alcohol, suggesting that epigenetic signatures in peripheral mononuclear cells can be used to monitor alcohol use when patients are effectively selected and grouped.
Future Work
The study of specific effects of AUD phases on neuroconnectivity has potential to usher in a more comprehensive understanding of neural adaptations and the value of targeting synaptic plasticity pathways for future addiction treatment. However, the recent evolution of epigenome-wide mapping continues to reveal novel gene network pathways perturbed by AUD, such as inflammation-related pathways, and more studies are needed in this direction.
To date, both systemic and brain-specific interventions with HDAC inhibitors and DNMT inhibitors decrease anxiety-like and alcohol-drinking behaviors and increase synaptic plasticity at the NAc and amygdala (Agudelo et al., 2011; Barbier et al., 2015; Pandey et al, 2008a; Sakharkar et al., 2016; Simon-O’Brien et al., 2015; Warnault et al., 2013) (Fig. 3), but HMT inhibitors have yet to be fully explored. These recent advances provide inspiration for future investigation into HMTs and the potential value of histone acetylation and demethylation induction. Furthermore, epigenetic mechanisms not discussed here, such as histone phosphorylation, ubiquitylation, ribosylation, and sumoylation, are as of yet poorly defined (Kouzarides, 2007), and contemporary AUD literature rarely considers these nebulous mechanisms. Moving forward, the elucidation of their mechanisms and subsequent study of their potential roles in AUD will likely advance.
Despite the complex nature of AUD and epigenetics, the robust indication of the importance of epigenetic mechanisms in AUD development, maintenance, and attenuation in multiple phases of the disorder presented here ultimately provides a favorable environment for achieving effective and innovative future treatments for those afflicted with AUD.
Acknowledgments
The work in the laboratory of SCP was supported by National Institute on Alcohol Abuse and Alcoholism Grants UO1AA-019971, U24AA-024605 (Neurobiology of Adolescent Drinking in Adulthood project), RO1AA-010005, RO1AA021662, P50AA-022538 (Center for Alcohol Research in Epigenetics) and by the Department of Veterans Affairs (I01BX000143, Merit Review Grant; Senior Research Career Scientist award). Authors would like to thank Diantha LaVine for her help in artwork and Luiza Kulikowska for editing and helping in the preparation of this article.
Footnotes
Conflict of interest:
SCP reports that a US patent application entitled “Histone acetyltransferase activators and histone deacetylase inhibitors in the treatment of alcoholism” (serial number 60/848237 filed on September 29th, 2006) is currently pending. TDMB reported no biomedical financial interests or potential conflicts of interest.
References
- Agudelo M, Gandhi N, Saiyed Z, Pichili V, Thangavel S, Khatavkar P, Yndart-Arias A, Nair M. Effects of alcohol on histone deacetylase 2 (HDAC2) and the neuroprotective role of trichostatin A (TSA) Alcohol Clin and Exp Res. 2011;35(8):1550–1556. doi: 10.1111/j.1530-0277.2011.01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Ameri M, Al Mansouri S, Al Maamari A, Bahi A. The histone deacetylase (HDAC) inhibitor valproic acid reduces ethanol consumption and ethanol-conditioned place preference in rats. Brain Res. 2014;1583:122–131. doi: 10.1016/j.brainres.2014.07.051. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 5th. Arlington, VA: American Psychiatric Publishing; 2013. [Google Scholar]
- Andersen AM, Dogan MV, Beach SR, Philibert RA. Current and future prospects for epigenetic biomarkers of substance use disorders. Genes. 2015;6(4):991–1022. doi: 10.3390/genes6040991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora DS, Nimitvilai S, Teppen TL, McElvain MA, Sakharkar AJ, You C, Pandey SC, Brodie MS. Hyposensitivity to gamma-aminobutyric acid in the ventral tegmental area during alcohol withdrawal: reversal by histone deacetylase inhibitors. Neuropsychopharmacology. 2013;38(9):1674–1684. doi: 10.1038/npp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier E, Tapocik JD, Juergens N, Pitcairn C, Borich A, Schank JR, Sun H, Schuebel K, Zhou Z, Yuan Q, Vendruscolo LF, Goldman D, Heilig M. DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. J Neurosci. 2015;35(15):6153–6164. doi: 10.1523/JNEUROSCI.4571-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier E, Johnstone A, Khomtchouk BB, Tapocik JD, Pitcairn C, Rehman F, Augier E, Borich A, Schank JR, Rienas CA, Van Booven DJ, Sun H, Nätt D, Wahlestedt C, Heilig M. Dependence-induced increase of alcohol self-administration and compulsive drinking mediated by the histone methyltransferase PRDM2. Mol Psychiatry. 2016 doi: 10.1038/mp.2016.131. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardag-Gorce F, French BA, Joyce M, Baires M, Montgomery RO, Li J, French S. Histone acetyltransferase p300 modulates gene expression in an epigenetic manner at high blood alcohol levels. Exp Mol Pathol. 2007;82(2):197–202. doi: 10.1016/j.yexmp.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavarajappa BS, Subbanna S. Epigenetic mechanisms in developmental alcohol-induced neurobehavioral deficits. Brain Sciences. 2016;6(2):E12. doi: 10.3390/brainsci6020012. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci U S A. 1998;95(24):14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekdash R, Zhang C, Sarkar D. Fetal alcohol programming of hypothalamic proopiomelanocortin system by epigenetic mechanisms and later life vulnerability to stress. Alcohol Clin Exp Res. 2014;38(9):2323–2330. doi: 10.1111/acer.12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biermann T, Reulbach U, Lenz B, Frieling H, Muschler M, Hillemacher T, Bleich S. N-methyl-D-aspartate 2b receptor subtype (NR2B) promoter methylation in patients during alcohol withdrawal. J Neural Transm (Vienna) 2009a;116(5):615–622. doi: 10.1007/s00702-009-0212-2. [DOI] [PubMed] [Google Scholar]
- Biermann T, Reulbach U, Lenz B, Muschler M, Sperling W, Hillemacher T, Kornhuber J, Bleich S. Herp mRNA expression in patients classified according to Lesch’s typology. Alcohol. 2009b;43(2):91–95. doi: 10.1016/j.alcohol.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Bleich S, Lenz B, Ziegenbein M, Beutler S, Frieling H, Kornhuber J, Bönsch D. Epigenetic DNA hypermethylation of the HERP gene promoter induces down-regulation of its mRNA expression in patients with alcohol dependence. Alcohol Clin Exp Res. 2006;30(4):587–591. doi: 10.1111/j.1530-0277.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- Bönsch D, Greifenberg V, Bayerlein K, Biermann T, Reulbach U, Hillemacher T, Kornhuber J, Bleich S. Alpha-synuclein protein levels are increased in alcoholic patients and are linked to craving. Alcohol Clin Exp Res. 2005a;29(5):763–765. doi: 10.1097/01.alc.0000164360.43907.24. [DOI] [PubMed] [Google Scholar]
- Bönsch D, Lenz B, Kornhuber J, Bleich S. DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. Neuroreport. 2005b;16(2):167–70. doi: 10.1097/00001756-200502080-00020. [DOI] [PubMed] [Google Scholar]
- Bönsch D, Lenz B, Fiszer R, Frieling H, Kornhuber J, Bleich S. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm (Vienna) 2006;113(9):1299–1304. doi: 10.1007/s00702-005-0413-2. [DOI] [PubMed] [Google Scholar]
- Bönsch D, Lenz B, Reulbach U, Kornhuber J, Bleich S. Homocysteine associated genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm (Vienna) 2004a;111(12):1611–1616. doi: 10.1007/s00702-004-0232-x. [DOI] [PubMed] [Google Scholar]
- Bönsch D, Reulbach U, Bayerlein K, Hillemacher T, Kornhuber J, Bleich S. Elevated alpha synuclein mRNA levels are associated with craving in patients with alcoholism. Biol Psychiatry. 2004b;56(12):984–986. doi: 10.1016/j.biopsych.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Brückmann C, Di Santo A, Karle KN, Batra A, Nieratschker V. Validation of differential GDAP1 DNA methylation in alcohol dependence and its potential function as a biomarker for disease severity and therapy outcome. Epigenetics. 2016;11(6):456–463. doi: 10.1080/15592294.2016.1179411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends in Neurosciences. 2005;28(8):436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Center for Behavioral Health Statistics and Quality. (HHS publication no. SMA 15-4927, NSDUH series H-50).Behavioral health trends in the United States: Results from the 2014 national survey on drug use and health. 2015 [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320(5880):1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Pan CH, Chen CC, Huang MC. Increased oxidative DNA damage in patients with alcohol dependence and its correlation with alcohol withdrawal severity. Alcohol Clin Exp Res. 2011;35(2):338–344. doi: 10.1111/j.1530-0277.2010.01349.x. [DOI] [PubMed] [Google Scholar]
- Cho Y, Cavalli V. HDAC signaling in neuronal development and axon regeneration. Curr Opin Neurobiol. 2014;27:118–126. doi: 10.1016/j.conb.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CS, Wang J, Wehman M, Rhoads DE. Severity of alcohol withdrawal symptoms depends on developmental stage of Long-Evans rats. Pharmacol Biochem Behav. 2008;89(2):137–144. doi: 10.1016/j.pbb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18(13):3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa F, De Laurentiis A, Franchi AM. Ethanol downregulates N-acyl phosphatidylethanolamine-phospholipase D expression in BV2 microglial cells via epigenetic mechanisms. Eur J Pharmacol. 2016;786:224–233. doi: 10.1016/j.ejphar.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Covington HE, 3rd, Maze I, Sun H, Bomze HM, DeMaio KD, Wu EY, Dietz DM, Lobo MK, Ghose S, Mouzon E, Neve RL, Tamminga CA, Nestler EJ. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;71(4):656–670. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Addario C, Caputi FF, Ekström TJ, Di Benedetto M, Maccarrone M, Romualdi P, Candeletti S. Ethanol induces epigenetic modulation of prodynorphin and pronociceptin gene expression in the rat amygdala complex. J Mol Neurosci. 2013;49(2):312–319. doi: 10.1007/s12031-012-9829-y. [DOI] [PubMed] [Google Scholar]
- Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C, Al-Sarraj S, Dobson R, Schalkwyk LC, Mill J. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13(6):R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatin T, Barbosa AD. Effect of isopregnanolone on rapid tolerance to the anxiolytic effect of ethanol. Rev Bras Psiquiatr. 2006;28(1):18–23. doi: 10.1590/s1516-44462006000100005. [DOI] [PubMed] [Google Scholar]
- Driessen M, Meier S, Hill A, Wetterling T, Lange W, Junghanns K. The course of anxiety, depression and drinking behaviours after completed detoxification in alcoholics with and without comorbid anxiety and depressive disorders. Alcohol Alcohol. 2001;36(3):249–255. doi: 10.1093/alcalc/36.3.249. [DOI] [PubMed] [Google Scholar]
- Ewald ER, Wand GS, Seifuddin F, Yang X, Tamashiro KL, Potash JB, Zandi P, Lee RS. Alterations in DNA methylation of Fkbp5 as a determinant of blood-brain correlation of glucocorticoid exposure. Psychoneuroendocrinology. 2014;44:112–122. doi: 10.1016/j.psyneuen.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13(9):673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- Farris SP, Harris RA, Ponomarev I. Epigenetic modulation of brain gene networks for cocaine and alcohol abuse. Frontiers in Neurosci. 2015;9:176. doi: 10.3389/fnins.2015.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79(6):734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Feng J, Nestler EJ. Epigenetic mechanisms of drug addiction. Curr Opin Neurobiol. 2013;23(4):521–528. doi: 10.1016/j.conb.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegersh A, Ferguson C, Maxwell S, Mazariegos D, Farrell D, Homanics GE. Repeated vapor ethanol exposure induces transient histone modifications in the brain that are modified by genotype and brain region. Front Mol Neurosci. 2015;8:39. doi: 10.3389/fnmol.2015.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegersh A, Homanics GE. Acute ethanol alters multiple histone modifications at model gene promoters in the cerebral cortex. Alcohol Clin Exp Res. 2014;38(7):1865–1873. doi: 10.1111/acer.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroud T, Wetherill LF, Liang T, Dick DM, Hesselbrock V, Kramer J, Numberger J, Schuckit M, Carr L, Porjesz B, Xuei X, Edenberg HJ. Association of alcohol craving with alpha-synuclein (SNCA) Alcohol Clin Exp Res. 2007;31(4):537–545. doi: 10.1111/j.1530-0277.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits fetal DNA methylation in mice: Implications for the fetal alcohol syndrome. Alcohol Clin Exp Res. 1991;15(3):395–398. doi: 10.1111/j.1530-0277.1991.tb00536.x. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Kusumo H, Zhang H, Guidotti A, Pandey SC. Role of growth arrest and DNA damage-inducible, beta in alcohol-drinking behaviors. Alcohol Clin Exp Res. 2016;40(2):263–272. doi: 10.1111/acer.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin NW, Henderson AN, Badia-Elder NE, Stewart RB. Effects of neuropeptide Y (NPY) and ethanol on arousal and anxiety-like behavior in alcohol-preferring (P) rats. Alcohol. 2011;45:137–145. doi: 10.1016/j.alcohol.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin NW, Stewart RB, Murphy JM, Li TK, Badia-Elder NE. Neuropeptide Y reduces oral ethanol intake in alcohol-preferring (P) rats following a period of imposed ethanol abstinence. Alcohol Clin Exp Res. 2003;27(5):787–794. doi: 10.1097/01.ALC.0000065723.93234.1D. [DOI] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8(7):764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Guertin MJ, Lis JT. Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet. 2010;6(9):e1001114. doi: 10.1371/journal.pgen.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Gavin DP, Veldic M, Zhao W, Bhaumik DK, Pandey SC, Grayson DR. DNA methylation/demethylation network expression in psychotic patients with a history of alcohol abuse. Alcohol Clin Exp Res. 2013;37(3):417–424. doi: 10.1111/j.1530-0277.2012.01947.x. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Crossey EL, Zhang L, Zucca S, George OL, Valenzuela CF, Zhao X. Alcohol exposure decreases CREB binding protein expression and histone acetylation in the developing cerebellum. PloS One. 2011;6(5):e19351. doi: 10.1371/journal.pone.0019351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerty SL, Bidwell LC, Harlaar N, Hutchison KE. An exploratory association study of alcohol use disorder and DNA methylation. Alcohol Clin Exp Res. 2016;40(8):1633–1640. doi: 10.1111/acer.13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon E, Lunnon K, Schalkwyk L, Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10(11):1024–1032. doi: 10.1080/15592294.2015.1100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkels CH, Khorasanizadeh S. Implications of a histone code mimic in epigenetic signaling. Mol Cell. 2007;27(4):521–522. doi: 10.1016/j.molcel.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Hillemacher T, Frieling H, Hartl T, Wilhelm J, Kornhuber J, Bleich S. Promoter specific methylation of the dopamine transporter gene is altered in alcohol dependence and associated with craving. J Psychiatr Res. 2009a;43(4):388–392. doi: 10.1016/j.jpsychires.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Hillemacher T, Frieling H, Luber K, Yazici A, Muschler MA, Lenz B, Wilhelm J, Kornhuber J, Bleich S. Epigenetic regulation and gene expression of vasopressin and atrial natriuretic peptide in alcohol withdrawal. Psychoneuroendocrinology. 2009b;34(4):555–560. doi: 10.1016/j.psyneuen.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Holliday R. Epigenetics: a historical overview. Epigenetics. 2006;1(2):76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15(2):R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasiewicz A, Rubis B, Samochowiec J, Małecka I, Suchanecka A, Jabłoński M, Grzywacz A. DAT1 methylation changes in alcohol-dependent individuals vs. controls. J Psychiatr Res. 2015;64:130–133. doi: 10.1016/j.jpsychires.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Jeanblanc J, Lemoine S, Jeanblanc V, Alaux-Cantin S, Naassila M. The class I-specific HDAC inhibitor MS-275 decreases motivation to consume alcohol and relapse in heavy drinking rats. Int J Neuropsychopharmacol. 2015;18(9):pyv029. doi: 10.1093/ijnp/pyv029. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38(11):e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19(2):187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kalivas PW, Volkow ND. New medications for drug addiction hiding in glutamatergic neuroplasticity. Mol Psychiatry. 2011;16(10):974–986. doi: 10.1038/mp.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkhoven E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68(6):1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- Kan PY, Lu X, Hansen JC, Hayes JJ. The H3 tail domain participates in multiple interactions during folding and self-association of nucleosome arrays. Mol Cell Biol. 2007;27(6):2084–2091. doi: 10.1128/MCB.02181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpyak VM, Winham SJ, Preuss UW, Zill P, Cunningham JM, Walker DL, Lewis KA, Geske JR, Colby CL, Abulseoud OA, Hall-Flavin DK, Loukianova LL, Schneekloth TD, Frye MA, Bazov I, Heit JA, Bakalikin G, Mrazek DA, Biernacka JM. Association of the PDYN gene with alcohol dependence and the propensity to drink in negative emotional states. Internl J Neuropsychopharmacol. 2013;16(5):975–985. doi: 10.1017/S1461145712001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna JM, Chau A, Shah G. Characterization of the phenomenon of rapid tolerance to ethanol. Alcohol. 1996;13(6):621–628. doi: 10.1016/s0741-8329(96)00083-3. [DOI] [PubMed] [Google Scholar]
- Khanna JM, Kalant H, Shah G, Weiner J. Rapid tolerance as an index of chronic tolerance. Pharmacol Biochem Behav. 1991;38(2):427–432. doi: 10.1016/0091-3057(91)90302-i. [DOI] [PubMed] [Google Scholar]
- Khanna JM, Kalant H, Weiner J, Shah G. Rapid tolerance and cross-tolerance as predictors of chronic tolerance and cross-tolerance. Pharmacol Biochem Behav. 1992;41(2):355–360. doi: 10.1016/0091-3057(92)90110-2. [DOI] [PubMed] [Google Scholar]
- Kim JT, Li J, Jang ER, Gulhati P, Rychahou PG, Napier DL, Wang C, Weiss HL, Lee EY, Anthony L, Townsend CM, Jr, Liu C, Evers BM. Deregulation of Wnt/β-catenin signaling through genetic or epigenetic alterations in human neuroendocrine tumors. Carcinogenesis. 2013;34(5):953–961. doi: 10.1093/carcin/bgt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35(1):217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Wall TL, Schafer J. Rapid induction of tolerance to the antipunishment effects of ethanol. Alcohol. 1987;4(6):481–484. doi: 10.1016/0741-8329(87)90090-5. [DOI] [PubMed] [Google Scholar]
- Koob GF. A role for brain stress systems in addiction. Neuron. 2008;59(1):11–34. doi: 10.1016/j.neuron.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF. Neuroadaptive mechanisms of addiction: studies on the extended amygdala. Eur Neuropsychopharmacol. 2003;13(6):442–452. doi: 10.1016/j.euroneuro.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krämer OH, Zhu P, Ostendorff HP, Golebiewski M, Tiefenbach J, Peters MA, Brill B, Groner B, Bach I, Heinzel T, Göttlicher M. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22(13):3411–3420. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HR, Sakharkar AJ, Teppen TL, Berkel TD, Pandey SC. The epigenetic landscape of alcoholism. Int Rev Neurobiology. 2014;115:75–116. doi: 10.1016/B978-0-12-801311-3.00003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwako LE, Momenan R, Litten RZ, Koob GF, Goldman D. Addictions neuroclinical assessment: a neuroscience-based framework for addictive disorders. Biol Psychiatry. 2016;80(3):179–189. doi: 10.1016/j.biopsych.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyzar EJ, Floreani C, Teppen TL, Pandey SC. Adolescent alcohol exposure: burden of epigenetic reprogramming, synaptic remodeling, and adult psychopathology. Front Neurosci. 2016a;10:222. doi: 10.3389/fnins.2016.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyzar EJ, Pandey SC. Molecular mechanisms of synaptic remodeling in alcoholism. Neuroscience Letters. 2015;601:11–19. doi: 10.1016/j.neulet.2015.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyzar EJ, Zhang H, Sakharkar AJ, Pandey SC. Adolescent alcohol exposure alters lysine demethylase 1 (LSD1) expression and histone methylation in the amygdala during adulthood. Addiction Biology. 2016b doi: 10.1111/adb.12404. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd-Acosta C. Epigenetic signatures as biomarkers of exposure. Curr Environ Health Rep. 2015;2(2):117–125. doi: 10.1007/s40572-015-0051-2. [DOI] [PubMed] [Google Scholar]
- Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harbor Perspect Biol. 2014;6(5):a019133. doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Thomas S, Sabo PJ, Eisen MB, Stamatoyannopoulos JA, Biggin MD. The role of chromatin accessibility in directing the widespread, overlapping patterns of drosophila transcription factor binding. Genome Biol. 2011;12(4):R34. doi: 10.1186/gb-2011-12-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Marioni RE, Hedman AK, Pfeiffer L, Tsai PC, Reynolds LM, Just AC, Duan Q, Boer CG, Tanaka T, Elks CE, Aslibekyan S, Brody JA, Kühnel B, Herder C, Almli LM, Zhi D, Wang Y, Huan T, Yao C, Mendelson MM, Joehanes R, Liang L, Love SA, Guan W, Shah S, McRae AF, Kretschmer A, Prokisch H, Strauch K, Peters A, Visscher PM, Wray NR, Guo X, Wiggins KL, Smith AK, Binder EB, Ressler KJ, Irvin MR, Absher DM, Hernandez D, Ferrucci L, Bandinelli S, Lohman K, Ding J, Trevisi L, Gustafsson S, Sandling JH, Stolk L, Uitterlinden AG, Yet I, Castillo-Fernandez JE, Spector TD, Schwartz JD, Vokonas P, Lind L, Li Y, Fornage M, Arnett DK, Wareham NJ, Sotoodehnia N, Ong KK, van Meurs JB, Conneely KN, Baccarelli AA, Deary IJ, Bell JT, North KE, Liu Y, Waldenberger M, London SJ, Ingelsson E, Levy D. A DNA methylation biomarker of alcohol consumption. Mol Psychiatry. 2016 doi: 10.1038/mp.2016.192. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CL, Zhou FC. Environmental alterations of epigenetics prior to the birth. Int Rev Neurobiol. 2014;115:1–49. doi: 10.1016/B978-0-12-801311-3.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Moreno JA, Marcos M, Calleja-Conde J, Echeverry-Alzate V, Bühler KM, Costa-Alba P, Bernardo E, Laso FJ, Rodríguez de Fonseca F, Nadal R, Viveros MP, Maldonado R, Giné E. Histone deacetylase gene expression following binge alcohol consumption in rats and humans. Alcohol Clin Exp Res. 2015;39(10):1939–1950. doi: 10.1111/acer.12850. [DOI] [PubMed] [Google Scholar]
- Ma DK, Guo JU, Ming GL, Song H. DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009;8(10):1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadev K, Vemuri MC. Effect of ethanol on chromatin and nonhistone nuclear proteins in rat brain. Neurochem Res. 1998;23(9):1179–1184. doi: 10.1023/a:1020778018149. [DOI] [PubMed] [Google Scholar]
- Maier SE, Cramer JA, West JR, Sohrabji F. Alcohol exposure during the first two trimesters equivalent alters granule cell number and neurotrophin expression in the developing rat olfactory bulb. J Neurobiology. 1999;41(3):414–423. doi: 10.1002/(sici)1097-4695(19991115)41:3<414::aid-neu9>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Mandrekar P. Epigenetic regulation in alcohol liver disease. World J Gastroenterol. 2011;17(20):2456–2464. doi: 10.3748/wjg.v17.i20.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzardo AM, Henkhaus RS, Butler MG. Global DNA promoter methylation in frontal cortex of alcoholics and controls. Gene. 2012;498(1):5–12. doi: 10.1016/j.gene.2012.01.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R, Roth SY. Histone acetyltransferases: function, structure, and catalysis. Curr Opin Genet Dev. 2001;11(2):155–161. doi: 10.1016/s0959-437x(00)00173-8. [DOI] [PubMed] [Google Scholar]
- Marutha Ravindran CR, Ticku MK. Changes in methylation pattern of NMDA receptor NR2B gene in cortical neurons after chronic ethanol treatment in mice. Brain Res Mol Brain Res. 2004;121(1–2):19–27. doi: 10.1016/j.molbrainres.2003.10.025. [DOI] [PubMed] [Google Scholar]
- Maze I, Covingtoni HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, Ren Y, Sampath SC, Hurd YL, Greengard P, Tarakhovsky A, Schaefer A, Nestler EJ. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327(5962):213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Nestler EJ. The epigenetic landscape of addiction. Ann N Y Acad Sci. 2011;1216:99–113. doi: 10.1111/j.1749-6632.2010.05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MJ, Duchemin AM, Neff NH, Hadjiconstantinou M. CREB involvement in the regulation of striatal prodynorphin by nicotine. Psychopharmacology (Berl) 2012;221(1):143–153. doi: 10.1007/s00213-011-2559-y. [DOI] [PubMed] [Google Scholar]
- Montesinos J, Pascual M, Rodríguez-Arias M, Miñarro J, Guerri C. Involvement of TLR4 in the long-term epigenetic changes, rewarding and anxiety effects induced by intermittent ethanol treatment in adolescence. Brain Behav Immunity. 2016;53:159–171. doi: 10.1016/j.bbi.2015.12.006. [DOI] [PubMed] [Google Scholar]
- Moonat S, Sakharkar AJ, Zhang H, Tang L, Pandey SC. Aberrant histone deacetylase2-mediated histone modifications and synaptic plasticity in the amygdala predisposes to anxiety and alcoholism. Biol Psychiatry. 2013;73(8):763–773. doi: 10.1016/j.biopsych.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonat S, Sakharkar AJ, Zhang H, Pandey SC. The role of amygdaloid brain-derived neurotrophic factor, activity-regulated cytoskeleton-associated protein and dendritic spines in anxiety and alcoholism. Addiction Biol. 2011;16(2):238–250. doi: 10.1111/j.1369-1600.2010.00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschler MA, Hillemacher T, Kraus C, Kornhuber J, Bleich S, Frieling H. DNA methylation of the POMC gene promoter is associated with craving in alcohol dependence. J Neural Transm (Vienna) 2010;117(4):513–519. doi: 10.1007/s00702-010-0378-7. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393(6683):386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Peña CJ, Kundakovic M, Mitchell A, Akbarian S. Epigenetic basis of mental illness. Neuroscientist. 2016;22(5):447–463. doi: 10.1177/1073858415608147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu MD, Zeisel SH. Diet, methyl donors and DNA methylation: Interactions between dietary folate, methionine and choline. J Nutr. 2002;132(8 Suppl):2333S–2335S. doi: 10.1093/jn/132.8.2333S. [DOI] [PubMed] [Google Scholar]
- Nieratschker V, Grosshans M, Frank J, Strohmaier J, von der Goltz C, El-Maarri O, Witt SH, Cichon S, Nöthen MM, Kiefer F, Rietschel M. Epigenetic alteration of the dopamine transporter gene in alcohol-dependent patients is associated with age. Addict Biol. 2014;19(2):305–311. doi: 10.1111/j.1369-1600.2012.00459.x. [DOI] [PubMed] [Google Scholar]
- Nixon K, McClain JA. Adolescence as a critical window for developing an alcohol use disorder: current findings in neuroscience. Curr Opin Psychiatry. 2010;23(3):227–232. doi: 10.1097/YCO.0b013e32833864fe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC. Anxiety and alcohol abuse disorders: a common role for CREB and its target, the neuropeptide Y gene. Trends in Pharmacol Sci. 2003;24(9):456–460. doi: 10.1016/S0165-6147(03)00226-8. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Roy A, Zhang H. The decreased phosphorylation of cyclic adenosine monophosphate (cAMP) response element binding (CREB) protein in the central amygdala acts as a molecular substrate for anxiety related to ethanol withdrawal in rats. Alcohol Clin Exp Res. 2003;27(3):396–409. doi: 10.1097/01.ALC.0000056616.81971.49. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Roy A, Zhang H, Xu T. Partial deletion of the cAMP response element-binding protein gene promotes alcohol-drinking behaviors. J Neuroscience. 2004;24(21):5022–5030. doi: 10.1523/JNEUROSCI.5557-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Sakharkar AJ, Tang L, Zhang H. Potential role of adolescent alcohol exposure-induced amygdaloid histone modifications in anxiety and alcohol intake during adulthood. Neurobiol Dis. 2015;82:607–619. doi: 10.1016/j.nbd.2015.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J of Neurosci. 2008a;28(14):3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Zhang H, Roy A, Xu T. Deficits in amygdaloid cAMP-responsive element–binding protein signaling play a role in genetic predisposition to anxiety and alcoholism. J Clin Invest. 2005;115(10):2762–2773. doi: 10.1172/JCI24381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Zhang H, Ugale R, Prakash A, Xu T, Misra K. Effector immediate-early gene arc in the amygdala plays a critical role in alcoholism. J Neurosci. 2008b;28(10):2589–2600. doi: 10.1523/JNEUROSCI.4752-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Boix J, Felipo V, Guerri C. Repeated alcohol administration during adolescence causes changes in the mesolimbic dopaminergic and glutamatergic systems and promotes alcohol intake in the adult rat. J Neurochem. 2009;108(4):920–931. doi: 10.1111/j.1471-4159.2008.05835.x. [DOI] [PubMed] [Google Scholar]
- Pascual M, Do Couto BR, Alfonso-Loeches S, Aguilar MA, Rodriguez-Arias M, Guerri C. Changes in histone acetylation in the prefrontal cortex of ethanol-exposed adolescent rats are associated with ethanol-induced place conditioning. Neuropharmacology. 2012;62(7):2309–2319. doi: 10.1016/j.neuropharm.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Pattaroni C, Jacob C. Histone methylation in the nervous system: functions and dysfunctions. Mol Neurobiol. 2013;47(2):740–756. doi: 10.1007/s12035-012-8376-4. [DOI] [PubMed] [Google Scholar]
- Philibert RA, Gunter TD, Beach SR, Brody GH, Madan A. MAOA methylation is associated with nicotine and alcohol dependence in women. Am J Med Genet. 2008;147B(5):565–570. doi: 10.1002/ajmg.b.30778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Penaluna B, White T, Shires S, Gunter T, Liesveld J, Erwin C, Hollenbeck N, Osborn T. A pilot examination of the genome-wide DNA methylation signatures of subjects entering and exiting short-term alcohol dependence treatment programs. Epigenetics. 2014;9(9):1212–1219. doi: 10.4161/epi.32252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Plume JM, Gibbons FX, Brody GH, Beach SR. The impact of recent alcohol use on genome wide DNA methylation signatures. Front Genet. 2012;3:54. doi: 10.3389/fgene.2012.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickens RW, Svikis DS, McGue M, Lykken DT, Heston LL, Clayton PJ. Heterogeneity in the inheritance of alcoholism: A study of male and female twins. Arch Gen Psychiatry. 1991;48(1):19–28. doi: 10.1001/archpsyc.1991.01810250021002. [DOI] [PubMed] [Google Scholar]
- Pirooznia SK, Elefant F. Targeting specific HATs for neurodegenerative disease treatment: Translating basic biology to therapeutic possibilities. Front Cell Neurosci. 2013;7:30. doi: 10.3389/fncel.2013.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32(5):1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang M, Denny A, Lieu M, Carreon S, Li J. Histone H3K9 modifications are a local chromatin event involved in ethanol-induced neuroadaptation of the NR2B gene. Epigenetics. 2011;6(9):1095–1104. doi: 10.4161/epi.6.9.16924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang M, Li JG, Denny AD, Yao JM, Lieu M, Zhang K, Carreon S. Epigenetic mechanisms are involved in the regulation of ethanol consumption in mice. Int J Neuropsychopharmacol. 2014;18(2) doi: 10.1093/ijnp/pyu072. pii.pyuo72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachdaoui N, Sarkar DK. Transgenerational epigenetics and brain disorders. Int Rev Neurobiol. 2014;115:51–73. doi: 10.1016/B978-0-12-801311-3.00002-0. [DOI] [PubMed] [Google Scholar]
- Rani CS, Qiang M, Ticku MK. Potential role of cAMP response element-binding protein in ethanol-induced N-methyl-D-aspartate receptor 2B subunit gene transcription in fetal mouse cortical cells. Mol Pharmacol. 2005;67(6):2126–2136. doi: 10.1124/mol.104.007872. [DOI] [PubMed] [Google Scholar]
- Renthal W, Nestler EJ. Epigenetic mechanisms in drug addiction. Trends in Mol Med. 2008;14(8):341–350. doi: 10.1016/j.molmed.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Nestler EJ. Histone acetylation in drug addiction. Semin Cell Dev Biol. 2009;20(4):387–394. doi: 10.1016/j.semcdb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggeri B, Nymberg C, Vuoksimaa E, Lourdusamy A, Wong CP, Carvalho FM, Jia T, Cattrell A, Macare C, Banaschewski T, Barker GJ, Bokde AL, Bromberg U, Büchel C, Conrod PJ, Fauth-Bühler M, Flor H, Frouin V, Gallinat J, Garavan H, Gowland P, Heinz A, Ittermann B, Martinot JL, Nees F, Pausova Z, Paus T, Rietschel M, Robbins T, Smolka MN, Spanagel R, Bakalkin G, Mill J, Sommer WH, Rose RJ, Yan J, Aliev F, Dick D, Kaprio J, Desrivières S, Schumann G, IMAGEN Consortium Association of protein phosphatase PPM1G with alcohol use disorder and brain activity during behavioral control in a genome-wide methylation analysis. Am J Psychiatry. 2015;172(6):543–552. doi: 10.1176/appi.ajp.2014.14030382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks JJ, Gonzales KR, Bouchery EE, Tomedi LE, Brewer RD. 2010 National and state costs of excessive alcohol consumption. Am J Prev Med. 2015;49(5):e73–9. doi: 10.1016/j.amepre.2015.05.031. [DOI] [PubMed] [Google Scholar]
- Sakharkar AJ, Tang L, Zhang H, Chen Y, Grayson DR, Pandey SC. Effects of acute ethanol exposure on anxiety measures and epigenetic modifiers in the extended amygdala of adolescent rats. Int J Neuropsychopharmacol. 2014a;17(12):2057–2067. doi: 10.1017/S1461145714001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar AJ, Vetreno RP, Zhang H, Kokare DM, Crews FT, Pandey SC. A role for histone acetylation mechanisms in adolescent alcohol exposure-induced deficits in hippocampal brain-derived neurotrophic factor expression and neurogenesis markers in adulthood. Brain Struct Funct. 2016;221(9):4691–4703. doi: 10.1007/s00429-016-1196-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar AJ, Zhang H, Tang L, Baxstrom K, Shi G, Moonat S, Pandey SC. Effects of histone deacetylase inhibitors on amygdaloid histone acetylation and neuropeptide Y expression: a role in anxiety-like and alcohol-drinking behaviours. Int J Neuropsychopharmacol. 2014b;17(8):1207–1220. doi: 10.1017/S1461145714000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar AJ, Zhang H, Tang L, Shi G, Pandey SC. Histone deacetylases (HDAC)-induced histone modifications in the amygdala: a role in rapid tolerance to the anxiolytic effects of ethanol. Alcohol Clin Exp Res. 2012;36(1):61–71. doi: 10.1111/j.1530-0277.2011.01581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25(8):781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SD, Lim RW. Epigenetic effects of ethanol on the liver and gastrointestinal system. Alcohol Res. 2013;35(1):47–55. [PMC free article] [PubMed] [Google Scholar]
- Simon-O’Brien E, Alaux-Cantin S, Warnault V, Buttolo R, Naassila M, Vilpoux C. The histone deacetylase inhibitor sodium butyrate decreases excessive ethanol intake in dependent animals. Addict Biol. 2015;20(4):676–689. doi: 10.1111/adb.12161. [DOI] [PubMed] [Google Scholar]
- Smolle M, Workman JL. Transcription-associated histone modifications and cryptic transcription. Biochim Biophys Acta. 2013;1829(1):84–97. doi: 10.1016/j.bbagrm.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear LP, Varlinskaya EI. Adolescence. Alcohol sensitivity, tolerance, and intake. Recent Dev Alcohol. 2005;17:143–159. [PubMed] [Google Scholar]
- Starkman BG, Sakharkar AJ, Pandey SC. Epigenetics-beyond the genome in alcoholism. Alcohol Res. 2012;34(3):293–305. [PMC free article] [PubMed] [Google Scholar]
- Subbanna S, Basavarajappa BS. Pre-administration of G9a/GLP inhibitor during synaptogenesis prevents postnatal ethanol-induced LTP deficits and neurobehavioral abnormalities in adult mice. Experimental Neurology. 2014;261:34–43. doi: 10.1016/j.expneurol.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbanna S, Shivakumar M, Umapathy NS, Saito M, Mohan PS, Kumar A, Nixon RA, Verin AD, Psychoyo D, Basavarajappa BS. G9a-mediated histone methylation regulates ethanol-induced neurodegeneration in the neonatal mouse brain. Neurobiol Dis. 2013;54:475–485. doi: 10.1016/j.nbd.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Maze I, Dietz DM, Scobie KN, Kennedy PJ, Damez-Werno D, Neve RL, Zachariou V, Shen L, Nestler EJ. Morphine epigenomically regulates behavior through alterations in histone H3 lysine 9 dimethylation in the nucleus accumbens. J Neurosci. 2012;32(48):17454–17464. doi: 10.1523/JNEUROSCI.1357-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levev AI, Vasanthakumar A, Godley LA, Chang Q, Cheng X, He C, Jin P. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14(12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Cornell N, Hoffman PL. Alcohol tolerance. Ann Emerg Med. 1986;15(9):1005–1012. doi: 10.1016/s0196-0644(86)80119-6. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16(14):1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008;27(20):2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taqi MM, Bazov I, Watanabe H, Sheedy D, Harper C, Alkass K, Druid H, Wentzel P, Nyberg F, Yakovleva T, Bakalkin G. Prodynorphin CpG-SNPs associated with alcohol dependence: elevated methylation in the brain of human alcoholics. Addict Biol. 2011;16(3):499–509. doi: 10.1111/j.1369-1600.2011.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylee DS, Kawaguchi DM, Glatt SJ. On the outside, looking in: a review and evaluation of the comparability of blood and brain “-omes”. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(7):595–603. doi: 10.1002/ajmg.b.32150. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67(3):946–50. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G, Pierce RC. Epigenetic inheritance of a cocaine-resistance phenotype. Nat Neurosci. 2013;16(1):42–47. doi: 10.1038/nn.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Koob G, Baler R. Biomarkers in substance use disorders. ACS Chem Neurosci. 2015;6(4):522–525. doi: 10.1021/acschemneuro.5b00067. [DOI] [PubMed] [Google Scholar]
- Wang F, Xu H, Zhao H, Gelernter J, Zhang H. DNA co-methylation modules in postmortem prefrontal cortex tissues of European Australians with alcohol use disorders. Sci Rep. 2016;6:19430. doi: 10.1038/srep19430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KS, Liu X, Zhang Q, Wu LY, Zeng M. Genome-wide association study identifies 5q21 and 9p24.1 (KDM4C) loci associated with alcohol withdrawal symptoms. J Neural Trans (Vienna) 2012;119(4):425–433. doi: 10.1007/s00702-011-0729-z. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnault V, Darcq E, Levine A, Barak S, Ron D. Chromatin remodeling–a novel strategy to control excessive alcohol drinking. Transl Psychiatry. 2013;19(3):e231. doi: 10.1038/tp.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11(9):607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuei X, Dick D, Flury-Wetherill L, Tian HJ, Agrawal A, Bierut L, Goate A, Bucholz K, Schuckit M, Nurnberger J, Jr, Tischfield J, Kuperman S, Porjesz B, Begleiter H, Foroud T, Edenberg HJ. Association of the kappa-opioid system with alcohol dependence. Mol Psychiatry. 2006;11(11):1016–1024. doi: 10.1038/sj.mp.4001882. [DOI] [PubMed] [Google Scholar]
- Yeh SH, Lin CH, Gean PW. Acetylation of nuclear factor-kappaB in rat amygdala improves long-term but not short-term retention of fear memory. Mol Pharmacol. 2004;65(5):1286–1292. doi: 10.1124/mol.65.5.1286. [DOI] [PubMed] [Google Scholar]
- You C, Zhang H, Sakharkar AJ, Teppen T, Pandey SC. Reversal of deficits in dendritic spines, BDNF and arc expression in the amygdala during alcohol dependence by HDAC inhibitor treatment. Int J Neuropsychopharacol. 2014;17(2):313–322. doi: 10.1017/S1461145713001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, Vishwanatha JK, Santella RM, Morabia A. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6(5):623–629. doi: 10.4161/epi.6.5.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gelernter J. DNA methylation and alcohol use disorders: progress and challenges. Am J Addict. 2016 doi: 10.1111/ajad.12465. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Herman AI, Kranzler HR, Anton RF, Zhao H, Zheng W, Gelernter J. Array-based profiling of DNA methylation changes associated with alcohol dependence. Alcohol Clin Exp Res. 2013;37(Suppl 1):E108–15. doi: 10.1111/j.1530-0277.2012.01928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Sakharkar AJ, Shi G, Ugale R, Prakash A, Pandey SC. Neuropeptide Y signaling in the central nucleus of amygdala regulates alcohol-drinking and anxiety-like behaviors of alcohol-preferring rats. Alcohol Clin Exp Res. 2010;34(3):451–461. doi: 10.1111/j.1530-0277.2009.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Kusumo H, Sakharkar AJ, Pandey SC, Guizzetti M. Regulation of DNA methylation by ethanol induces tissue plasminogen activator expression in astrocytes. J Neurochem. 2014;128(3):344–349. doi: 10.1111/jnc.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Zhang R, Li W, Liao Y, Tang J, Miao Q, Hao W. Genome-wide DNA methylation patterns in discordant sib pairs with alcohol dependence. Asia Pac Psychiatry. 2013;5(1):39–50. doi: 10.1111/appy.12010. [DOI] [PubMed] [Google Scholar]
- Zhu ZZ, Hou L, Bollati V, Tarantini L, Marinelli B, Cantone L, Yang AS, Vokonas P, Lissowska J, Fustinoni S, Pesatori AC, Bonzini M, Apostoli P, Costa G, Bertazzi PA, Chow WH, Schwartz J, Baccarelli A. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol. 2012;41(1):126–139. doi: 10.1093/ije/dyq154. [DOI] [PMC free article] [PubMed] [Google Scholar]