The Sympathetic Neurotransmitter Switch Depends on the Nuclear Matrix Protein Satb2 (original) (raw)
Abstract
Sympathetic neurons can switch their neurotransmitter phenotype from noradrenergic to cholinergic on exposure to neuropoietic cytokines in vitro and in vivo. Here, we provide evidence that this transspecification is regulated by the chromatin architecture protein Satb2. Treatment with the neuropoietic cytokines ciliary neurotrophic factor (CNTF) and leukemia inhibitory factor rapidly and strongly increases Satb2 transcript and protein levels in cultures of rat superior cervical ganglia neurons. Knockdown of endogenous Satb2 by short interfering RNA prevents the upregulation of choline acetyltransferase (Chat) and vesicular acetylcholine transporter (Vacht) by CNTF as well as the loss of norepinephrine transporter (Net). Conversely, overexpression of Satb2 in the noradrenergic sympathetic phenotype results in a marked increase of Chat and Vacht expression and reduced Net mRNA levels in the absence of neuropoietic cytokines. Chromatin immunoprecipitation analysis in primary sympathetic neurons reveals that Satb2 binds to matrix attachment regions (MARs) within the Chat locus. In vivo, in the rat stellate ganglion, Satb2 is expressed exclusively in sudomotor cholinergic neurons innervating the sweat glands and only after establishment of contact between neurons and target. These findings demonstrate a function of the MAR-binding protein Satb2 in growth factor-dependent neurotransmitter plasticity in postmitotic neurons.
Introduction
The vast majority of mature sympathetic neurons uses noradrenaline as a neurotransmitter and has vasoconstrictor, secretomotor, or pilomotor functions. A minor subpopulation secretes acetylcholine and innervates sweat glands, vasculature, and periosteum.
The acquisition of noradrenergic phenotype by neural crest precursor cells occurs early in development, before target contact, and is mediated via a transcription factor network induced by bone morphogenetic proteins. The concerted action of Mash1, Phox2b/2a, Hand2, and Gata2/3 has been shown to activate both the expression of the noradrenalin biosynthetic machinery and pan-neuronal markers in autonomic precursors (Goridis and Rohrer, 2002; Rohrer, 2003; Sarkar and Howard, 2006).
The cholinergic sudomotor neurons, innervating the sweat glands, develop much later by a transspecification process from fully functional noradrenergic neurons. This neurotransmitter plasticity occurs at postnatal stages in individual postmitotic neurons under the influence of neuropoietic cytokines, released by the target tissue after innervation (Landis, 1990, 1996; Stanke et al., 2006). The noradrenergic-to-cholinergic neurotransmitter switch can be induced in vitro in cultured sympathetic neurons by members of the neuropoietic cytokine family, such as ciliary neurotrophic factor (CNTF) or leukemia inhibitory factor (LIF) (Fann and Patterson, 1994). Although this neurotransmitter switch has served as an intensively studied model of neuronal diversification, little is known about the nuclear molecules and processes that act as a counterpart of the well characterized noradrenergic transcription factor network in controlling the downregulation of noradrenergic markers and the specification of the cholinergic transmitter phenotype.
The DNA-binding proteins Satb1 and Satb2 represent a novel class of gene regulators, which affect transcription by binding to AT-rich sequences of matrix attachment regions (MARs), tethering them to the nuclear matrix and forming distinct chromatin loops (Cai et al., 2003, 2006; Britanova et al., 2005; Szemes et al., 2006). These higher-order chromatin changes coupled with recruitment of chromatin remodeling complexes at the anchorage sites ensure precise, coordinated regulation of many close and distant genes (Yasui et al., 2002; Galande et al., 2007; Gyorgy et al., 2008). Previous studies have elegantly demonstrated a crucial role of Satb2 in cell specification and differentiation processes in non-neuronal cells. For example, Satb2 directs B-cell differentiation by binding to the MARs of the Ig μ locus and enhancing its expression (Dobreva et al., 2003). Satb2 also functions as a molecular determinant of skeletal development and osteoblast differentiation (Dobreva et al., 2006). Recently, it has been shown that Satb2 is a part of cell-intrinsic genetic mechanisms that control the projection neuron identity in the developing cortex (Alcamo et al., 2008; Britanova et al., 2008).
Here, we show for the first time that Satb2 can also be regulated acutely by neurotrophic growth factors. Members of the gp130 cytokine family strongly induce Satb2 in postmitotic sympathetic neurons undergoing neurotransmitter transspecification. We find that Satb2 binds to MARs of the cholinergic locus and is both required and sufficient for the activation of its transcription. Moreover, Satb2 is specifically expressed in sudomotor cholinergic neurons only after target contact, suggesting that it may regulate target-induced noradrenergic-to-cholinergic phenotype switch after sweat gland innervation.
Materials and Methods
Primary cell culture.
Superior cervical ganglia (SCGs) were dissected from newborn rats [postnatal day 0 (P0) to P2]. Ganglia were trypsinized and dissociated by trituration, and the resulting cell suspension was preplated for 2–3 h in noncoated tissue culture dishes to allow adherence of the non-neuronal cells (Friedel et al., 1997). Neurons were plated in either 35 mm tissue culture dishes (immunoblotting assay) or 12-well plates [reverse transcription (RT)-PCR assay], previously coated with poly-dl-ornithine (Sigma-Aldrich) and laminin (Invitrogen). Cells were incubated for different time periods (2, 6, 8, 24, and 72 h) in Ham's F-14 medium (Invitrogen), containing 0.1 mg/ml apotransferrin, 16 mg/ml putrescine, 6 ng/ml progesterone, 8 ng/ml sodium selenite, 5 μg/ml insulin, 100 mg/ml penicillin G, 60 mg/ml streptomycin sulfate (all supplements from Sigma-Aldrich), in the presence of either NGF (20 ng/ml; PeproTech) or CNTF (25 ng/ml; PeproTech) and NGF (5 ng/ml). In some experiments, LIF (ESGRO LIF; 1000 U/ml; Millipore), glial cell line-derived neurotrophic factor (GDNF) (25 ng/ml; PeproTech), or Neurturin (25 ng/ml; PeproTech) were used instead of CNTF. The cultures were maintained in a humidified atmosphere of 5% CO2 in air at 37°C. The medium of the long-term cultures was changed 1 or 2 d after plating. Glial cell proliferation was inhibited with Aphidicolin (5 μg/ml; Sigma-Aldrich).
Electroporation.
For Satb2 knockdown experiments, 1–3 × 106 dissociated rat SCG neurons were centrifuged at 80 × g for 5 min at room temperature, resuspended in 100 μl of Nucleofector Solution (Amaxa Rat Neuron Nucleofector kit), and mixed with either stealth small interfering RNA (siRNA) duplexes against Satb2 (three duplexes; 2 μg each; Invitrogen) or 6 μg of stealth siRNA Negative Control (Medium GC; Invitrogen). Cells were electroporated with Amaxa Nucleofector device (program G13) according to the manufacturer's instructions. Immediately after nucleofection, prewarmed culture medium (900 μl) was added to the cell suspension and the neurons were plated in 35 mm tissue culture dishes, previously coated with poly-dl-ornithine/laminin at a density of 300,000 cells per dish. The cultures were incubated for 2 d in Ham's F-14 medium, supplemented as above, in the presence of CNTF (25 ng/ml) and NGF (5 ng/ml). The medium was changed 1 d after plating. Cells were directly harvested in lysis/binding buffer for mRNA isolation (100 mm Tris-HCl, pH 7.5, 500 mm LiCl, 10 mm EDTA, 1% LiDS, 5 mm dithiothreitol) or in 2× Roti-Load 1 sample buffer (Roth) for immunoblotting, since the transfection efficiency in knockdown experiments was close to 100% (tested by electroporation of Cy3-conjugated siRNAs). The sequences of the siRNAs against rat Satb2 (Invitrogen) were as follows: stealth_734, GCUGAAGAUCCAAUUACAAAGUUGU; stealth_995, CGAGAGAACCUUUCAGACUAUUGU; stealth_1721, CGUCAACAUCACAGCUGCCAUUUAU.
For overexpression experiments, neurons were nucleofected (program G13) with expression plasmids—either 3.5 μg of pmaxGFP (Lonza) and 5 μg of pcDNA3.1-mSatb2 or, as control, 3.5 μg of pmaxGFP and 5 μg of empty pcDNA3.1 vector (Invitrogen). The expression plasmid pcDNA3.1-mSatb2 was constructed by subcloning the coding sequence of Mus musculus Satb2 (cDNA clone MGC:170250, IMAGE:8861645) into the EcoRI site of pcDNA3.1. After nucleofection, the neurons were plated in either 35 mm poly-dl-ornithine/laminin-coated dishes for real-time RT-PCR analysis or in precoated glass-bottom dishes (MatTek) for immunocytochemistry. The cultures, grown on 35 mm dishes, were incubated for 2 d in supplemented F14 medium, containing 5 ng/ml NGF. Since the transfection efficiency in overexpression experiments (10–20%) was lower than in siRNA knockdown experiments, only green fluorescent protein (GFP)-positive nucleofected neurons were used for analysis—three to five pools of ∼150–200 GFP-positive neurons per condition were aspirated by using a glass microcapillary (inner diameter, 15–20 μm) and transferred into Microfuge tubes, containing 20 μl of lysis/binding buffer for RNA extraction. The neurons, plated on glass-bottom dishes, were cultured for 3 d in the presence of NGF (10 ng/ml) and CNTF (10 ng/ml). The caspase inhibitor Q-VD-OPH (10 μm; Calbiochem) was added to the culture medium to prevent neuronal death.
RNA isolation and real-time PCR.
mRNA was isolated using Dynabeads Oligo-dT25 (Invitrogen) following the manufacturer's protocol. Briefly, mRNA was annealed to the beads by incubation in lysis/binding buffer on a rotating mixer for 10 min at room temperature. After several washing steps, mRNA was eluted from the beads by adding 16 μl of 10 mm Tris-HCl and incubating for 3 min at 85°C. The eluted mRNA was quickly transferred to a new tube, chilled on ice, and immediately used for cDNA synthesis according to High-Capacity cDNA Reverse Transcription Kit protocol (Applied Biosystems).
The levels of mRNAs were assessed by quantitative real-time PCR using Fast SYBRMaster Mix (Applied Biosystems). All amplification reactions were conducted in triplicate. Relative expression ratios were calculated by the ΔΔ_Ct_ method (Pfaffl, 2001). The Gapdh gene was used for normalization.
The following primers were used: rat Gapdh, forward, 5′-CTGGAGAAACCTGCCAAGTATG-3′; rat Gapdh, reverse, 5′-ACAACCTGGTCCTCAGTGTAG-3′; rat Satb2, forward, 5′-CCGCACACAGGGATTATTGTC-3′; rat Satb2, reverse, 5′-TCCACTTCAGGCAGGTTGAG-3′; rat Vacht, forward, 5′-CTCACCACTTGTAACATTCCCC-3′; rat Vacht, reverse, 5′-AAACCATGCCCATCTCCCAC-3′; rat Net, forward, 5′-CAGCACCATCAACTGTGTTACC-3′; rat Net, reverse, 5′-GGCTTCTGGATACAGGACAAATAC-3′; rat VIP, forward, 5′-CACGCCCTATTATGATGTGTCC-3′; rat VIP, reverse, 5′-TCCGAGATGCTACTGCTGATTC-3′; rat Chat, forward, 5′-CAGCCAATTGGGTCTCTGAATAC-3′; rat Chat, reverse, 5′-GAGGCAGGCTGCAAATCTTAG-3′; rat NF160, forward, 5′-AGCATTGAGCTCGAGTCGGTG-3′; rat NF160, reverse, 5′-CTGCTGGATGGTGTCCTGGTAG-3′.
Immunocytochemistry.
Primary SCG neurons were grown on coverslips or glass-bottom dishes precoated with poly-dl-ornithine (Sigma-Aldrich) and laminin (Invitrogen) in Ham's F-14 medium, supplemented as above for 3 d. Cells were washed in PBS (0.01 m), pH 7.4, fixed in 4% paraformaldehyde in PBS for 15 min, and permeabilized in 0.3% Triton X-100 in PBS (PBST) for 10 min. After blocking with serum (10% normal serum, 1% BSA in PBST), cells were incubated overnight at 4°C with the appropriate primary antibodies (rabbit anti-Tau, 1:300, Dako; mouse anti-Satb2, 1:100, Abcam; rabbit anti-Vacht, 1:1000, Sigma-Aldrich) diluted in 1% BSA in PBST. After several rinses in PBST, cells were incubated for 1 h with Alexa Fluor secondary antibodies (Alexa 488-conjugated goat anti-rabbit, Alexa 555-conjugated goat anti-mouse or goat anti-rabbit; 1:2000; Invitrogen) and washed three times in PBS. The coverslips were incubated for 2 min with the DNA stain 4′,6-diamidino-2-phenylindole (DAPI) (300 nm in PBS), rinsed in PBS and deionized water, and mounted on slides with Mowiol (Sigma-Aldrich). Pictures were taken with an ApoTome Imaging System based on Axio Observer.Z1 (Zeiss) using AxioVision software.
Immunoblotting.
Total protein extracts from cultured primary SCG neurons were prepared by direct lysis of the cells in 2× Roti-Load 1 sample buffer (Roth). The lysates were briefly sonicated, centrifuged for 15 min at 13,000 rpm to remove the cellular debris, and subjected to 10% SDS-PAGE. The separated proteins were transferred to a polyvinylidene difluoride membrane (Millipore), which was first blocked with 5% milk powder in TBST (0.1% Tween 20 in TBS) for 1 h and then incubated overnight at 4°C with the corresponding primary antibodies, diluted in blocking solution: mouse anti-Satb2 (1:100; Abcam), goat anti-Erk2 (1:2000; Santa Cruz), or mouse anti-α-tubulin (1:100,000; Sigma-Aldrich). After incubation with HRP-coupled secondary antibodies, the blots were developed using ECL reagent (GE Healthcare). For quantification of protein expression, ImageJ software was used (http://rsb.info.nih.gov/ij).
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation (ChIP) assay was performed using the EZ-Magna ChIP G kit (Millipore) following the manufacturer's instructions. Briefly, primary SCG neurons, cultured for 2 d in the presence of CNTF (25 ng/ml) and NGF (5 ng/ml), were fixed with 1% formaldehyde in growth medium for 10 min at room temperature. After cell and nuclear lysis, the cross-linked DNA was sheared to ∼200–600 bp lengths by sonication on ice in a Fisher 200 sonifier (six to seven 10-s-long bursts at power output 2; duty cycle, 90%). The sheared cross-linked chromatin was diluted with dilution buffer and incubated overnight at 4°C with 5 μg of mouse anti-Satb2 antibody (Abcam) or 1 μg of normal mouse IgG as a negative control. Protein G magnetic beads (20 μl) were added to the samples to collect the immune complexes. After washing the beads in several wash buffers (low salt, high salt, LiCl, and TE buffer), protein–DNA complexes were eluted and reverse cross-linked at 62°C for 2 h with shaking by incubation in ChIP elution buffer, containing proteinase K. The precipitated genomic DNA was purified using spin columns and used as a template in real-time PCRs. The input DNA (an aliquot of the sheared chromatin, not subjected to immunoprecipitation, but reverse-cross-linked and purified) was diluted to 1% and used to validate the performance of different primer pairs, designed to amplify MARs within the Chat locus (for primer sequences, see supplemental Methods, available at www.jneurosci.org as supplemental material). The fold enrichment was estimated using the following equation: 2∧(_Ct_mock − _Ct_specific), where _Ct_mock and _Ct_specific are the mean threshold cycles of PCR on DNA samples from mock (IgG) and Satb2 immunoprecipitations (Nelson et al., 2006). In one of the technical replications, semiquantitative PCR was used to quantify the fold change after ChIP. After resolving the PCR products in gel electrophoresis, band intensities were quantified using ImageJ software and the fold change after ChIP was calculated as the ratio of the intensity of the Satb2 IP band to that of the mock (IgG) IP band.
Tissue preparation and immunohistochemistry.
Stellate ganglia from newborn or 30-d-old rats were dissected and fixed in 4% paraformaldehyde in PBS for 20 min. After several washes in PBS, the ganglia were cryoprotected by immersion in 25% sucrose solution in PBS at 4°C and mounted in TissueTek medium (Sakura Finetek). Fourteen-micrometer-thick sections were cut with a cryostat, mounted on gelatin-coated slides, and air-dried for 1 h at 37°C.
The sections were fixed for 5 min in precooled acetone (−20°C), washed two times, 5 min each, in Tris-buffered saline (TBS) and permeabilized in 0.25% Triton X-100 in TBS (TBST) for 10 min. After 1 h preincubation in blocking solution (10% normal serum, 1% BSA in TBST), primary antibodies were applied, diluted in blocking solution, and then the sections were incubated at 4°C overnight. The following antibodies were used: mouse anti-Satb2 (1:100; Abcam); rabbit anti-Vacht (1:1000; Sigma-Aldrich); rabbit anti-rat calcitonin gene-related peptide (CGRP) (1:300; AbD Serotec). Subsequently, the sections were rinsed three times, 5 min each in TBST, incubated in the corresponding Alexa Fluor secondary antibodies (1:2000 dilution in blocking solution; Invitrogen) for 1 h at room temperature. Finally, the sections were incubated for 2 min with the DNA stain DAPI (300 nm in PBS), rinsed again in TBS and deionized water, and mounted with Mowiol (Sigma-Aldrich). Images were obtained with a Zeiss Axio Observer.Z1 fluorescence microscope and AxioVision software.
Results
Satb2 mRNA and protein levels are strongly induced by members of the gp130 cytokine family in primary rat SCG neurons
We previously analyzed the global transcriptome changes accompanying the sympathetic neurotransmitter transspecification by using Affymetrix GeneChip Rat Genome 230 2.0 microarrays. Gene expression was compared between primary SCG neurons cultured for 7 d in the presence of either NGF or CNTF and NGF (unpublished observations). Among the Affymetrix features most strongly induced by CNTF, we identified the probe set 1390838_at (18-fold upregulation), which was located on chromosome 9, immediately downstream of the Satb2 locus.
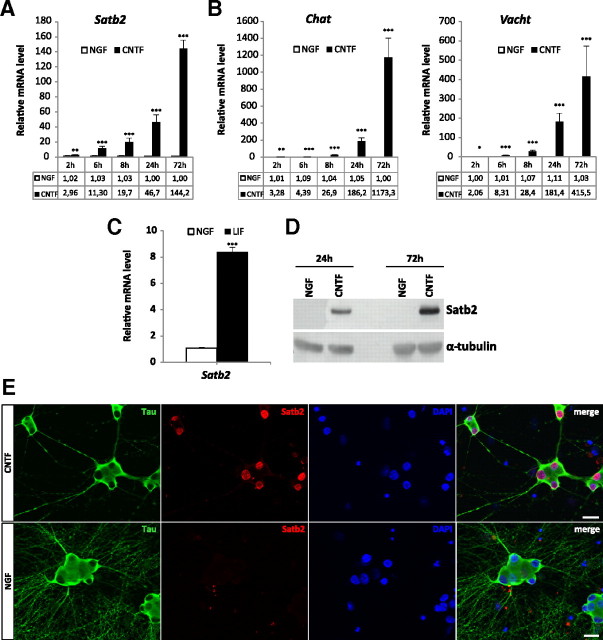
Although not part of the current annotation for Satb2 mRNA sequence (ENSRNOT0000001378; NM_001109306), this feature most likely represents the 3′-untranslated region of the Satb2 transcript, based on alignment with a cluster of expressed sequence tag sequences associated with this locus. Since the nuclear matrix protein Satb2 was previously implicated in various developmental processes, we tested for Satb2 differential expression. Satb2 mRNA levels were determined in primary SCG neurons at different time points after CNTF treatment by quantitative RT-PCR. The results show that Satb2 mRNA expression was significantly increased after CNTF application, starting from ∼3-fold in 2 h and reaching ∼140-fold in 72 h, similarly to the expression of the cholinergic marker genes Chat and Vacht (Fig. 1A,B). Furthermore, the comparison of Satb2, Chat, and Vacht time course revealed that Satb2 expression preceded that of the cholinergic markers by several hours. The cholinergic differentiation factor LIF, another member of the gp130 cytokine family, had the same effect on Satb2 transcript levels as CNTF (Fig. 1C), whereas GDNF family ligands GDNF and Neurturin did not induce Satb2 expression in primary SCG neurons (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). As shown by immunoblotting, Satb2 protein levels were also strongly increased in CNTF- versus NGF-treated primary cultures (Fig. 1D). Double immunostaining with antibodies to Satb2 and the neuron-specific marker Tau confirmed the induction of Satb2 in virtually all CNTF-treated neurons, demonstrating strong nuclear Satb2 staining (Fig. 1E). In neurons cultured with NGF, Satb2 expression was very low and could not be detected either by immunostaining or by Western blot. Satb2 was also not detected in non-neuronal cells (Fig. 1E).
Figure 1.
Neuropoietic cytokines induce Satb2 in primary SCG neurons. A, Quantitative RT-PCR analysis reveals a rapid and significant increase in Satb2 mRNA level on CNTF stimulation. B, The induction of the cholinergic locus by CNTF follows a similar but slightly delayed time course. C, Satb2 mRNA level is also markedly induced 24 h after treatment with LIF. Data represent mean ± SEM of three independent experiments; unpaired two-tailed t test, comparison with NGF-treated neurons: *p < 0.05, **p < 0.01, ***p < 0.001. D, Immunoblotting shows strong Satb2 protein expression in CNTF-treated SCG neurons and undetectable Satb2 protein levels in NGF-treated cultures. Tubulin was used as a loading control. E, Images of primary SCG neurons, cultured in the presence or absence of CNTF, costained for Tau and Satb2. Nuclei were stained with DAPI. The expression of Satb2 is virtually absent from the nuclei of NGF-treated neurons. Scale bar, 20 μm.
Satb2 siRNA knockdown prevents cholinergic gene locus induction by CNTF
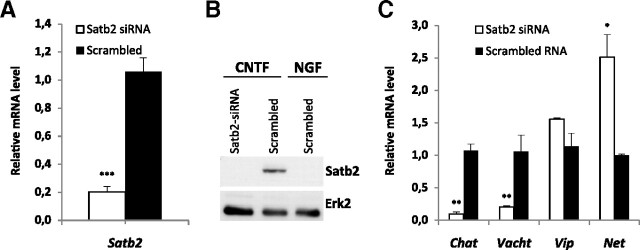
The upregulation of both Satb2 and the cholinergic marker genes in response to CNTF raised the possibility that Satb2 is required for transcriptional regulation of cholinergic or noradrenergic markers by CNTF. To test this hypothesis, we used siRNAs to knock down Satb2 expression in SCG primary neurons. We first assessed the extent of Satb2 gene silencing in CNTF-treated cultures by quantitative RT-PCR and Western blotting. Dissociated ganglion cells were electroporated with either Satb2 siRNAs or control siRNA before plating and analyzed for Satb2 mRNA and protein expression at day 2 and day 3 in culture, respectively. The combined application of three siRNA duplexes against Satb2 caused ∼80% reduction in the Satb2 mRNA level relative to the control siRNA transfected sample, both in neuronal mass cultures (Fig. 2A) and in microcapillary aspirated neurons (data not shown). Western blot analysis confirmed the effectiveness of siRNA duplexes in knocking down Satb2 also on the protein level. In lysates from Satb2 siRNA-transfected CNTF-treated cultures, Satb2 protein could not be detected, similarly to NGF-treated cultures, whereas in neurons transfected with control siRNA and treated with CNTF, Satb2 protein levels were strongly induced (Fig. 2B). Next, we measured Chat and Vacht mRNA expression in CNTF-treated sympathetic neurons after Satb2 siRNA knockdown. Strikingly, the elimination of Satb2 resulted in a drastic reduction in Chat and Vacht transcript levels relative to control siRNA (10-fold and 5-fold decrease, respectively), whereas the expression of the neuropeptide vasoactive intestinal peptide (VIP), which serves as an additional cholinergic marker, was unaffected. Additionally, Satb2 knockdown produced a significant increase in the expression of the noradrenergic marker Net.
Figure 2.
Satb2 is required for induction of cholinergic gene locus by CNTF. A, B, Quantitative RT-PCR (A) and immunoblotting (B) experiments demonstrate effective Satb2 knockdown on mRNA and protein level by siRNA in primary SCG neurons. C, Satb2 gene silencing in CNTF-treated SCG neurons greatly reduces Chat and Vacht transcript levels, without affecting VIP levels. Data shown represent mean ± SEM (n = 4–6); unpaired two-tailed t test, comparison with control siRNA-transfected neurons: *p < 0.05, **p < 0.01, ***p < 0.001.
These results show that Satb2 is necessary for the induction of the cholinergic gene locus by CNTF in primary sympathetic neurons. Furthermore, Satb2 functions in antagonizing the expression of the noradrenergic marker gene Net after CNTF application.
Satb2 overexpression results in increased Chat and Vacht mRNA levels and strong Vacht immunoreactivity in noradrenergic neurons
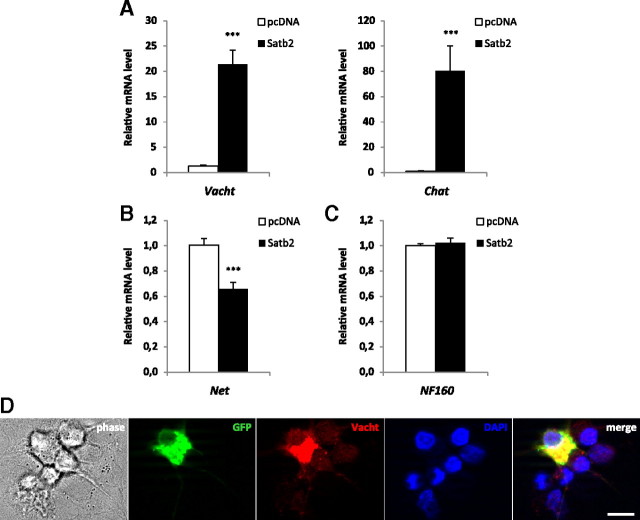
To investigate whether Satb2 can promote cholinergic gene expression in noradrenergic neurons, we transfected primary SCG neurons with a Satb2 expression construct to achieve overexpression of Satb2. GFP plasmid was coelectroporated to detect transfected cells. Almost all GFP-positive, nucleofected neurons strongly expressed Satb2, whereas the nonelectroporated neurons had undetectable levels of Satb2 under both culturing conditions (supplemental Fig. 2, available at www.jneurosci.org as supplemental material): 5 ng/ml NGF (RT-PCR analysis) and 10 ng/ml NGF plus 10/ng ml CNTF (immunocytochemistry). After 2 d in culture, GFP-positive neurons were aspirated by a glass microcapillary and analyzed for marker gene mRNA expression. Quantitative RT-PCR revealed that forced expression of Satb2 caused not only a strong increase in both Chat and Vacht mRNA levels (80.3-fold and 21.4-fold, respectively, relative to mock-transfected samples) but also a significant reduction in the level of Net mRNA (Fig. 3A,B). At the same time, the expression of the neuronal marker NF160 was unaltered (Fig. 3C). These findings suggest a specific function of Satb2 in driving cholinergic and suppressing Net gene expression without affecting a pan-neuronal marker in postmitotic sympathetic neurons. Furthermore, immunocytochemistry analysis revealed strong Vacht immunostaining in neurons electroporated with Satb2 expression plasmid as opposed to undetectable Vacht immunoreactivity in GFP- and Satb2-negative nontransfected neurons (Fig. 3D). In control experiments, in which only GFP/mock plasmids were electroporated, there was no difference in Vacht immunofluorescence of GFP-positive and GFP-negative neurons (data not shown). The increase in Vacht immunostaining after Satb2 overexpression was similar to the induction of Vacht immunoreactivity after CNTF treatment (supplemental Fig. 3, available at www.jneurosci.org as supplemental material).
Figure 3.
Satb2 promotes cholinergic gene expression, suppresses Net expression, and induces Vacht immunoreactivity. A–C, Quantitative RT-PCR experiment shows a marked increase in Vacht and Chat transcript levels (A) after Satb2 overexpression in NGF-treated SCG neurons. The mRNA level of the noradrenergic marker Net is reduced on Satb2 nucleofection (B), whereas the expression of the generic neuronal marker, NF160, remains unaltered (C). Data in A and C represent mean ± SEM of seven separate experiments; three to five pools of ∼150–200 GFP-positive nucleofected neurons were analyzed per experiment and per condition (Satb2 or mock transfection); unpaired two-tailed t test, comparison with mock-transfected neurons: ***p < 0.0001. Data in B represent mean ± SEM of three independent experiments; two pools of ∼150–200 GFP-positive nucleofected neurons were analyzed per experiment and per condition; unpaired two-tailed t test, comparison with mock-transfected neurons: ***p < 0.001. D, Immunofluorescence for Vacht (red) demonstrates that overexpression of Satb2 results in increased Vacht immunoreactivity in transfected, GFP-positive neurons compared with nonelectroporated, GFP-negative SCG primary neurons. Scale bar, 10 μm.
Together, the results of gain- and loss-of-function experiments demonstrate that Satb2 is both necessary and sufficient for stimulation of cholinergic locus gene expression in primary sympathetic neurons. In addition, Satb2 has a function in reducing the levels of the noradrenergic marker Net during the cholinergic switch.
Satb2 binds to MAR sequences in the Chat locus
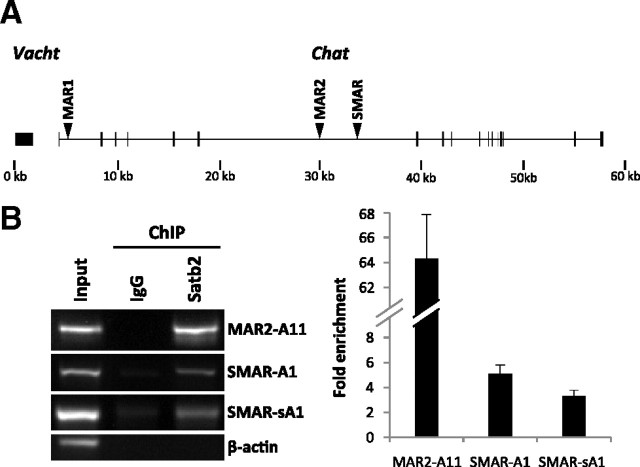
Previous studies have shown that Satb2 interacts with different AT-rich DNA sequences, part of MARs/SARs (scaffold attachment regions), and is capable of either activating or repressing the transcription of the corresponding gene loci (Dobreva et al., 2003, 2006; Alcamo et al., 2008; Britanova et al., 2008; Savarese et al., 2009). To examine whether the positive transcriptional effect of Satb2 on the cholinergic locus correlates with in vivo binding of Satb2 to MARs within the locus, we performed ChIP experiments in CNTF-treated rat primary sympathetic neurons. First, we tested whether the commercially available mouse monoclonal Satb2 antibody can immunoprecipitate Satb2-bound chromatin fragments. Sheared chromatin from lysates of cultured sympathetic neurons was immunoprecipitated with anti-Satb2 antibody or normal mouse IgG, and the immunoprecipitates were subjected to Western blotting. As shown on supplemental Figure 4 (available at www.jneurosci.org as supplemental material), Satb2 was detected in the sample immunoprecipitated with anti-Satb2 antibody, demonstrating the usability of this antibody in the ChIP assay. Next, we used the available computer algorithms, MAR-Wiz (http://genomecluster.secs.oakland.edu/marwiz/) and SMARTest (http://www.genomatix.de) to search for putative MARs within the 60 kb genomic sequence encompassing the rat cholinergic locus. The MAR-Wiz algorithm identified two MARs elements, ∼1000 bp long (termed MAR1 and MAR2), whereas the Genomatix software predicted only one potential MAR, ∼400 bp long (termed SMAR) (Fig. 4A). We designed primer pairs that allow for the amplification of overlapping or neighboring regions within the in silico predicted MAR elements (three amplicons per MAR element), as well as regions located directly upstream or downstream of the MARs. ChIP assay followed by PCR analysis of the immunoprecipitated chromatin fragments showed that in vivo Satb2 occupies some (MAR2 and SMAR) but not all predicted MARs within the rat Chat locus (Fig. 4B) and do not bind to coding sequences of β-actin (negative control). The level of enrichment in Satb2 immunoprecipitations was lower for fragments amplified with primers located upstream or downstream of the SMAR element (data not shown).
Figure 4.
Satb2 occupies MAR elements within the Chat locus. A, Schematic representation of the putative MARs (arrowheads) within the rat Chat locus. B, ChIP assay shows Satb2 binding to some of the MARs (MAR2 and SMAR) within the Chat locus in primary SCG neurons. At day 2 in culture, CNTF-treated primary SCG neurons were cross-linked with formaldehyde and immunoprecipitated with mouse monoclonal Satb2 antibody or normal mouse IgG as a negative control. Input DNA represents sheared chromatin not subjected to immunoprecipitation. ChIP assay was followed by amplification of DNA fragments within the putative MARs (MAR1: amplicons A6, A7, A8; MAR2: amplicons A9, A10, A11; SMAR: amplicons A1, sA1), and β-actin coding region (negative control). The fold enrichment of the ChIP target was calculated relative to IgG control. Error bars represent the SD of four immunoprecipitations (two cell culture experiments and preparations of cross-linked chromatin, respectively, each with two technical replications of the immunoprecipitation reaction).
Satb2 expression in vivo labels a subpopulation of cholinergic neurons in the sympathetic stellate ganglion
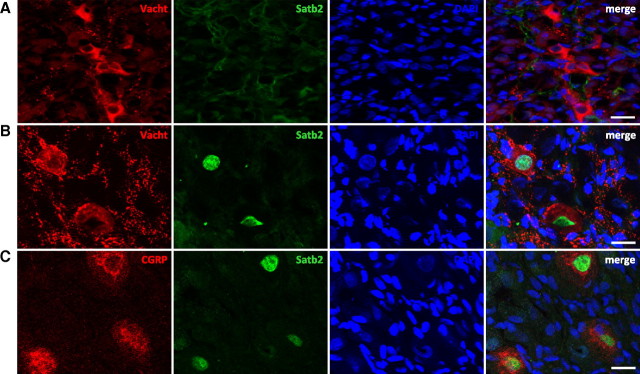
Our in vitro functional studies demonstrate that Satb2 is required for CNTF-triggered cholinergic transspecification of cultured sympathetic neurons. To find in vivo correlates for the proposed function of Satb2 as a cholinergic transmitter phenotype determinant, we applied immunohistochemistry to examine the expression of Satb2 relative to cholinergic markers in sympathetic stellate ganglion. In mature paravertebral sympathetic ganglia of rodents, cholinergic postganglionic neurons are a minority (i.e., they only constitute ∼3–5%) (Masliukov and Timmermans, 2004). The stellate ganglion, in particular, contains the highest number of cholinergic neurons. They fall into two classes with different morphological and neurochemical properties: large, scattered, CGRP-positive sudomotor neurons that project to the forelimb sweat glands, and significantly smaller CGRP-negative cholinergic neurons that form loose clusters within the ganglion and innervate the periosteum (Anderson et al., 2006). Both populations initially express noradrenergic markers and require interactions with their target tissues to acquire cholinergic phenotype (Landis, 1990; Asmus et al., 2000).
We analyzed Satb2 and Vacht coexpression in sections of rat stellate ganglion at three developmental stages, embryonic day 18 (E18), P1/P2, and P30, that is, before (E18, P1/P2) and after (P30) the innervation of one of the cholinergic target tissues, the forepaw sweat glands (Guidry and Landis, 1998). At E18 and P1/P2, we found a small number of postganglionic sympathetic neurons positive for Vacht immunoreactivity, as previously reported (Schäfer et al., 1997). At these developmental stages, however, we could not detect Satb2-positive nuclei in sections of stellate ganglia from five animals (Fig. 5A; supplemental Fig. 5, available at www.jneurosci.org as supplemental material). In contrast, at P30 several Satb2-positive nuclei were detected in virtually every section (Fig. 5B; supplemental Fig. 6, available at www.jneurosci.org as supplemental material). Notably, all Satb2-labeled neurons were also immunoreactive for Vacht. But only 63% of the examined 336 Vacht-immunoreactive cells coexpressed Vacht and Satb2, whereas the remaining cells were single Vacht-positive. To characterize the Satb2-expressing subpopulation of cholinergic neurons more precisely, we analyzed the colocalization of Satb2 with CGRP, a marker of the sudomotor cholinergic neurons, which project to the sweat glands (Cane and Anderson, 2009). We examined a total of 120 Satb2-positive neurons on sections from several animals. Ninety-eight percent of these neurons also expressed CGRP (Fig. 5C; supplemental Fig. 6, available at www.jneurosci.org as supplemental material). The difference from 100% double-positive neurons might be attributable to the difficulty of precisely distinguishing the cytoplasmic CGRP staining from the abundant CGRP-immunoreactive preganglionic terminals.
Figure 5.
Satb2 is expressed by sudomotor cholinergic neurons in rat stellate ganglia after target contact. A–C, Sections from P1/P2 (A) and P30 (B, C) rat stellate ganglia were coimmunostained for Satb2 and Vacht (A, B), and Satb2 and CGRP (C). Scale bar, 20 μm. Nuclei were stained with DAPI. At P1/P2, no Satb2-positive cells were detected, although clusters of Vacht-expressing cells were readily seen (A). At P30, Satb2 is expressed in Vacht-positive (B), CGRP-positive neurons (C).
Collectively, these data indicate that in vivo Satb2 marks a distinct subpopulation of cholinergic sympathetic neurons, the large CGRP-positive sudomotor neurons that project to the sweat glands—the source of the cholinergic differentiation factor. Satb2 is not present in the stellate ganglion before the innervation of the sweat glands, but the expression is high after the establishment of the target contact.
Discussion
Here, we demonstrate that the nuclear matrix protein Satb2 has an important function in controlling the neurotransmitter switch in primary sympathetic neurons. Satb2 is acutely upregulated on activation of neuropoietic cytokine receptors; it reduces Net expression, occupies MARs within the cholinergic locus, and is both necessary and sufficient for driving its expression. We provide evidence that Satb2 acts with high target gene specificity in postmitotic neurons only after they have established contacts with a specific target organ, the sweat gland. Our finding that a nuclear architecture protein, engaged in chromatin regulation, is induced downstream of growth factor signaling, provides a novel concept for the role of chromatin modifications in growth factor responses of fully differentiated postmitotic neurons. Future experiments with Satb2 conditional mutants will be required to confirm in vivo the proposed functional role of Satb2 in the acquisition of the cholinergic phenotype of the sudomotor neurons.
Satb2 regulates the cholinergic locus but not neuropeptides
The cholinergic phenotype is functionally defined by the expression of Chat, the acetylcholine-synthesizing enzyme, and Vacht, the vesicular membrane transporter that translocates cytoplasmic acetylcholine into secretory vesicles. The genes encoding the two proteins have a close chromosomal location, with Vacht being embedded into the first intron of Chat (Eiden, 1998). This arrangement, called the cholinergic gene locus (or Chat locus), is considered to permit coordinate regulation of the two cholinergic markers (Berrard et al., 1995; Misawa et al., 1995). In addition, as part of their chemical phenotype, all cholinergic sympathetic neurons also express VIP and in the case of the sudomotor neurons, CGRP. It appears that the establishment of a mature “chemical code” is a rather complex process, involving heterogeneous signals and also modes of regulation. Our data demonstrate that, even though Satb2 is an essential candidate player, it is not the only regulator. For example, in vitro the expression of CGRP is triggered by activin A and not by neuropoietic cytokines (Fann and Patterson, 1994; Cheng et al., 1997). In vivo, in rat sweat glands, a close homolog of activin A (activin B) is detected (Fann and Patterson, 1995). Thus, it seems unlikely that Satb2, acting downstream of gp130 cytokines, is implicated in the control of CGRP phenotype.
With regard to VIP, although both VIP and the Chat locus are strongly induced by gp130 cytokines (Lewis et al., 1994), our RNA interference experiments reveal that only the induction of Chat and Vacht selectively depends on Satb2 and not that of VIP. The neuropoietic signaling that triggers the neurotransmitter switch is quite diverse; it comprises several pathways downstream of the gp130 receptor (Heinrich et al., 2003), and consequently multiple genes appear to be affected (unpublished observations). CNTF induces VIP expression through the activation of signal transducer and activator of transcription (STAT) and AP-1 proteins, which bind to distinct sites within a 180 bp element in the VIP promoter, termed cytokine response element (CyRE) (Symes et al., 1997). Even though two perfect matches and two CyRE homologous sequences have also been identified in the Chat locus (Berse and Blusztajn, 1995), our knockdown results suggest that the presence of these elements is not sufficient to ensure robust transcriptional activation in response to CNTF and that Satb2 is required as well. In analogy with the role of Satb1 in the regulation of cytokine genes (Cai et al., 2006), it is plausible that Satb2 by binding to MARs in the Chat locus forms transcriptionally active chromatin structure (with specific chromatin “loopscape”) and recruits chromatin-modifying or remodeling enzymes, thus conferring inducibility of the Chat locus and permitting binding of STAT and AP-1 to CyREs. This is consistent with previous findings demonstrating that some transcription factors bind target promoters only at open chromatin states (Maier et al., 2004).
Alternatively, Satb2 itself might have several activities depending on the individual locus. Our overexpression results, showing that Satb2 is capable of activating Vacht expression in the absence of CNTF, indicate a more direct role in augmenting cholinergic gene expression, not simply ensuring favorable transcriptional environment. One possible mechanism is that Satb2 may cooperate with REST4 to counteract the repressor activity of REST (RE-1 silencing transcription factor), a transcriptional repressor that has previously been shown to bind Chat promoter and downregulate cholinergic gene transcription in both cholinergic and noncholinergic cells (Lonnerberg et al., 1995; Hahm et al., 1997; Hersh and Shimojo, 2003).
Satb2 expression in the rat stellate ganglion
Several arguments point to a highly specific effect of Satb2 in sudomotor neuron development. First, our colocalization experiments demonstrate that, in rat stellate ganglia, Satb2 is exclusively restricted to this subpopulation of cholinergic sympathetic neurons. Furthermore, it appears that the expression of Satb2 in these neurons is strictly developmentally regulated in a target-dependent manner. At late embryonic (E18) and early postnatal stages (P1/P2), before establishing the contact with the target tissue, neurons of stellate or thoracic sympathetic chain ganglia do not express Satb2, whereas at P30, after contacting the forepaw sweat glands, all CGRP+ cholinergic neurons also express Satb2. These findings, together with the in vitro functional data, strongly suggest an important role of Satb2 in the target-dependent cholinergic transspecification of sweat gland innervation.
The postnatal target-induced neurotransmitter switch is not the only mechanism to generate cholinergic neurons (Apostolova and Dechant, 2009). Some neurons in rodent sympathetic ganglia are known to acquire cholinergic properties before birth at early embryonic stages (Schäfer et al., 1997; Huber and Ernsberger, 2006). Studies in mice deficient for gp130, LIFRβ (LIF receptor β), or CNTFR (CNTF receptor) have demonstrated that the embryonic cholinergic differentiation is not controlled by gp130 cytokines (Stanke et al., 2000, 2006). Consistently, our results show a lack of Satb2 expression during late embryonic or early postnatal stages. At the same time, growth factors of the GDNF family, acting through the ret tyrosine kinase receptor, have been implicated in the maturation of cholinergic sympathetic neurons during prenatal development (Burau et al., 2004). Since GDNF family ligands cannot induce Satb2 expression—at least _in vitro_—and Satb2 is not expressed during this time period, it is unlikely that the embryonic cholinergic differentiation follows mechanisms that require Satb2.
It seems plausible that the expression of cholinergic markers in early sympathetic precursors or newborn postmitotic neurons occurs in a more naive, relaxed chromatin state, in which Satb2 is dispensable. During the course of development, the chromatin structure becomes more and more restricted and nonpermissive to transcription. Thus, the function of Satb2 in the late target-dependent neurotransmitter switch might be to return the chromatin into a more plastic state that enables transspecification, otherwise not possible in more restrictive chromatin state.
Growth factor-induced Satb2 expression
Previous studies have implicated Satb2 in various cell-intrinsic differentiation programs, in which it functions to couple changes in chromatin structure with gene transcription. For instance, it has been shown that Satb2 represses Hoxa2 during skeletogenesis and also acts with other regulatory proteins to promote osteoblast differentiation (Dobreva et al., 2006); in the developing cerebral cortex, Satb2 inhibits corticospinal motor neuron fate and promotes the establishment of callosal projection neurons (Alcamo et al., 2008; Britanova et al., 2008). Notably, in both developmental paradigms, the expression of Satb2 appears to be cell-intrinsically regulated. Our data show that, in postmitotic sympathetic neurons, Satb2 is rapidly and strongly induced by members of the neuropoietic cytokine family. To our knowledge, this is the first evidence supporting a novel mode of Satb2 regulation (i.e., induction by cell-extrinsic cues as opposed to cell-intrinsically programmed expression).
What could be the implications of such growth factor-induced expression of a chromatin remodeling protein in differentiated postmitotic neurons? Recent studies provide evidence that target-derived growth factors not only control neuronal survival but also the acquisition of late aspects of neuronal fate, such as mature axonal projections, neurotransmitter identity, and terminal position of the neuron cell bodies (Hippenmeyer et al., 2004). These fine adjustments in neuronal connectivity and function matching the target needs are considered to be mediated trough transcriptional events. Thus, one would speculate that the execution of the target-induced programs of terminal differentiation requires not only recruitment of classical, stimuli-induced transcriptional regulators but also alterations in higher-order chromatin structure and composition of chromatin-modifying platforms to render postmitotic neurons responsive to the target-derived cues. Our results, demonstrating that the nuclear architecture protein Satb2 is essential for the target-driven cholinergic differentiation of sympathetic neurons provide support for this hypothesis. It will be of future interest to determine whether Satb2, or its close homolog Satb1, is involved in other growth factor- or neuronal activity-driven plastic changes affecting postmitotic neurons.
At the same time, our finding that Satb2 is activated downstream of CNTF or LIF signaling raises an interesting possibility that Satb2 participates in another developmental switch (i.e., from neurogenesis to gliogenesis) known to be regulated by neuropoietic cytokines and to involve epigenetic modifications (Allen, 2008).
In conclusion, our results reveal a novel function for Satb2 in gp130 cytokine-dependent specification of the cholinergic neurotransmitter phenotype. Our data also give strong support for the idea that changes in chromatin architecture in addition to other epigenetic modifications are important for postmitotic neuron plasticity and target-mediated terminal neuronal differentiation.
Footnotes
This study was supported by a grant from the Fonds zur Förderung der Wissenschaftlichen Forschung (Signal Processing in Neurons W1206-B05). We thank M. Waldner and T. Massimo for expert technical assistance.
References
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Fariñas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Allen ND. Temporal and epigenetic regulation of neurodevelopmental plasticity. Philos Trans R Soc Lond B Biol Sci. 2008;363:23–38. doi: 10.1098/rstb.2006.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CR, Bergner A, Murphy SM. How many types of cholinergic sympathetic neuron are there in the rat stellate ganglion? Neuroscience. 2006;140:567–576. doi: 10.1016/j.neuroscience.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Apostolova G, Dechant G. Development of neurotransmitter phenotypes in sympathetic neurons. Auton Neurosci. 2009;151:30–38. doi: 10.1016/j.autneu.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Asmus SE, Parsons S, Landis SC. Developmental changes in the transmitter properties of sympathetic neurons that innervate the periosteum. J Neurosci. 2000;20:1495–1504. doi: 10.1523/JNEUROSCI.20-04-01495.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrard S, Varoqui H, Cervini R, Israël M, Mallet J, Diebler MF. Coregulation of two embedded gene products, choline acetyltransferase and the vesicular acetylcholine transporter. J Neurochem. 1995;65:939–942. doi: 10.1046/j.1471-4159.1995.65020939.x. [DOI] [PubMed] [Google Scholar]
- Berse B, Blusztajn JK. Coordinated up-regulation of choline acetyltransferase and vesicular acetylcholine transporter gene expression by the retinoic acid receptor alpha, cAMP, and leukemia inhibitory factor/ciliary neurotrophic factor signaling pathways in a murine septal cell line. J Biol Chem. 1995;270:22101–22104. doi: 10.1074/jbc.270.38.22101. [DOI] [PubMed] [Google Scholar]
- Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci. 2005;21:658–668. doi: 10.1111/j.1460-9568.2005.03897.x. [DOI] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, Sestan N, Molnár Z, Tarabykin V. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Burau K, Stenull I, Huber K, Misawa H, Berse B, Unsicker K, Ernsberger U. c-ret regulates cholinergic properties in mouse sympathetic neurons: evidence from mutant mice. Eur J Neurosci. 2004;20:353–362. doi: 10.1111/j.1460-9568.2004.03500.x. [DOI] [PubMed] [Google Scholar]
- Cai S, Han HJ, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Cane KN, Anderson CR. Generating diversity: mechanisms regulating the differentiation of autonomic neuron phenotypes. Auton Neurosci. 2009;151:17–29. doi: 10.1016/j.autneu.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Cheng JG, Pennica D, Patterson PH. Cardiotrophin-1 induces the same neuropeptides in sympathetic neurons as do neuropoietic cytokines. J Neurochem. 1997;69:2278–2284. doi: 10.1046/j.1471-4159.1997.69062278.x. [DOI] [PubMed] [Google Scholar]
- Dobreva G, Dambacher J, Grosschedl R. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 2003;17:3048–3061. doi: 10.1101/gad.1153003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Fariñas I, Karsenty G, Grosschedl R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Eiden LE. The cholinergic gene locus. J Neurochem. 1998;70:2227–2240. doi: 10.1046/j.1471-4159.1998.70062227.x. [DOI] [PubMed] [Google Scholar]
- Fann MJ, Patterson PH. Neuropoietic cytokines and activin A differentially regulate the phenotype of cultured sympathetic neurons. Proc Natl Acad Sci U S A. 1994;91:43–47. doi: 10.1073/pnas.91.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fann MJ, Patterson PH. Activins as candidate cholinergic differentiation factors in vivo. Int J Dev Neurosci. 1995;13:317–330. doi: 10.1016/0736-5748(94)00075-e. [DOI] [PubMed] [Google Scholar]
- Friedel RH, Schnürch H, Stubbusch J, Barde YA. Identification of genes differentially expressed by nerve growth factor- and neurotrophin-3-dependent sensory neurons. Proc Natl Acad Sci U S A. 1997;94:12670–12675. doi: 10.1073/pnas.94.23.12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galande S, Purbey PK, Notani D, Kumar PP. The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr Opin Genet Dev. 2007;17:408–414. doi: 10.1016/j.gde.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Goridis C, Rohrer H. Specification of catecholaminergic and serotonergic neurons. Nat Rev Neurosci. 2002;3:531–541. doi: 10.1038/nrn871. [DOI] [PubMed] [Google Scholar]
- Guidry G, Landis SC. Target-dependent development of the vesicular acetylcholine transporter in rodent sweat gland innervation. Dev Biol. 1998;199:175–184. doi: 10.1006/dbio.1998.8929. [DOI] [PubMed] [Google Scholar]
- Gyorgy AB, Szemes M, de Juan Romero C, Tarabykin V, Agoston DV. SATB2 interacts with chromatin-remodeling molecules in differentiating cortical neurons. Eur J Neurosci. 2008;27:865–873. doi: 10.1111/j.1460-9568.2008.06061.x. [DOI] [PubMed] [Google Scholar]
- Hahm SH, Chen L, Patel C, Erickson J, Bonner TI, Weihe E, Schäfer MK, Eiden LE. Upstream sequencing and functional characterization of the human cholinergic gene locus. J Mol Neurosci. 1997;9:223–236. doi: 10.1007/BF02800504. [DOI] [PubMed] [Google Scholar]
- Heinrich PC, Behrmann I, Haan S, Hermanns HM, Müller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh LB, Shimojo M. Regulation of cholinergic gene expression by the neuron restrictive silencer factor/repressor element-1 silencing transcription factor. Life Sci. 2003;72:2021–2028. doi: 10.1016/s0024-3205(03)00065-1. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Kramer I, Arber S. Control of neuronal phenotype: what targets tell the cell bodies. Trends Neurosci. 2004;27:482–488. doi: 10.1016/j.tins.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Huber K, Ernsberger U. Cholinergic differentiation occurs early in mouse sympathetic neurons and requires Phox2b. Gene Expr. 2006;13:133–139. doi: 10.3727/000000006783991854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis SC. Target regulation of neurotransmitter phenotype. Trends Neurosci. 1990;13:344–350. doi: 10.1016/0166-2236(90)90147-3. [DOI] [PubMed] [Google Scholar]
- Landis SC. The development of cholinergic sympathetic neurons: a role for neuropoietic cytokines? Perspect Dev Neurobiol. 1996;4:53–63. doi: 10.1080/0907676x.1996.9961274. [DOI] [PubMed] [Google Scholar]
- Lewis SE, Rao MS, Symes AJ, Dauer WT, Fink JS, Landis SC, Hyman SE. Coordinate regulation of choline acetyltransferase, tyrosine hydroxylase, and neuropeptide mRNAs by ciliary neurotrophic factor and leukemia inhibitory factor in cultured sympathetic neurons. J Neurochem. 1994;63:429–438. doi: 10.1046/j.1471-4159.1994.63020429.x. [DOI] [PubMed] [Google Scholar]
- Lonnerberg P, Lendahl U, Funakoshi H, Arhlund-Richter L, Persson H, Ibanez CF. Regulatory region in choline acetyltransferase gene directs developmental and tissue-specific expression in transgenic mice. Proc Natl Acad Sci U S A. 1995;92:4046–4050. doi: 10.1073/pnas.92.9.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H, Ostraat R, Gao H, Fields S, Shinton SA, Medina KL, Ikawa T, Murre C, Singh H, Hardy RR, Hagman J. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol. 2004;5:1069–1077. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- Masliukov PM, Timmermans JP. Immunocytochemical properties of stellate ganglion neurons during early postnatal development. Histochem Cell Biol. 2004;122:201–209. doi: 10.1007/s00418-004-0692-y. [DOI] [PubMed] [Google Scholar]
- Misawa H, Takahashi R, Deguchi T. Coordinate expression of vesicular acetylcholine transporter and choline acetyltransferase in sympathetic superior cervical neurones. Neuroreport. 1995;6:965–968. doi: 10.1097/00001756-199505090-00004. [DOI] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer H. The role of bone morphogenetic proteins in sympathetic neuron development. Drug News Perspect. 2003;16:589–596. doi: 10.1358/dnp.2003.16.9.829341. [DOI] [PubMed] [Google Scholar]
- Sarkar AA, Howard MJ. Perspectives on integration of cell extrinsic and cell intrinsic pathways of signaling required for differentiation of noradrenergic sympathetic ganglion neurons. Auton Neurosci. 2006;126–127:225–231. doi: 10.1016/j.autneu.2006.02.029. [DOI] [PubMed] [Google Scholar]
- Savarese F, Dávila A, Nechanitzky R, De La Rosa-Velazquez I, Pereira CF, Engelke R, Takahashi K, Jenuwein T, Kohwi-Shigematsu T, Fisher AG, Grosschedl R. Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev. 2009;23:2625–2638. doi: 10.1101/gad.1815709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer MK, Schütz B, Weihe E, Eiden LE. Target-independent cholinergic differentiation in the rat sympathetic nervous system. Proc Natl Acad Sci U S A. 1997;94:4149–4154. doi: 10.1073/pnas.94.8.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Geissen M, Götz R, Ernsberger U, Rohrer H. The early expression of VAChT and VIP in mouse sympathetic ganglia is not induced by cytokines acting through LIFRbeta or CNTFRalpha. Mech Dev. 2000;91:91–96. doi: 10.1016/s0925-4773(99)00275-0. [DOI] [PubMed] [Google Scholar]
- Stanke M, Duong CV, Pape M, Geissen M, Burbach G, Deller T, Gascan H, Otto C, Parlato R, Schütz G, Rohrer H. Target-dependent specification of the neurotransmitter phenotype: cholinergic differentiation of sympathetic neurons is mediated in vivo by gp 130 signaling. Development. 2006;133:141–150. doi: 10.1242/dev.02189. [DOI] [PubMed] [Google Scholar]
- Symes A, Gearan T, Eby J, Fink JS. Integration of Jak-Stat and AP-1 signaling pathways at the vasoactive intestinal peptide cytokine response element regulates ciliary neurotrophic factor-dependent transcription. J Biol Chem. 1997;272:9648–9654. doi: 10.1074/jbc.272.15.9648. [DOI] [PubMed] [Google Scholar]
- Szemes M, Gyorgy A, Paweletz C, Dobi A, Agoston DV. Isolation and characterization of SATB2, a novel AT-rich DNA binding protein expressed in development- and cell-specific manner in the rat brain. Neurochem Res. 2006;31:237–246. doi: 10.1007/s11064-005-9012-8. [DOI] [PubMed] [Google Scholar]
- Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]