Chromatin Remodeling at a DNA Double-Strand Break Site in Saccharomyces cerevisiae (original) (raw)
. Author manuscript; available in PMC: 2006 Mar 6.
Published in final edited form as: Nature. 2005 Nov 17;438(7066):379–383. doi: 10.1038/nature04148
Abstract
The repair of DNA double-strand breaks (DSBs) is critical for maintaining genome stability. Eukaryotic cells repair DSBs using both non-homologous end joining (NHEJ) and homologous recombination (HR). How chromatin structure is altered in response to DSBs and how such alterations influence DSB repair processes are important questions. In vertebrates, phosphorylation of the histone variant H2A.X (γ-H2A) occurs rapidly after formation of DSBs1, spreads over megabase chromatin domains, and is required for stable accumulation of DNA repair proteins at DNA damage foci2. In Saccharomyces cerevisiae, phosphorylation of the two major H2A species is also signaled by DSB formation, spreading ∼40 Kb in either direction from a DSB3. Here we show that near a DSB γ-H2A is followed by loss of histones H2B and H3 and increased sensitivity of chromatin to digestion by micrococcal nuclease. However, γ-H2A and nucleosome loss occur independently of one another. The DNA damage sensor MRX (Mre11-Rad50-Xrs2)4 is required for histone eviction, which additionally depends on the ATP-dependent nucleosome-remodeling complex, INO805. The repair protein Rad516 shows delayed recruitment to a DSB in the absence of histone loss, suggesting that MRX-dependent nucleosome remodeling regulates the accessibility of factors with direct roles in DNA damage repair by HR. We propose that MRX regulates two pathways of chromatin changes, including nucleosome displacement, required for efficient recruitment of HR proteins, and γ-H2A, which modulates checkpoint responses to DNA damage2.
To elucidate the chromatin pathways leading to DSB repair in Saccharomyces cerevisiae, we employed a _MAT_α haploid strain that lacks HMR and HML donor sequences and carries a galactose-inducible HO gene7. In this strain, HO endonuclease introduces a DSB at MAT that can only be repaired by NHEJ, although the major HR proteins are recruited to the break site6. We analyzed chromatin structure along 12-20 Kb encompassing the DSB by chromatin immunoprecipitation (ChIP) followed by real-time PCR, which provided sensitive measurement of the kinetics and spatial distribution of chromatin changes and recruitment of repair proteins around the break site.
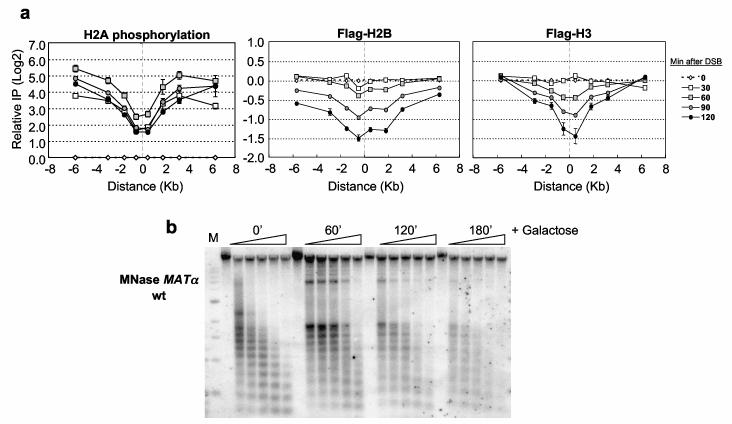
Budding yeast H2A is phosphorylated on serine 129 by the ATM/ATR homologs Tel1/Mec18. In agreement with a recent report3, we found that γ-H2A accumulated rapidly and extensively on either side of the DSB, and that γ-H2A levels were lower close to the DSB relative to 6 Kb distant (Figure 1a; Supplementary Figure 3a). These latter results suggested a loss in nucleosome integrity near the DSB. The nucleosome consists of 146 bp of DNA wrapped ∼two times around a histone octamer comprising an (H3/H4)2 tetramer and two H2A/H2B dimers. To determine if nucleosome stability was altered at the DSB, we performed ChIP in strains expressing either Flag-H2B or Flag-H3. The levels of both histones decreased 60-90 min after HO induction and were reduced three-fold by 120 min (Figure 1a). The comparable loss of both histones suggests that entire nucleosomes were displaced from chromatin near the DSB. Although no histone loss occurred within the first 30 min of DSB induction, γ-H2A levels were ∼4-fold lower near the DSB compared to distal sites at this time (Figure 1a). This may reflect phosphatase activity near the DSB or enhanced phosphorylation at distal sites.
Figure 1.
Chromatin changes at the MATαDSB.(a) A DSB was induced at _MAT_α in Flag-H2B or Flag-H3 expressing strains, and ChIP was performed with γ-H2A or anti-Flag antibodies. DNA was analyzed by real-time PCR using primers corresponding to sequences on the left (“−”) or right (“+”) side of the DSB (“0”), and results were normalized to the 0′ IP/Input DNA. Data in graphs are means +/− s.e.m. (b) Nuclei were prepared after DSB induction, and chromatin was digested with MNase and subjected to Southern blot analysis using a _MAT_α DNA probe. M=1 Kb DNA ladder.
To confirm that nucleosomes were remodeled near the DSB, we analyzed the micrococcal nuclease (MNase) sensitivity of _MAT_α chromatin (Figure 1b). Prior to DSB induction, a strong MNase ladder reflected positioned nucleosomes9. Following break formation, the nucleosome ladder became progressively less organized with time (Figure 1b; Supplementary Figures 1, 2). The alteration in the nucleosome pattern closely paralleled histone depletion, indicating that nucleosome integrity is compromised around the DSB.
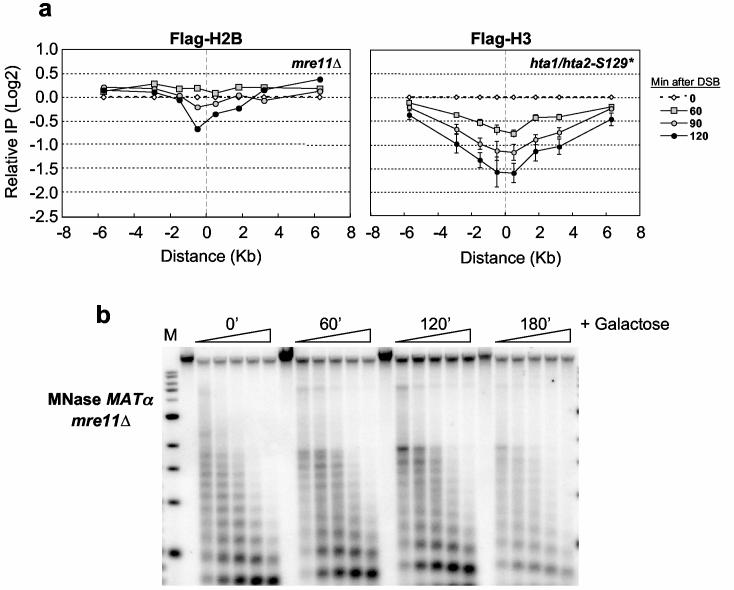
Histone eviction during transcription often depends on histone modifying or ATP-dependent nucleosome remodeling factors10-12. Because γ-H2A temporally preceded histone loss, we first examined its role in nucleosome displacement by eliminating the MRX complex. H2A.X phosphorylation by ATM in vertebrates requires the homologous MRN (Mre11-Rad50-Nbs1) complex13. MRX is one of the earliest factors recruited to yeast DSBs, and regulates the ATM homolog, Tel114. In _mre11_Δ, γ-H2A was not abolished at the DSB, although its overall levels were reduced 2-3-fold (Supplementary Figure 3c). However, histone loss was significantly impeded (Figure 2a), with Flag-H2B present 3 hours after DSB induction (data not shown). Additionally, the nucleosome ladder at _MAT_α did not change over this time period, indicating that chromatin structure remained intact (Figure 2b; Supplementary Figures 1, 2). Because these data suggested that γ-H2A and histone loss might not be directly coupled, we examined histone occupancy in an H2A mutant lacking the serine 129 phosphorylation site (hta1/hta2-S129*) (Figure 2a). Although γ-H2A could not be detected at the DSB in this mutant (data not shown), Flag-H3 was lost to the same extent as in a wild type strain. Thus, histone eviction depends on MRX but not γ-H2A.
Figure 2.
MRX plays a role in histone loss at the MATαDSB.(a) A DSB was induced at _MAT_α, and ChIP was performed with anti-Flag antibodies in an mre11::Kan-MX strain expressing Flag-H2B and an hta1/hta2-S129* strain expressing Flag-H3. DNA was analyzed by realtime PCR on both the left and right sides of the break site (“0”), and data are means +/− s.e.m.. (b) MNase analysis was performed on nuclei isolated from the mre11::Kan-MX strain by Southern blot analysis as described in Figure 1c.
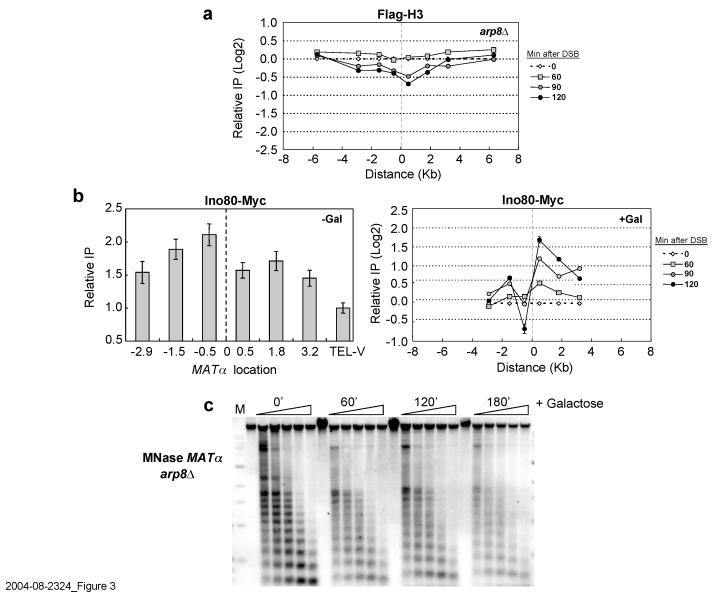
Next, we determined if an ATP-dependent nucleosome remodeling activity was required for histone loss. We focused on the INO80 complex because an _ino80_Δ mutant shows sensitivity to agents that cause DSBs, INO80 can move nucleosomes in vitro5,15, and Ino80 is enriched at MAT after DSB induction16-18. Because _ino80_Δ is inviable in our strain background, we used an _arp8_Δ mutant, which is defective in the Ino80 ATPase and chromatin remodeling, and is sensitive to DNA damage15. In _arp8_Δ Flag-H3 and Flag-H2B were retained at the DSB for two hours, and were evicted only three hours after break formation (Figure 3a; Supplementary Figure 3b). Consistent with delayed histone eviction, there was a similar delay in nucleosome disruption (Figure 3b; Supplementary Figures 1, 2). Thus, we conclude that _arp8_Δ significantly slows but does not eliminate histone loss. Together, the results indicate that INO80-dependent chromatin remodeling increases the kinetics of nucleosome displacement at a DSB, and that this activity depends on MRX but not γ-H2A.
Figure 3.
**The INO80 complex is required for histone eviction at the MAT**α DSB.(a) A DSB was induced at _MAT_α in an arp8::Kan-MX mutant expressing Flag-H3, and ChIP was performed with antibodies against the Flag epitope. Precipitated DNA was quantitated as described in Figure 1a. (b) ChIP was performed with anti-Myc antibodies in a wild type strain that contained Ino80-Myc before (−Gal) and after (+Gal) DSB induction. Ino80-Myc association was normalized to histone H3 occupancy. (c) MNase digestion was performed on nuclei isolated from an arp8::Kan-MX mutant as described in Figure 1c. Data in graphs are means +/− s.e.m.
In wild-type cells, histone loss begins ∼60 min after DSB induction, which temporally coincides with 5′ to 3′ resection of DNA ends19. Moreover, an _mre11_Δ mutant, which is defective in histone eviction, is also defective in end processing7. We tested whether the slower rate of histone eviction in _arp8_Δ results from a slower rate of DSB formation or end processing, and found that both cleavage and resection were similar in wild-type and _arp8_Δ (Supplementary Figure 4), which contrasts to a recent report17. This also indicates that nucleosomes can associate in vivo with single-stranded DNA, as observed in vitro20, and that nucleases can resect DNA that is nucleosomal. Thus, single-stranded DNA is necessary but not sufficient for histone eviction, and the INO80 ATPase is required for chromatin remodeling at the DSB.
γ-H2A–independent histone displacement was puzzling in light of recent reports that Ino80 physically associates with γ-H2A and that γ-H2A recruits Ino80 to the MAT DSB16-18. To address this issue, we measured Ino80 association with _MAT_α before DSB induction and found it to be present even in the absence of the break (Figure 3b, −Gal). As shown by DNA microarray studies, this pool of Ino80 likely contributes to MAT_α transcription21, and we confirmed that MAT_α_1 and α_2 transcript levels are 2-3 fold lower in _arp8_Δ (data not shown). After DSB induction (Figure 3b, +Gal), Ino80 preferentially accumulated on the right side of the DSB, and in contrast to the pre-existing pool of Ino80, this association required γ-H2A16,17 (Supplementary Figure 5). Additionally, 2 hours after break induction ∼50% of the pre-existing Ino80 pool was lost 0.5 Kb from the left side of the DSB, a region corresponding to the _MAT_α promoter (Figure 3b, +Gal). Together, the data suggest that a pre-existing, γ-H2A-independent pool of Ino80 related to _MAT_α transcription is responsible for histone displacement, while a newly recruited pool performs another function. Ino80 binds to both DNA and histones5,15, and interestingly, DNA sequences to the right of the MAT DSB initially participate in strand invasion during HR, suggesting a role for newly recruited Ino80 in this process.
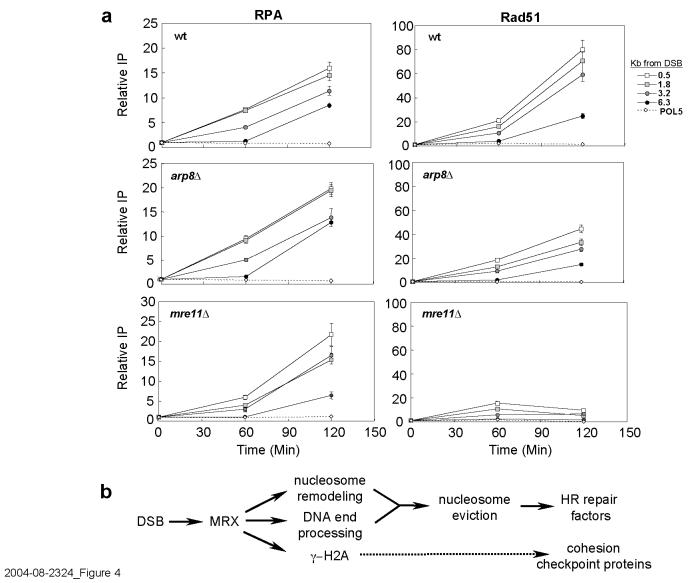
The INO80 complex is associated with repair of DSBs by HR17 (Supplementary Figure 6), suggesting that histone depletion could affect the recruitment of HR proteins to the MAT DSB. HO-generated DSBs are stable for almost 60 min19, and recruitment of HR proteins such as Rad51 and Rad54 is delayed until broken ends are processed to 3′ single-stranded tails6. RPA binds first, spreads along DNA, and is subsequently displaced by Rad5122,23. RPA was efficiently recruited to the DSB in wild-type cells, and with slightly faster kinetics in _arp8_Δ (Figure 4a). In both strains, RPA eventually spread 6.3 Kb from the DSB, confirming that strand resection occurs normally in _arp8_Δ. However, recruitment and spreading of Rad51 were delayed in _arp8_Δ (Figure 4a) and correlated precisely with the delay in histone eviction in this mutant. In _mre11_Δ there was a pronounced delay in the binding of both RPA and Rad51 (Figure 4a), consistent with the delayed resection of DNA ends7,24 (data not shown) and extended period of nucleosome retention in this strain. In striking contrast, Rad51 recruitment was indistinguishable from wild-type in the H2A-S129* mutant (data not shown), supporting the observation that histone displacement proceeds with normal timing in the absence of γ-H2A. Together, the data suggest that nucleosome eviction controls the rate at which Rad51 displaces RPA from resected DNA during HR repair, and that this displacement depends on MRX and the INO80 complex.
Figure 4.
**MRX and INO80 are required for recruitment of Rad51 to the MAT**α DSB.(a) ChIP was performed in wild type, _arp8_Δ, or _mre11_Δ with RPA (left) or Rad51 (right) antibodies after DSB induction at _MAT_α. DNA on the right side of the DSB was analyzed by real-time PCR, and data are means +/− s.e.m.. (b) MRX controls chromatin remodeling at DSBs. MRX is recruited to DSBs, regulating DNA end processing and Tel1-depenedent γ-H2A formation. MRX regulates nucleosome-remodelingthrough INO80, leading to nucleosome eviction and efficient recruitment of HR proteins. γ-H2A is independent of nucleosome displacement and controls accumulation of checkpoint proteins, as well as cohesin, at DSBs.
The results indicate that a temporal sequence of events occurs to alter chromatin at DSBs. First, γ-H2A spreads over a large domain around the break. Next, nucleosomes are displaced near the DSB through the remodeling activity of the INO80 complex, which increases the kinetics of histone loss. Nucleosome loss at MAT requires MRX, one of the earliest factors to be recruited to DSBs in vivo. We propose that MRX regulates nucleosome displacement through its role in two pathways (Figure 4b). The first pathway involves MRX-dependent resection of DNA ends. When strand resection is prevented, or significantly delayed as in _mre11_Δ, nucleosome displacement cannot occur efficiently. However, resection per se is not sufficient for nucleosome loss, and MRX also functions in a second pathway that controls INO80-dependent nucleosome remodeling. These two MRX-dependent pathways then converge to promote nucleosome eviction. This could account for the more severe defect in nucleosome displacement in _mre11_Δ compared to _arp8_Δ, although we cannot rule out that another nucleosome remodeling factor plays a redundant role with INO8025.
Histone displacement occurs in a region of the MAT_α locus that includes the promoter and α_1/α_2_ coding regions, raising the interesting possibility that transcription itself plays a role in nucleosome loss through INO80-dependent chromatin remodeling. At highly transcribed genes elevated levels of RNA polymerase II (Pol II) correlate with decreased histone occupancy11,12. Our preliminary data indicate that MAT_α_1 transcripts transiently accumulate upon DSB induction, coinciding with increased Pol II levels and loss of nucleosomes at the α_1_ TATA; these effects are significantly reduced in _arp8_Δ (unpublished data). Thus, the DSB might signal through MRX to the MAT transcription machinery, which then employs the remodeling activity of INO80 to displace histones. This might reflect a mechanism coupling transcription-dependent chromatin remodeling to DSB repair in active promoter regions.
INO80-promoted nucleosome displacement regulates recruitment of proteins with direct roles in HR. Rad51 does not efficiently replace RPA in the absence of nucleosome loss, and this is likely the result of delayed recruitment of Rad52 (unpublished data), which co-operates with Rad51 to displace RPA6,26. In agreement with a role for INO80 in HR repair, NHEJ is relatively unaffected in _arp8_Δ, while _arp8_Δ and HR mutants show similar sensitivities to DNA damaging agents17 (Supplementary Figure 6) and a 2-3-fold decrease in allelic recombination (S. Krishna and J.A. N., unpublished data). In addition to nucleosome displacement, the cellular response to DNA damage also requires formation of γ-H2A. Both forms of chromatin remodeling are ultimately dependent on MRX but independent of one another, comprising two parallel pathways of chromatin changes that have different but complementary roles in the DSB repair response (Figure 4b). MRX-dependent nucleosome remodeling by INO80 is required for efficient recruitment of HR proteins, while MRX-dependent H2A phosphorylation through Tel1 mediates accumulation of cohesin and checkpoint proteins at DSB sites14,24,27.
Methods
ChIP and PCR analysis. Fifty mls of cells were fixed with 1% formaldehyde for either 60 min or 15 min (histones) and lysed in FA buffer28. Immunoprecipitation was performed with 5 OD600 units for histones and 20 OD600 units for all other proteins. Five μl of anti-Myc antibody (9E10, Upstate Biotechnology), 45 μl of anti-Flag agarose beads (M2, Sigma), 5 μl of anti-phospho-H2A.X(Ser139) antibody (Upstate Biotechnology), and 2 μl of anti-RPA or 4 μl of anti-Rad51 antibodies (gift from W-D. Heyer) were used. Extracted DNA was analyzed by real-time PCR using a SYBR Green master mix (ABI) in an ABI 7000 Sequence Detection System. DNA primers were designed to span a 12-20 Kb region around the HO cut site at _MAT_α as well as a region of the POL5 gene; primer sequences are available upon request. Dissociation curve analysis of the amplified DNA melting temperature showed that the each primer set gave a single and specific product. The IP data were normalized to the POL5 gene, where DSB does not occur, to correct for experimental variation and loss of DNA at the DSB site6. Most experiments were repeated at least two times, and in each experiment, PCR reactions were performed in triplicate. The relative IP value represents the ratio of IP DNA/POL5 input DNA after HO induction upon normalization to the IP DNA/POL5 input ratio before HO induction. Standard errors were calculated using error analysis for more than one measurement29. This analysis finds the largest error source by combining standard errors of two measurements, in this case, measurement of IP DNA and input DNA.
Micrococcal nuclease digestion. One-liter cultures were collected just prior to, and at 60 min intervals after DSB induction. Nuclei were prepared under conditions maintaining HO induction (+ 2% galactose), and 1.5 units of MNase (Worthington) were added to 0.25 mls of nuclei as described30. Samples were removed at 1, 2, 4, 8, and 16 min intervals, DNA was purified, and electrophoresed on a 1.25% agarose-TBE gel. DNA was blotted to Genescreen (Dupont-NEN) and hybridized to an 800 bp _MAT_α fragment that is ∼200 bp from the right side of the HO-induced DSB. Radioactive images were captured on a Storm Model 860 Phosphoimager and band intensities were quantitated using ImageQuant TL software.
Supplementary Material
Supplemental Methods
Supplemental Figure Legends
Supplemental Figure 1
Supplemental Figure 2
Supplemental Figure 3
Supplemental Figure 4a
Supplemental Figure 4b-c
Supplemental Figure 5
Supplemental Figure 6
Acknowledgements
Jim Haber, Xuetong Shen, Jessica Downs, Michael Christman, and Virginia Zakian are acknowledged for strains or plasmids, and Wolf-Dietrich Heyer is thanked for antibodies against RPA and Rad51. Cory Hillyer and Nicholas Clark are thanked for technical assistance, and Cheng-Fu Kao is acknowledged for insightful comments. Supported by grants from the N.I.H. to M.A.O. (GM40118), J.A.N. (CA55302), and a supplement to CA55302 to J.A.N. and M.A.O.
References
- 1.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 2.Arkady C, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 3.Shroff R, et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Usui T, Ogawa H, Petrini JH. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 5.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 6.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 7.Lee SE, et al. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 8.Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 9.Weiss K, Simpson RT. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLα. Mol. Cell. Biol. 1998;18:5392–5403. doi: 10.1128/mcb.18.9.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 11.Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uziel T, et al. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 15.Shen X, Ranallo R, Choi E, Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- 16.Morrison AJ, et al. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 17.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–88. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 18.Downs JA, et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Frank-Vaillant M, Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell. 2002;10:1189–1199. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- 20.Palter KB, Foe VE, Alberts BM. Evidence for the formation of nucleosome-like histone complexes on single-stranded DNA. Cell. 1979;18:451–467. doi: 10.1016/0092-8674(79)90064-3. [DOI] [PubMed] [Google Scholar]
- 21.Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Haber JE. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol. 2004;2:0104–0112. doi: 10.1371/journal.pbio.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kantake N, Sugiyama T, Kolodner RD, Kowalczykowski SC. The recombination-deficient mutant RPA (rfa1-t11) is displaced slowly from single-stranded DNA by Rad51 protein. J. Biol. Chem. 2003;278:23410–23417. doi: 10.1074/jbc.M302995200. [DOI] [PubMed] [Google Scholar]
- 24.Unal E, et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 25.Chai B, Huang J, Cairns B, Laurent BC. Distinct roles for the Rsc and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyazaki T, Bressan DA, Shinohara M, Haber JE, Shinohara A. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair. EMBO J. 2004;23:939–949. doi: 10.1038/sj.emboj.7600091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura TM, Du LL, Redon C, Russell P. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol. Cell. Biol. 2004;24:6215–6230. doi: 10.1128/MCB.24.14.6215-6230.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 29.Lichten W. Data and error analysis in the introductory physics laboratory. Allyn and Bacon, Inc.; Newton, MA: 1988. [Google Scholar]
- 30.Fleming AB, Pennings S. Antagonistic remodelling by Swi-Snf and Tup1-Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. EMBO J. 2001;20:5219–5231. doi: 10.1093/emboj/20.18.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Methods
Supplemental Figure Legends
Supplemental Figure 1
Supplemental Figure 2
Supplemental Figure 3
Supplemental Figure 4a
Supplemental Figure 4b-c
Supplemental Figure 5
Supplemental Figure 6