Generation and Characterization of Telomere Length Maintenance in Tankyrase 2-Deficient Mice (original) (raw)
Abstract
Telomere length and function are crucial factors that determine the capacity for cell proliferation and survival, mediate cellular senescence, and play a role in malignant transformation in eukaryotic systems. The telomere length of a specific mammalian species is maintained within a given range by the action of telomerase and telomere-associated proteins. TRF1 is a telomere-associated protein that inhibits telomere elongation by its binding to telomere repeats, preventing access to telomerase. Human TRF1 interacts with tankyrase 1 and tankyrase 2 proteins, two related members of the tankyrase family shown to have poly(ADP-ribose) polymerase activity. Human tankyrase 1 is reported to ADP-ribosylate TRF1 and to down-regulate the telomeric repeat binding activity of TRF1, resulting in telomerase-dependent telomere elongation. Human tankyrase 2 is proposed to have activity similar to that of tankyrase 1, although tankyrase 2 function has been less extensively characterized. In the present study, we have assessed the in vivo function of mouse tankyrase 2 by germ line gene inactivation and show that inactivation of tankyrase 2 does not result in detectable alteration in telomere length when monitored through multiple generations of breeding. This finding suggests that either mouse tankyrases 1 and 2 have redundant functions in telomere length maintenance or that mouse tankyrase 2 differs from human tankyrase 2 in its role in telomere length maintenance. Tankyrase 2 deficiency did result in a significant decrease in body weight sustained through at least the first year of life, most marked in male mice, suggesting that tankyrase 2 functions in potentially telomerase-independent pathways to affect overall development and/or metabolism.
Telomeres are structures at the ends of linear chromosomes in all eukaryotic cells and consist of tandem DNA repeats and associated proteins (4, 8, 16). Telomere length is critical for aspects of chromosome stability that affect cell proliferation and survival (4, 8). The determination and maintenance of species-specific telomere length settings are controlled by a complex system of regulation that is not completely understood. As a consequence of primer dependence and incomplete replication of chromosomal termini during cell division, eukaryotic telomeres shorten as cells replicate (4, 8). Telomerase, an enzyme consisting of the _t_elomerase _R_NA template (TR) and _te_lomerase _r_everse _t_ranscriptase (TERT) (4, 8), is capable of compensating for this loss by adding telomeric repeats to chromosome ends. Numerous reports have described additional telomere-associated proteins, including telomeric repeat binding factors 1 and 2 (TRF1 and TRF2) (7), TRF1-interacting protein 2 (TIN2) (20), tankyrases 1 (34) and 2 (18), protein interacting with NIMA-interacting factor 1 (PINX1) (38), Rap1 (25), Rad50 (24), Mre11 (29), Nbs1 (26), ATM (21), WRN (28), ERCC1/XPF (39), and POT1 (2), which individually and through interactions with one another play important roles in telomere length maintenance.
Both TRF1 and TRF2 bind directly to telomeric DNA repeats through their carboxyl-terminal Myb-like DNA binding motifs, contributing to formation of a specific T-loop structure, which may protect telomere ends from fusion and may modulate access to telomerase (6, 7). However, TRF1 and TRF2 appear to have quite different functions despite their similarities in protein structure and DNA binding, TRF1 negatively regulating telomere elongation whereas TRF2 plays a protective role at the telomere end (6, 7). Overexpression of TRF1 in telomerase-expressing cell lines results in telomere shortening, while expression of a dominant-negative (DN) TRF1 causes telomere elongation (3, 36). In vitro studies in human cells revealed that the inhibitory activity of TRF1 on telomere elongation is controlled by tankyrase 1 (34), tankyrase 2 (18), and TIN2 (20, 37). Tankyrases 1 and 2 share overall domain structure and high amino acid sequence similarity. Tankyrases contain 24 copies of the ankyrin (ANK) repeat that is responsible for their interaction with TRF1. The carboxy terminus of tankyrases contains a region with homology to the catalytic domain of poly(ADP-ribose) polymerases (18, 34). Overexpression of wild-type (WT) human tankyrase 1 in human cell lines induced telomere elongation while mutated tankyrase 1 that lacked poly(ADP-ribose) polymerase activity did not have this effect (12). These and other findings suggest that tankyrases act to ADP-ribosylate TRF1. As a result of this modification, ADP-ribosylated TRF1 is either released from telomere complexes or degraded, leading to increased access of telomerase to the telomere complex. This hypothesized mechanism of telomere length regulation mediated by the collaboration of TRF1 and tankyrases was derived largely from observations in human fibroblasts or immortalized cells, and to date there has been no direct experimental assessment of the in vivo function of tankyrases.
In most human somatic cells, telomeres shorten progressively with cell division, under conditions in which telomerase activity is repressed. In contrast to normal somatic cells, germ line cells express telomerase, and telomere length is maintained within a given size range from generation to generation (4, 8, 16). However, only limited studies have been performed to assess the mechanism mediating telomere maintenance in vivo in mammalian germ cell and somatic lineages. In the most seminal of these studies, progressive telomere shortening was observed in telomerase (TERT or TR) knockout mice (5, 10). The lethality of TRF1 (19) and TIN2 (11) inactivation in knockout mice has hindered efforts to uncover the physiological roles of TRF1 and TIN2 in telomere length maintenance in vivo. In studies reported here, we have analyzed the effect of germ line tankyrase 2 inactivation on telomere length maintenance. We found that there is no effect on telomere length in tankyrase 2 knockout mice through as many as seven generations, suggesting that the function of tankyrase 2 in telomere length maintenance may be redundant, e.g., with that of tankyrase 1, or that mouse tankyrases do not play an essential role in maintenance of mouse telomeres. We identified a significant decrease in body weight of tankyrase 2-deficient mice relative to littermate controls, most notably in male mice, persisting through at least 1 year of life and suggesting that tankyrase may play a telomere-independent role in development and/or metabolism.
MATERIALS AND METHODS
Targeting vector, electroporation, and selection.
A tankyrase 2 (TANK2) gene targeting vector was constructed from an 8.5-kb EcoRI/HindIII fragment (18) of mouse genomic DNA containing the first exon of tankyrase 2. A loxP sequence-flanked neo gene, employed as a positive selectable marker, was inserted upstream of exon 1, and a third loxP sequence was inserted downstream of exon 1 (Fig. 1A). The thymidine kinase cassette (TK) was used as a negative selectable marker. Electroporation and selection were performed with the CJ7 embryonic stem (ES) cell line as described by Tessarollo (35).
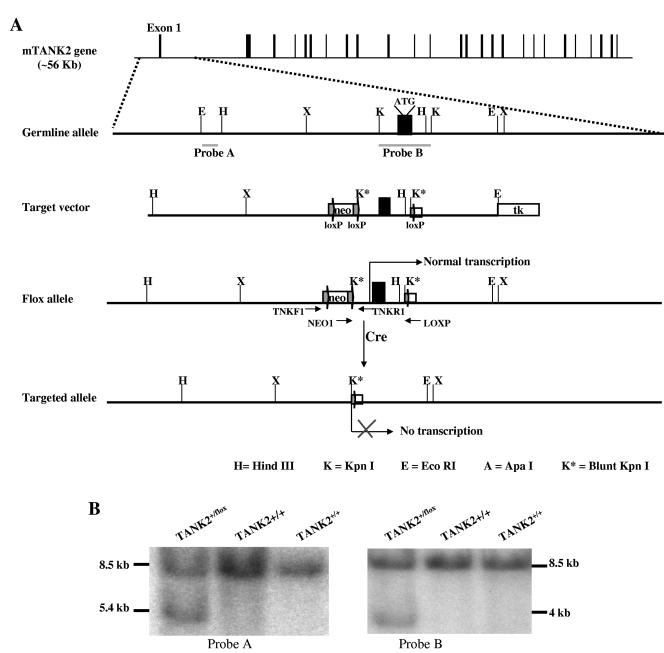
FIG.1.
Generation of TANK2-deficient mice by gene targeting. (A) Gene targeting strategy and restriction map of TANK2 gene. Filled boxes indicate exons, and labeled boxes indicate neomycin (neo) resistance or herpesvirus thymidine kinase (tk) genes. loxP sites are indicated. (B) Southern blot analysis of ES cell DNA. The 8.5-kb band represents the germ line allele. The 5.4- and 4-kb bands represent the targeted alleles. (C) PCR analysis for the conditional TANK2 knockout mouse genotype. The 270- and 340-bp PCR products represent the wild-type and floxed alleles, respectively. (D) PCR analysis for the constitutive TANK2 knockout mouse genotype. The 270- and 240-bp PCR products represent the wild-type and knockout alleles, respectively. (E) RT-PCR analysis for TANK2 mRNA expression. 5′-TANK2 and 3′-TANK2 PCR products represent upstream and downstream cDNA of TANK2, respectively. Actin cDNA was the RT-PCR loading control.
Generation of tankyrase 2 conditional and constitutive knockout mice.
Two independent TANK2 targeted ES cell clones were injected into C57BL/6 blastocysts, generating chimeras that transmitted the targeted allele to progeny. The heterozygous offspring were bred to generate homozygous conditional TANK2 knockout mice or floxed mice (TANK2flox/flox). The floxed mice were bred with C57BL/6 EIIa-cre mice that express the loxP-specific DNA recombinase, cre. EIIa drives expression of cre on embryonic days 1 and 2 and therefore results in cre-mediated deletion of germ line cells, generating a heritable constitutive gene knockout. Mice bearing a TANK2 exon 1-deleted allele were selected from the offspring of TANK2flox/flox × EIIa-cre (22) crosses. Mice homozygous for the TANK2 exon deletion were constitutive TANK2 knockout mice (TANK2−/−). TANK2−/− mice were subsequently bred and selected to eliminate the EIIa-cre gene.
Mice.
TANK2 knockout mice were maintained by intercrosses of heterozygous TANK2 knockout mice. The resulting homozygous mice were designated TANK2−/−G1 mice. Intercrosses among TANK2−/−G1 mice produced TANK2−/−G2 mice; intercrosses among TANK2−/−G2 mice produced TANK2−/−G3 mice, and so on. All animals were housed at Bioqual (Rockville, MD).
Southern blot analysis.
The DNA for Southern blot analysis was isolated from ES cells and mouse tails. DNA isolation and Southern blot analysis procedures are described elsewhere (11). DNA was digested with EcoRI, electrophoresed on a 0.7% agarose gel, transferred to a nylon membrane, and probed with probe A and probe B (Fig. 1A and B).
PCR genotyping.
Tail DNA was purified with the Dneasy tissue kit (QIAGEN, Valencia, CA). PCR primers were TNKF1 (5′-TCT CCT AAC CCC TTT CTC CC-3′), NEO1 (5′-TTC TGG ATT CAT CGA CTG TG-3′), TNKR1 (5′-GCA TAC ACA TCA AAG TTT TCC G-3′), LOXP (5′-GAC GTA AAC TCC TCT TCA GAC GTA ATA AC-3′). PCRs were carried out in 50 μl of PCR mix containing 25 μl of PCR buffer (Supermix PCR kit; Life Technologies, Rockville, MD) and 1 to 100 ng of tail DNA and in the presence of either the NEO1 primer set (0.2 μM each of TNKKF1, TNKR1, and NEO1 primers) or the LOXP primer set (0.2 μM each of TNKF1, TNKR1, and LOXP primers). PCR was carried out at 95°C for 30 s, 58°C for 30 s, and 72°C for 1 min. PCR products were revealed by agarose gel electrophoresis and ethidium bromide staining. The 270-bp band corresponds to the wild-type allele, the 340-bp band corresponds to the conditional knockout allele, and the 240-bp band corresponds to both conditional and constitutive knockout alleles.
RT-PCR.
Spleen lymphocytes were stimulated with concanavalin A (5 μg/ml), lipopolysaccharide (15 μg/ml), and recombinant interleukin-2 (150 IU/ml) for 48 h. A NucleoSpin kit (BD Biosciences) was used to isolate total RNA from spleen cells for reverse transcription-PCR (RT-PCR) analysis. Two primer pairs were used to detect TANK2 expression: for detection of 5′ tankyrase 2 cDNA, TNK393F (5′-GCC AAC CAT CCG AAA TAC AG-3′), starting at position 393 of TANK2 cDNA sequence, and TNK615R (5′-TCT GTT TAT GTA TCC TGC TGC C-3′), ending at position 615 of TANK2 cDNA sequence; for detection of 3′ tankyrase 2 cDNA, TNK2998F (5′-GAA GAA AAC CAC AAC ACT GC-3′), starting at position 2998 of TANK2 cDNA sequence, and TNK3179R (5′-CCA CCT CCA ATT CCA TAC AC-3′), ending at position 3179 of TANK2 cDNA sequence. RT-PCRs were carried out in 50 μl of PCR mix containing 25 μl of PCR buffer (One-step RT-PCR kit; Life Technologies), 1 μg of total RNA, and 0.2 μM primers. RT-PCR was carried out at 42°C for 30 min, with 35 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 1 min. RT-PCR products were visualized by agarose gel electrophoresis with ethidium bromide staining. The 5′-TANK2 cDNA (222-bp) PCR product was visualized when the primer pair TNK393F and TNK615R was used. The 3′-TANK2 cDNA (161-bp) PCR product was visualized when TNK2998F and TNK3179 were used.
Q-FISH.
Spleen lymphocytes were stimulated with concanavalin A (5 μg/ml), lipopolysaccharide (15 μg/ml), and recombinant interleukin-2 (150 IU/ml) for 48 h. Activated cells were treated with Colcemid for 2.5 h and fixed with 3:1 methanol:acetic acid. Metaphase cells were dropped on slides that were then hybridized with Cy3-labeled (CCCTAA)3 oligonucleotides, and chromosomes were stained with DAPI (4′,6′-diamidino-2-phenylindole). Fluorescent signals were collected and analyzed as previously described (10, 23). Using FluoreSphere beads (Molecular Probes) as standards, fluorescent signal intensities were calculated by using quantitative fluorescence in situ hybridization (Q-FISH) software from the Lansdorp laboratory (23). At least 20 metaphases and 600 telomeres each from TANK2+/+, TANK2−/−G1, TANK2−/−G2, TANK2−/−G3, TANK2−/−G5, and TANK2−/−G7 mice were used to collect telomere fluorescent signal data.
Flow-FISH.
The protocol for flow cytometric analysis of fluorescence in situ hybridization (Flow-FISH) was modified from the work of Baerlocher et al. (1). Spleen lymphocytes were prepared and incubated with fluorescein isothiocyanate-labeled (CCCTAA)3 oligonucleotides in buffer containing 20 mM Tris, 75% formamide, 1% bovine serum albumin, and 20 mM NaCl at 86°C for 15 min. The cells were then incubated at room temperature for 2 h. Cells were used for fluorescence-activated cell sorting analysis after being washed four times with buffer containing 20 mM Tris, 75% formamide, 1% bovine serum albumin, and 0.1% Tween 20 (14).
RESULTS
Generation of TANK2 conditional knockout mice.
It has been reported that overexpression of tankyrase 2 in vitro results in rapid cell death (18) and that knockdown of tankyrase 1 causes mitotic arrest in primary human cells (13), suggesting that TANK2 inactivation might cause mouse embryonic lethality. We therefore used cre/loxP conditional knockout technology (15) to study the physiological roles of tankyrase 2. Figure 1A illustrates the strategy for tankyrase 2 conditional gene targeting. TANK2 conditional knockout ES cell clones were identified by Southern blot analysis. For Southern analysis, ES cell genomic DNA was digested with EcoRI, electrophoresed, and hybridized with probes A and B (Fig. 1A). Figure 1B shows the results of a representative Southern blot in which wild-type ES cell DNA generated a single 8.5-kb band with both probe A and probe B, while heterozygous conditional knockout ES cell DNA generated an additional 5.4-kb band with probe A and a 4-kb band with probe B. Three heterozygous ES cell clones were identified from screening of 96 clones, and two of these clones were used to generate conditional TANK2 knockout mice. Mouse genotypes were identified using genomic PCR with primers TNKF1, NEO1, and TNKR1. Figure 1C shows that DNA from wild-type mice (TANK2+/+) gave one band (270 bp), DNA from homozygous floxed mice (TANK2flox/flox) gave one band at about 340 bp, and DNA from heterozygous mice (TANK2+/flox) gave bands of both 270 bp and 340 bp. Intercrosses of TANK2+/flox mice resulted in 5 TANK2flox/flox, 10 TANK2+/flox, and 6 TANK2+/+ offspring. As expected, TANK2flox/flox mice had normal expression of tankyrase 2 as assessed by RT-PCR using two primer sets, TNK393F/TNK615R and TNK2998F/3179R. Both 5′-TANK2 cDNA (222-bp) and 3′-TANK2 cDNA (161-bp) PCR products were detected in TANK2+/+ and TANK2flox/flox mice (Fig. 1E).
Generation of TANK2 constitutive knockout mice from conditional knockout mice.
To determine the effect of TANK2 inactivation in vivo, we next generated constitutive TANK2 knockouts from TANK2flox/flox parental mice. Conditional TANK2 knockout mice (TANK2flox/flox) were bred with EIIa-cre transgenic mice to generate constitutive knockouts, as described in Materials and Methods (Fig. 1A). The resulting knockout mice were identified by genomic DNA PCR with primers TNKF1, LOX, and TNKR1. As shown in Fig. 1D, wild-type mice (TANK2+/+) gave one 270-bp band, homozygous knockout mice (TANK2−/−) gave one 240-bp band, and heterozygous mice (TANK2+/−) gave both 270-bp and 240-bp bands. Expression of tankyrase 2 was determined by RT-PCR with primer sets TNK393F/TNK615R and TNK2998F/3179R as described above. The 5′-TANK2 cDNA (222-bp) and 3′-TANK2 cDNA (161-bp) PCR products were detected in both wild-type and heterozygous TANK2 knockout mice while neither 5′-TANK2 cDNA nor 3′-TANK2 cDNA from RT-PCR products was detected in homozygous (TANK2−/−) knockout mice (Fig. 1E). Twenty-nine TANK2+/+, 63 TANK2+/−, and 27 TANK2−/− mice were observed in 109 offspring from TANK2+/− mouse intercrosses, consistent with Mendelian segregation and indicating that no lethality was associated with complete TANK2 inactivation.
Telomere length is not altered in TANK2 knockout mice.
Previous in vitro studies have indicated that tankyrases influence telomere length in a telomerase-dependent manner and that interfering with tankyrase function can lead to decreasing telomere length, possibly through decreased access to telomerase (17, 32). Telomere shortening has been reported to occur over successive generations of mTERT- (10) and mTR-deficient (5) mice that lack telomerase activity. Telomere shortening might similarly occur in successive generations of TANK2 knockout mice; therefore, TANK2−/− mice were intercrossed up to seven generations for telomere length analysis. In Fig. 2A, spleen cells from seven TANK2−/−G1 (generation 1) knockout mice and the same number of their wild-type littermates were analyzed for telomere length by Flow-FISH. This analysis demonstrated that there was no detectable telomere shortening in TANK2−/−G1 knockout mice in comparison to littermate controls (TANK2+/+); telomere length was in fact slightly increased in TANK2−/−G1 knockout mice. Additional analysis using three TANK2−/−G5 and four TANK2−/−G7 knockout mice (Fig. 2A) demonstrated no difference in telomere length between TANK2−/−G1 and TANK2−/−G5 or TANK2−/−G7 knockout mice, although the telomeres of G5 and G7 mice were again slightly longer than those of wild-type mice.
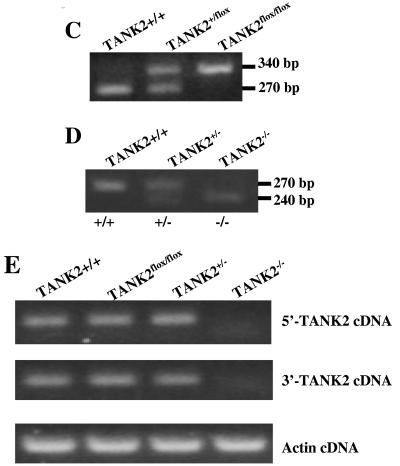
FIG. 2.
Telomere length was not altered in TANK2 knockout mice. (A) Spleen cells were isolated from TANK2+/+ (n = 7), TANK2−/−G1 (n = 7), and TANK2−/−G5/G7 (n = 7) mice, and relative telomere length was determined by Flow-FISH. The fluorescein isothiocyanate fluorescent signal of the cell binding telomeric probe was converted to arbitrary units of molecule equivalents of soluble fluorescence (MESF). (B and C) Histograms, based on Q-FISH data from the comparison of one TANK2+/+ mouse with one TANK2−/−G1 mouse (B) and the comparison of one TANK2+/+ mouse with one TANK2−/−G7 mouse (C), depict the distribution of fluorescent intensities corresponding to individual telomeres of different lengths present in metaphase chromosomes. (D) Spleen cells were isolated from TANK2+/+ (n = 8), TANK2−/−G1 (n = 8), and TANK2−/−G5/G7 (n = 9) mice, and relative telomere length was determined by Q-FISH. The average of fluorescent intensities from each mouse was normalized to that of a TANK2+/+ mouse (defined as 100). The relative telomere length of each strain of mice is plotted.
We utilized the Q-FISH technique, with its ability to assess the distribution of individual telomere lengths, to further analyze telomere length. Figures 2B and C show representative Q-FISH analysis of telomere length of TANK2+/+, TANK2−/−G1, and TANK2−/−G7 mice. There was no significant difference in telomere lengths among TANK2+/+, TANK2−/−G1, and TANK2−/−G7 mice. Overall, telomere lengths of eight TANK2+/+ mice, eight littermate TANK2−/−G1 mice, five TANK2−/−G5 mice, and four TANK2−/−G7 mice were analyzed with Q-FISH. Average telomere fluorescent signals were normalized and plotted in Fig. 2D. Statistical analysis indicated that there was no significant difference in telomere length among TANK2+/+, TANK2−/−G1, and TANK2−/−G5/G7 mice. We also did not observe chromosome abnormalities in over 300 metaphases from TANK2−/−G1 and TANK2−/−G5/G7 mice, and there was no difference in the number of telomere-free ends in tankyrase 2-deficient and control mice (data not shown).
Body weight of tankyrase 2 knockout mice was reduced.
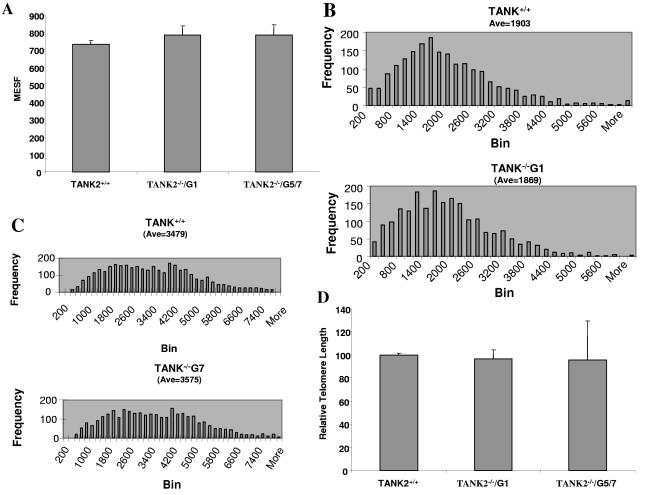
Tankyrase 2 knockout mice were observed for approximately 1 year, with no apparent disease or defect in survival. However, it was noted that compared to WT littermates the body weight of TANK2−/− mice was reduced. Figure 3A shows body weight changes in female TANK2−/− mice and WT controls followed over 42 weeks. The body weight of female TANK2−/− mice was slightly decreased in comparison to control mice, most notably at ages between 5 weeks and 13 weeks. Statistical analysis indicated that there was a significant difference in body weight between female TANK2−/− and control mice at each time point measured except at 42 weeks. There was a more dramatic decrease of body weight in male TANK2−/− mice, with weight approximately 20% less than WT control mice over a 42-week period of observation (Fig. 3B). Differences between male TANK2+/+ and TANK2−/− mice were statistically significant at all time points.
FIG. 3.
The body weights of tankyrase 2 knockout mice were reduced. (A) Body weight changes in female TANK+/+ (n = 5) and TANK−/− (n = 6) mice. (B) Body weight changes in male TANK+/+ (n = 6) and TANK−/− (n = 7) mice.
DISCUSSION
Tankyrase 2 is a member of the tankyrase family, and human tankyrase 2 has the capacity to bind human TRF1 (18), a telomere-binding protein that negatively regulates telomere elongation (33). Overexpression of tankyrase 2 induces rapid cell death in primary human cells (18). In contrast to tankyrase 2, cell survival and growth were normal in cells that overexpress tankyrase 1 (12). Overexpression of tankyrase 1 in fact induced telomere elongation, suggesting that tankyrases (potentially tankyrase 2 as well as tankyrase 1) are able to ADP-ribosylate TRF1, releasing ADP-ribosylated TRF1 from telomeres and allowing telomerase access and telomere elongation (12). To test the hypothesized function of tankyrase 2 in vivo, we generated TANK2 knockout mice and analyzed the effect of TANK2 inactivation on telomere length maintenance. Knockout mice were viable. Contrary to expectation, we found neither telomere shortening nor chromosomal abnormalities even across multiple generations of TANK2 knockout mice. Phenotyping of TANK2 knockout mice did reveal a significant decrease in body weight through nearly a year of follow-up. Preliminary assessment indicates that TANK2 knockout mice have substantially reduced fat pads, suggesting that TANK2 plays a potentially telomere-independent role in vivo in overall metabolic regulation.
Human tankyrase 2 has been demonstrated to have TRF1 binding activity indistinguishable from that of tankyrase 1 (31). Moreover, it has been shown that tankyrase 2 can oligomerize with tankyrase 1 (31), raising the possibility that tankyrases 1 and 2 collaborate or are redundant in vivo. It was striking that there was no telomere shortening in early or late generations of TANK2 knockout mouse in the present study (Fig. 2), while normal expression of tankyrase 1 was detected in multiple tissues of TANK2−/− mice in comparison to wild-type controls (data not shown). This suggests that tankyrase 1 and tankyrase 2 could have a redundant function in telomere length maintenance rather than functioning as obligate heterodimers or oligomers. It will therefore be important to determine the consequences of inactivating both tankyrase genes. To this end, we are in the process of generating TANK1 and TANK1/2 double knockout mice.
It has been reported that mouse TRF1 lacks the motif (RXXPDG) of human TRF1 that is required for binding to tankyrases (30). The failure to find an effect of TANK2 inactivation on telomere length is thus also consistent with the possibility that mouse tankyrases do not bind mouse TRF1 and therefore are not involved in telomere length maintenance in mice. Characterization of TANK1- and TANK1/TANK2-deficient mice will help to further elucidate the role of tankyrases in mouse models.
The observed decrease in body weight of TANK2 knockout mice was unexpected (Fig. 3). Initial observations suggest that body fat is reduced in TANK2 knockout mice, contributing at least in part to the observed decrease in total body weight (data not shown). It is of interest that previous reports indicate that the majority of tankyrase protein is nonnuclear and is located in the cytosol (9). It has also been reported that tankyrases associate with the vesicular protein IRAP (insulin response aminopeptidase) (30) and with the Src homology 2-containing adaptor protein Grb14 (27), which regulates the activity of glucose transporter GLUT4 in adipocytes. We therefore speculate that inactivation of tankyrase 2 results in altered glucose/fat metabolism due to disruptions in these pathways, a hypothesis that is now under investigation.
Acknowledgments
We are grateful to Nanping Weng and Karen S. Hathcock for their critical reading of the manuscript. We thank Eileen Southon and Susan Reid for their help in generating mutant mice. We thank Genevieve Sanchez-Howard and staff at Bioqual for expert animal care and breeding.
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute.
REFERENCES
- 1.Baerlocher, G. M., J. Mak, T. Tien, and P. M. Lansdorp. 2002. Telomere length measurement by fluorescence in situ hybridization and flow cytometry: tips and pitfalls. Cytometry 47**:**89-99. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292**:**1171-1175. [DOI] [PubMed] [Google Scholar]
- 3.Bianchi, A., S. Smith, L. Chong, P. Elias, and T. de Lange. 1997. TRF1 is a dimer and bends telomeric DNA. EMBO J. 16**:**1785-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackburn, E. H. 2005. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 579**:**859-862. [DOI] [PubMed] [Google Scholar]
- 5.Blasco, M. A., H. W. Lee, M. P. Hande, E. Samper, P. M. Lansdorp, R. A. DePinho, and C. W. Greider. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91**:**25-34. [DOI] [PubMed] [Google Scholar]
- 6.Broccoli, D., L. Chong, S. Oelmann, A. A. Fernald, N. Marziliano, B. van Steensel, D. Kipling, M. M. Le Beau, and T. de Lange. 1997. Comparison of the human and mouse genes encoding the telomeric protein, TRF1: chromosomal localization, expression and conserved protein domains. Hum. Mol. Genet. 6**:**69-76. [DOI] [PubMed] [Google Scholar]
- 7.Broccoli, D., A. Smogorzewska, L. Chong, and T. de Lange. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17**:**231-235. [DOI] [PubMed] [Google Scholar]
- 8.Bryan, T. M., and T. R. Cech. 1999. Telomerase and the maintenance of chromosome ends. Curr. Opin. Cell Biol. 11**:**318-324. [DOI] [PubMed] [Google Scholar]
- 9.Chi, N. W., and H. F. Lodish. 2000. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 275**:**38437-38444. [DOI] [PubMed] [Google Scholar]
- 10.Chiang, Y. J., M. T. Hemann, K. S. Hathcock, L. Tessarollo, L. Feigenbaum, W. C. Hahn, and R. J. Hodes. 2004. Expression of telomerase RNA template, but not telomerase reverse transcriptase, is limiting for telomere length maintenance in vivo. Mol. Cell. Biol. 24**:**7024-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang, Y. J., S. H. Kim, L. Tessarollo, J. Campisi, and R. J. Hodes. 2004. Telomere-associated protein TIN2 is essential for early embryonic development through a telomerase-independent pathway. Mol. Cell. Biol. 24**:**6631-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook, B. D., J. N. Dynek, W. Chang, G. Shostak, and S. Smith. 2002. Role for the related poly(ADP-ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol. 22**:**332-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dynek, J. N., and S. Smith. 2004. Resolution of sister telomere association is required for progression through mitosis. Science 304**:**97-100. [DOI] [PubMed] [Google Scholar]
- 14.Fritsch, R. D., X. Shen, G. P. Sims, K. S. Hathcock, R. J. Hodes, and P. E. Lipsky. 2005. Stepwise differentiation of CD4 memory T cells defined by expression of CCR7 and CD27. J. Immunol. 175**:**6489-6497. [DOI] [PubMed] [Google Scholar]
- 15.Gu, H., J. D. Marth, P. C. Orban, H. Mossmann, and K. Rajewsky. 1994. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science 265**:**103-106. [DOI] [PubMed] [Google Scholar]
- 16.Hathcock, K. S., Y. J. Chiang, and R. J. Hodes. 2005. In vivo regulation of telomerase activity and telomere length. Immunol. Rev. 205**:**104-113. [DOI] [PubMed] [Google Scholar]
- 17.Houghtaling, B. R., L. Cuttonaro, W. Chang, and S. Smith. 2004. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 14**:**1621-1631. [DOI] [PubMed] [Google Scholar]
- 18.Kaminker, P. G., S. H. Kim, R. D. Taylor, Y. Zebarjadian, W. D. Funk, G. B. Morin, P. Yaswen, and J. Campisi. 2001. TANK2, a new TRF1-associated poly(ADP-ribose) polymerase, causes rapid induction of cell death upon overexpression. J. Biol. Chem. 276**:**35891-35899. [DOI] [PubMed] [Google Scholar]
- 19.Karlseder, J., L. Kachatrian, H. Takai, K. Mercer, S. Hingorani, T. Jacks, and T. de Lange. 2003. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol. Cell. Biol. 23**:**6533-6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, S. H., P. Kaminker, and J. Campisi. 1999. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23**:**405-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kishi, S., X. Z. Zhou, Y. Ziv, C. Khoo, D. E. Hill, Y. Shiloh, and K. P. Lu. 2001. Telomeric protein Pin2/TRF1 as an important ATM target in response to double strand DNA breaks. J. Biol. Chem. 276**:**29282-29291. [DOI] [PubMed] [Google Scholar]
- 22.Lakso, M., J. G. Pichel, J. R. Gorman, B. Sauer, Y. Okamoto, E. Lee, F. W. Alt, and H. Westphal. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 93**:**5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lansdorp, P. M., N. P. Verwoerd, F. M. van de Rijke, V. Dragowska, M. T. Little, R. W. Dirks, A. K. Raap, and H. J. Tanke. 1996. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 5**:**685-691. [DOI] [PubMed] [Google Scholar]
- 24.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152**:**143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, B., S. Oestreich, and T. de Lange. 2000. Identification of human Rap1: implications for telomere evolution. Cell 101**:**471-483. [DOI] [PubMed] [Google Scholar]
- 26.Lombard, D. B., and L. Guarente. 2000. Nijmegen breakage syndrome disease protein and MRE11 at PML nuclear bodies and meiotic telomeres. Cancer Res. 60**:**2331-2334. [PubMed] [Google Scholar]
- 27.Lyons, R. J., R. Deane, D. K. Lynch, Z. S. Ye, G. M. Sanderson, H. J. Eyre, G. R. Sutherland, and R. J. Daly. 2001. Identification of a novel human tankyrase through its interaction with the adaptor protein Grb14. J. Biol. Chem. 276**:**17172-17180. [DOI] [PubMed] [Google Scholar]
- 28.Opresko, P. L., C. von Kobbe, J. P. Laine, J. Harrigan, I. D. Hickson, and V. A. Bohr. 2002. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 277**:**41110-41119. [DOI] [PubMed] [Google Scholar]
- 29.Ritchie, K. B., and T. D. Petes. 2000. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155**:**475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sbodio, J. I., and N. W. Chi. 2002. Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner. J. Biol. Chem. 277**:**31887-31892. [DOI] [PubMed] [Google Scholar]
- 31.Sbodio, J. I., H. F. Lodish, and N. W. Chi. 2002. Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase). Biochem. J. 361**:**451-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, S., and T. de Lange. 2000. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10**:**1299-1302. [DOI] [PubMed] [Google Scholar]
- 33.Smith, S., and T. de Lange. 1997. TRF1, a mammalian telomeric protein. Trends Genet. 13**:**21-26. [DOI] [PubMed] [Google Scholar]
- 34.Smith, S., I. Giriat, A. Schmitt, and T. de Lange. 1998. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282**:**1484-1487. [DOI] [PubMed] [Google Scholar]
- 35.Tessarollo, L. 2001. Gene knockouts: manipulating mouse embryonic stem cells. Methods Mol. Biol. 158**:**47-63. [DOI] [PubMed] [Google Scholar]
- 36.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385**:**740-743. [DOI] [PubMed] [Google Scholar]
- 37.Ye, J. Z., J. R. Donigian, M. van Overbeek, D. Loayza, Y. Luo, A. N. Krutchinsky, B. T. Chait, and T. de Lange. 2004. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 279**:**47264-47271. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, X. Z., and K. P. Lu. 2001. The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell 107**:**347-359. [DOI] [PubMed] [Google Scholar]
- 39.Zhu, X. D., L. Niedernhofer, B. Kuster, M. Mann, J. H. Hoeijmakers, and T. de Lange. 2003. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12**:**1489-1498. [DOI] [PubMed] [Google Scholar]