A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis (original) (raw)
Abstract
Plant recognition of pathogens leads to rapid activation of MPK3 and MPK6, two Arabidopsis mitogen-activated protein kinases (MAPKs), and their orthologs in other species. Here, we report that synthesis of camalexin, the major phytoalexin in Arabidopsis, is regulated by the MPK3/MPK6 cascade. Activation of MPK3/MPK6 by expression of active upstream MAPK kinase (MAPKK) or MAPKK kinase (MAPKKK) was sufficient to induce camalexin synthesis in the absence of pathogen attack. Induction of camalexin by Botrytis cinerea was preceded by MPK3/MPK6 activation, and compromised in mpk3 and mpk6 mutants. Genetic analysis placed the MPK3/MPK6 cascade upstream of PHYTOALEXIN DEFICIENT 2 (PAD2) and PAD3, but independent or downstream of PAD1 and PAD4. Camalexin induction after MPK3/MPK6 activation was preceded by rapid and coordinated up-regulation of multiple genes encoding enzymes in the tryptophan (Trp) biosynthetic pathway, in the conversion of Trp to indole-3-acetaldoxime (IAOx, a branch point between primary and secondary metabolism), and in the camalexin biosynthetic pathway downstream of IAOx. These results indicate that the MPK3/MPK6 cascade regulates camalexin synthesis through transcriptional regulation of the biosynthetic genes after pathogen infection.
Keywords: defense signaling, fungal resistance, camalexin biosynthesis, Botrytis cinerea
Plant active defense is initiated by host recognition of pathogen effectors or by the binding of non-host-specific pathogen-associated molecular patterns (PAMPs) to host receptors (1–4). Mitogen-activated protein kinase (MAPK) cascades are conserved eukaryotic signaling modules that function downstream of sensors/receptors (5). Studies from a number of laboratories demonstrated that the tobacco MAPKs SIPK and WIPK and their orthologs in other species including Arabidopsis MPK6 and MPK3 are activated after PAMP treatment or pathogen infection (reviewed in refs. 6–10). In tobacco, SIPK and WIPK share a common upstream MAPK kinase (MAPKK), NtMEK2 (11). There are two NtMEK2 orthologs in Arabidopsis, MKK4 and MKK5 (10, 12, 13). The MAPKK kinases (MAPKKKs) upstream of NtMEK2/MKK4/MKK5 include MEKK1 and MAPKKKα (13, 14).
Phytoalexins are defined as low-molecular-weight antimicrobial compounds produced by plants after exposure to pathogens (15, 16). Phytoalexins are an integral part of induced plant disease resistance. Disruption of pathogen genes that encode enzymes known to detoxify phytoalexins can lead to loss of pathogenicity, and the virulence of a pathogen on a specific host sometimes coevolves with the generation of enzymes that are capable of degrading phytoalexins (17). Mutations that cause reduced phytoalexin can lead to increased susceptibility of plants to pathogens including Botrytis cinerea (18–20). Engineering-enhanced phytoalexin production can lead to increased disease resistance (15). Phytoalexin induction is associated with the activation of genes encoding biosynthetic enzymes (15, 16, 21, 22). However, the signaling pathway(s) leading to activation of these genes are poorly understood.
In this article, we demonstrate that a pathogen-responsive MAPK cascade, MAPKKKα/MEKK1-MKK4/MKK5-MPK3/MPK6, plays a positive role in regulating the biosynthesis of camalexin [3-thiazol-2′-yl-indole (23)] in Arabidopsis. Previous mutant screens identified PAD1 (PHYTOALEXIN DEFICIENT 1), PAD2, PAD3, and PAD4 as components of the biosynthetic or regulatory pathways that lead to pathogen-induced camalexin production (24, 25). Genetic analysis placed this MAPK cascade upstream of PAD2 and PAD3 but independent or downstream of PAD1 and PAD4. Activation of the MPK3/MPK6 cascade leads to rapid and coordinated up-regulation of multiple genes encoding enzymes in the camalexin biosynthetic pathway. Mutations in MPK3 and MPK6 compromise _B. cinerea_-induced camalexin accumulation, which is associated with reduced resistance, providing loss-of-function evidence supporting a positive role of this MAPK cascade in signaling camalexin biosynthesis in plants challenged by pathogens.
Results
Plant recognition of pathogens leads to the activation of multiple signaling pathways. To identify the defense responses regulated by the MPK3/MPK6 cascade, we used a conditional gain-of-function transgenic system, in which constitutively active mutants of the upstream MAPKKs (tobacco NtMEK2DD and Arabidopsis MKK4DD or MKK5DD) were expressed under the control of a steroid-inducible promoter (11, 12, 26). Upon treatment with the steroid dexamethasone (DEX), induction of the active MAPKKs activates the endogenous MAPKs, which in turn induce the responses further downstream. Extracts from DEX-treated GVG-NtMEK2DD Arabidopsis seedlings, prepared by using an established camalexin extraction procedure (21, 23, 24), showed excitation and emission spectra characteristic of camalexin. Comparison of this compound with camalexin standard by using thin-layer chromatography confirmed its identity [supporting information (SI) Fig. S1]. Induction of camalexin was detectable in GVG-NtMEK2DD Arabidopsis 6 h after DEX treatment, preceded by MPK3/MPK6 activation (Fig. 1). DEX treatment of GVG-MKK4 DD or GVG-MKK5DD Arabidopsis plants also led to camalexin induction (Fig. S2). However, the levels of camalexin accumulation were lower and variable, because of transgene silencing in some of the T4 seedlings. For this reason, we used the GVG-NtMEK2DD Arabidopsis line, which did not show transgene silencing, for further analyses. Expression of wild-type NtMEK2, MKK4, MKK5, or kinase-inactive NtMEK2KR, MKK4KR, MKK5KR did not activate MPK3/MPK6 or induce camalexin (12) (Fig. S2).
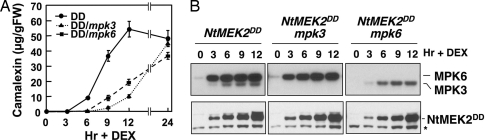
Fig. 1.
Activation of MPK3 and MPK6 induces camalexin in conditional gain-of-function GVG-NtMEK2 DD transgenic plants. (A) Endogenous MPK3 and MPK6 are required for the full induction of camalexin in GVG-NtMEK2 DD plants. Two-week-old GVG-NtMEK2 DD, GVG-NtMEK2DD/mpk3, and GVG-NtMEK2DD/mpk6 seedlings were treated with 1 μM DEX. Camalexin was quantitated by fluorospectrometry at the indicated times. Error bars indicate standard deviations (n = 3). (B) MPK3 and/or MPK6 activation (Upper), as determined by the in-gel kinase assay, and NtMEK2DD induction (Lower), as determined by immunoblot analysis using anti-Flag antibody. The asterisk indicates a nonspecific band that is recognized by the secondary antibody.
Loss of either MPK3 or MPK6 partially compromised camalexin induction, as indicated by delayed and reduced camalexin induction in GVG-NtMEK2DD/mpk3 and GVG-NtMEK2DD/mpk6 double mutants (Fig. 1A). At 24 h, camalexin accumulation in GVG-NtMEK2 DD seedlings declined, and that in GVG-NtMEK2DD/mpk3 and GVG-NtMEK2DD/mpk6 caught up, perhaps a result of earlier cell death in GVG-NtMEK2 DD seedlings. NtMEK2DD protein induction was comparable in all three genotypes (Fig. 1B Lower). In GVG-NtMEK2DD/mpk3 plants, only MPK6 activation was detected, and in GVG-NtMEK2DD/mpk6 plants, only MPK3 activation was detected (Fig. 1B Upper). These results suggest that MPK3 and MPK6 have overlapping functions, and both are required for full induction of camalexin biosynthesis.
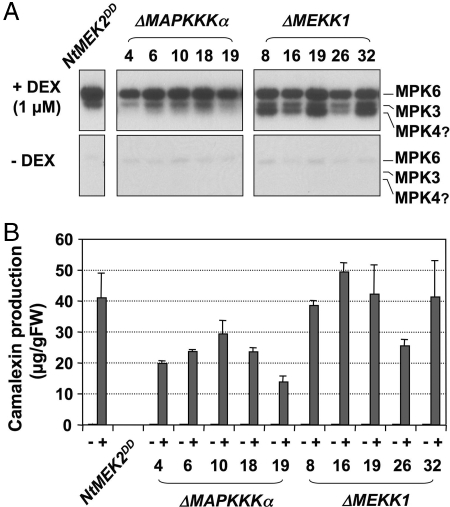
MEKK1 and LeMAPKKKα were implicated in PAMPs and gene-for-gene-mediated defense responses, respectively (13, 14). To determine whether either or both MAPKKKs act upstream of the MKK4/MKK5-MPK3/MPK6 module in regulating camalexin biosynthesis, we generated transgenic plants expressing the constitutively active forms of Arabidopsis MAPKKKα and MEKK1 (ΔMAPKKKα and ΔMEKK1) under the control of the DEX-inducible promoter. Camalexin production was examined in five independent T2 lines that showed transgene induction in the T1 generation. High levels of MPK3 and MPK6 activities and camalexin production were detected only after DEX treatment (Fig. 2). The identities of these two MAPKs were determined by using an immune-complex kinase assay using member-specific antibodies (data not shown). ΔMEKK1 also activated another MAPK (Fig. 2A), possibly MPK4 based on its size and the fact that MEKK1 functions upstream of MPK4 (27). Taken together, our results show that, in the conditional gain-of-function MAPKK and MAPKKK plants, the activation of endogenous downstream MPK3/MPK6 is sufficient to induce camalexin biosynthesis.
Fig. 2.
Camalexin induction after MPK3/MPK6 activation in conditional gain-of-function MAPKKK transgenic plants. (A) Activation of endogenous MAPKs by constitutively active ΔMEKK1 and ΔMAPKKKα. Five-day-old hygromycin-resistant T2 seedlings were transferred to GC vials. When the seedlings were 2 weeks old, three vials were treated with 1 μM DEX, and three were treated with an equal volume of ethanol as controls (−DEX). Seedlings were collected 24 h later, and MAPK activation was determined by the in-gel kinase assay. (B) Activation of MPK3/MPK6 by ΔMEKK1 and ΔMAPKKKα leads to camalexin accumulation. After the seedlings were collected, camalexin accumulation in the medium was quantitated by fluorospectrometry. Bars represent means and standard deviations (n = 3).
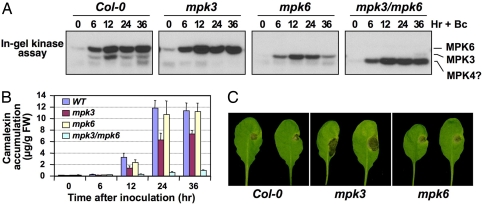
To test for a role of this MAPK cascade in pathogen-induced camalexin biosynthesis, we measured camalexin accumulation in mpk3 and mpk6 mutants infected with B. cinerea. Activation of MPK3 and MPK6 was observed in Arabidopsis seedlings concurrently with B. cinerea spore germination (Fig. S3), followed by accumulation of camalexin (Fig. 3). Neither response was induced by inoculation of boiled spores (data not shown). Camalexin accumulation was reduced by ≈50% in mpk3 plants and delayed in mpk6 plants. Importantly, in partially rescued double mpk3/mpk6 mutant (28), induction of camalexin was almost abolished, demonstrating that both MPK3 and MPK6 are involved in fungus-induced camalexin production. Consistent with reports that camalexin is required for resistance to B. cinerea (19), compromised camalexin induction in mpk3 was associated with increased susceptibility (Fig. 3C). In addition to MPK3 and MPK6, B. cinerea infection also activated a smaller MAPK, possibly MPK4. Activation of this MAPK was elevated in mpk3/mpk6 plants, possibly a result of overabundant stomata, where MPK4 is highly expressed (28, 29) and/or loss of cross-talk between the two MAPK cascades. Camalexin induction by B. cinerea was not compromised in mpk4 plants (data not shown). These data demonstrate that MPK3 and MPK6 are required for _B. cinerea_-induced camalexin synthesis and the consequent limitation of fungal growth.
Fig. 3.
Function of MPK3 and MPK6 in pathogen-induced camalexin biosynthesis and resistance against B. cinerea. (A) MAPK activation in Arabidopsis after B. cinerea inoculation. Two-week-old WT (Col-0), mpk3, mpk6, and rescued mpk3/mpk6 double-mutant seedlings were inoculated with Botrytis spores. At the indicated times, seedlings were collected for in-gel kinase assay. (B) Camalexin in the medium was quantitated by fluorospectrometry. Error bars indicate standard deviations (n = 3). (C) Five-week-old soil-grown Arabidopsis plants were inoculated with 10 μl of spore suspension (1 × 105 spores per milliliter). Disease symptoms were scored 3 days later. More than 20 plants per genotype were assayed in each of three independent experiments. Representative images are shown.
Genetic screens identified several important components in the camalexin biosynthetic and regulatory pathways. Mutations in PAD1, PAD2, PAD3, or PAD4 abolish or reduce camalexin accumulation in Arabidopsis after bacterial infection (24, 25). However, PAD1 and PAD4 are not required for camalexin induction by fungal pathogens (18, 20). PAD4 encodes a protein with sequence similarity to lipases, and is required for induced expression of many genes after Psm ES4326 infection (30, 31). PAD3 encodes cytochrome P450 monooxygenase CYP71B15 that catalyzes the last step of camalexin biosynthesis (22, 32). PAD2 encodes γ-glutamylcysteine synthetase (GSH1), one of the two enzymes in the glutathione biosynthesis pathway (33). PAD1 remains to be identified.
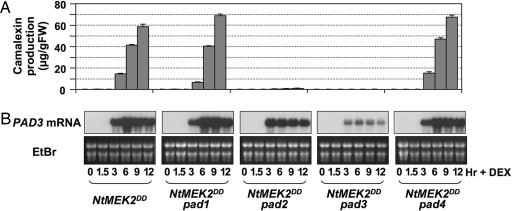
To determine the genetic relationships between the MPK3/MPK6 cascade and PAD genes, we crossed GVG-NtMEK2 DD into pad1, pad2, pad3, and pad4 backgrounds. Double-homozygous plants were analyzed for the production of camalexin after DEX treatment. As shown in Fig. 4A, PAD2 and PAD3 were required for MPK3/MPK6-induced camalexin production, whereas PAD1 and PAD4 were not. In DEX-treated double mutants, NtMEK2DD induction and the activation of downstream MPK3/MPK6 were comparable to those in the GVG-NtMEK2 DD plants (Fig. S4). Consistent with previous reports (18, 20), we found that PAD2 and PAD3, but not PAD1 and PAD4, were required for _Botrytis_-induced camalexin biosynthesis (Fig. S5). These results suggest that PAD2 and PAD3 act downstream of the MPK3/MPK6 cascade in B. cinerea induction of camalexin, whereas PAD1 and PAD4 function in a pathway that leads to camalexin induction by other pathogens.
Fig. 4.
PAD2 and PAD3, but not PAD1 and PAD4, are required for camalexin induction in GVG-NtMEK2 DD plants. (A) Two-week-old seedlings homozygous for the GVG-NtMEK2 DD transgene and pad1, pad2, pad3, or pad4 were treated with 1 μM DEX, samples were taken at the indicated times, and camalexin levels in the culture medium were determined by fluorospectrometry. Bars represent means and standard deviations (n = 3). (B) Induction of PAD3 expression in pad mutants after MPK3/MPK6 activation, as determined by RNA blot analysis (Upper). Equal loading was confirmed by ethidium bromide (EtBr)-staining of the gel (Lower).
PAD3 expression was highly induced in GVG-NtMEK2 DD plants after DEX treatment. In mpk3 and mpk6 plants, PAD3 induction was reduced and delayed, correlating with the reduced and delayed induction of camalexin (Fig. 1A and Fig. S6). In GVG-NtMEK2 DD/pad3 seedlings, the accumulation of pad3 mutant transcript was drastically reduced (Fig. 4B), consistent with previous analysis of pad3 plants (19, 22). In GVG-NtMEK2 DD/pad1 and GVG-NtMEK2 DD/pad4 seedlings, PAD3 mRNA levels were similar to GVG-NtMEK2 DD (Fig. 4B), consistent with the conclusion that these two genes function either upstream or independent of the MPK3/MPK6 cascade. In GVG-NtMEK2 DD/pad2 seedlings, which only synthesized trace amounts of camalexin (Fig. 4A), induced expression of PAD3 was ≈3-fold lower than in GVG-NtMEK2 DD seedlings. Also, PAD3 expression peaked at 3 h and then decreased in contrast to the peak at 6 h in GVG-NtMEK2 DD seedlings. These results suggest that PAD2 might play a regulatory role downstream of the MPK3/MPK6 cascade.
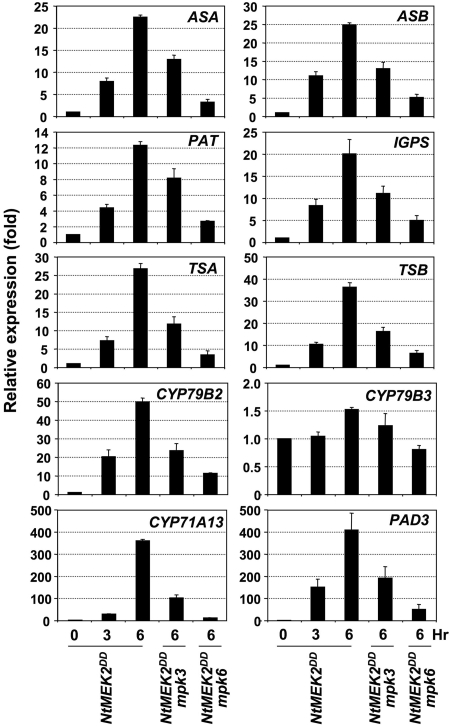
Camalexin is synthesized from tryptophan (Trp) via indole-3-acetaldoxime (IAOx), a key branching point between primary and secondary metabolism in Arabidopsis (34). Camalexin induction in Arabidopsis after pathogen infection is associated with the activation of genes encoding enzymes in the Trp biosynthetic pathway and P450 enzymes in the camalexin branch (21, 22). Camalexin induction by B. cinerea (Fig. 3B) is also associated with activation of these genes (Table S1). To determine whether camalexin induction in GVG-NtMEK2 DD plants is associated with the induction of these biosynthetic genes, we examined their expression using real-time qRT-PCR. As shown in Fig. 5, Trp biosynthetic genes encoding anthranilate synthase α and β subunits (ASA and ASB), phosphoribosylanthranilate transferase (PAT), indole-3-glycerolphosphate synthase (IGPS), and tryptophan synthase α and β subunits (TSA and TSB) were all highly induced. The relative levels of induction ranged from 12-fold for PAT to >35-fold for TSB within 6 h after DEX treatment.
Fig. 5.
Coordinated induction of Trp biosynthetic genes and cytochrome P450 genes in the camalexin pathway after MPK3/MPK6 activation. Total RNA was extracted from GVG-NtMEK2 DD seedlings before (0 h) and at 3 and 6 h after DEX treatment and from GVG-NtMEK2DD/mpk3 and GVG-NtMEK2DD/mpk6 seedlings 6 h after DEX treatment. After reverse transcription, the levels of each gene were determined by real-time qRT-PCR analysis. The comparative Ct method was used to calculate the levels of transcripts relative to that in GVG-NtMEK2 DD plants without DEX treatment (0 h), which was set at 1. Levels of _EF1_α transcript were used to normalize different samples. Bars represent means and standard deviations (n = 3).
The P450 enzymes showed more dramatic increases in gene expression (Fig. 5). Of the two P450 enzymes that catalyze the conversion of Trp to IAOx (34), CYP79B2 expression was highly induced (≈50-fold), whereas the expression of CYP79B3 showed minimal increase. In addition to CYP71B15 (PAD3), CYP71A13 was recently found to be in the camalexin biosynthetic pathway and is also pathogen inducible (35). Within 6 h of DEX treatment, CYP71A13 and CYP71B15 (PAD3) expression increased by >350- and 400-fold, respectively. High-level PAD3 induction detected by using qRT-PCR is consistent with RNA blot results (Fig. 4B and Fig. S6). The induction of all of these genes was partially compromised in GVG-NtMEK2DD/mpk3 and GVG-NtMEK2DD/mpk6 plants, consistent with the conclusion that the transgenic MAPKK functions through the redundant downstream MPK3 and MPK6. Based on these results, we conclude that the MPK3/MPK6 cascade coordinates the induction of multiple genes in the Trp pathway, the conversion of Trp to IAOx, and the camalexin biosynthetic pathway, thereby driving metabolic flow to camalexin synthesis (Fig. 6).
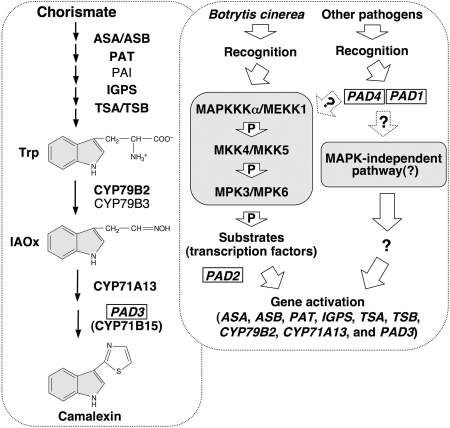
Fig. 6.
A model of the role of the MAPKKKα/MEKK1–MKK4/MKK5–MPK3/MPK6 cascade in regulating camalexin biosynthesis in plants challenged by pathogens. A simplified camalexin biosynthetic pathway and its regulatory pathway are placed in separate rectangular boxes with dashed outlines. Genes identified by genetic screens (PAD1 to PAD4) are boxed. Enzymes whose encoding genes are induced by pathogen infection and MPK3/MPK6 activation are marked by bold font. One arrow may represent multiple steps because of unknown components.
Discussion
Production of phytoalexins in plants after pathogen invasion is an integral part of induced plant disease resistance. The biosynthetic pathways of a number of phytoalexins have been defined (15, 16). However, the signaling pathways that regulate their biosynthesis are largely unknown. In this article, we have demonstrated that the Arabidopsis MAPKKKα/MEKK1-MKK4/MKK5-MPK3/MPK6 cascade is an important regulatory pathway controlling camalexin biosynthesis in Arabidopsis.
Pathogen-induced phytoalexin production is associated with the activation of multiple genes encoding enzymes in the biosynthetic pathway (15, 16). Camalexin induction in GVG-NtMEK2 DD plants and in wild-type plants infected with pathogens including B. cinerea is associated with up-regulation of multiple genes in the camalexin biosynthetic pathway, which can drive the metabolic flow from primary metabolism to the formation of camalexin (refs. 21, 22, and 34, Fig. 5, and Table S1). As depicted in our model (Fig. 6), the coordinated up-regulation of multiple biosynthetic genes in Trp biosynthesis and P450s in the IAOx and camalexin pathways is likely a result of activation of an unidentified transcription factor(s), which may be a substrate(s) of MPK3/MPK6. In animal and yeast systems, stress-responsive MAPKs regulate gene expression by direct phosphorylation of transcription factors (5). We could not identify a simple shared _cis_-element motif in the promoters of these biosynthetic genes, suggesting the involvement of multiple _cis_- and _trans_-acting factors, which is consistent with the current knowledge that most gene regulation is mediated by composite elements.
The effectors and PAMPs in B. cinerea and the host receptors in Arabidopsis involved in triggering MPK3/MPK6 activation and camalexin induction are unknown. Known fungal PAMPs such as chitin, cell wall glucans, and oligogalacturonides activate MPK3/MPK6 only transiently and do not induce camalexin (refs. 36 and 37 and data not shown). The strong and long lasting activation of MPK3/MPK6 after B. cinerea infection (Fig. 3A) suggests the involvement of additional effectors and host sensors. It is known that B. cinerea produces multiple effectors, and plant resistance is controlled by multiple quantitative trait loci (38).
Based on genetic analysis, we place PAD2 and PAD3 downstream of the MPK3/MPK6 cascade in camalexin regulatory or biosynthetic pathways (Fig. 6). PAD3 is an essential enzyme in the camalexin biosynthetic pathway (22, 32) (Fig. 4). PAD2 encodes γ-glutamylcysteine synthetase (GSH1) (33). We found that supplementing the GVG-NtMEK2 DD/pad2 seedlings with glutathione up to 1 mM failed to restore camalexin production (data not shown), suggesting that PAD2/GSH1 has an additional function besides its role in glutathione biosynthesis. It is also possible that the exogenously added glutathione could not mimic the right concentration or location of endogenously produced glutathione. PAD4 is required for bacterial, but not fungal, induction of camalexin (20, 30). We found that PAD4 is not required for camalexin induction in GVG-NtMEK2 DD plants, suggesting that PAD4 functions in a pathogen-specific pathway either upstream of the MPK3/MPK6 cascade or in a parallel signaling pathway (Fig. 6). PAD1, which remains to be identified, may act similarly.
Camalexin is essential for resistance to B. cinerea in Arabidopsis (19). In mpk3 plants, camalexin accumulation after B. cinerea infection was reduced by ≈50%, which may explain the compromised resistance in mpk3 plants (Fig. 3). In mpk6 plants, camalexin levels were reduced only at early time points. In the mpk3/mpk6 double mutant, camalexin induction was almost abolished. These results suggest that MPK3 and MPK6 play redundant, yet differential, roles in regulating camalexin biosynthesis, with MPK6 being more important early, and MPK3 being more important late. This speculation is consistent with the observations that mpk6 has a stronger effect on gene expression at early time points and a weaker effect on later camalexin levels. At later times, mpk3 plants continue to have reduced gene expression, whereas gene expression levels in mpk6 plants approach wild-type levels (Fig. S6). The partially rescued double mpk3/mpk6 mutant seedlings are arrested at the cotyledon stage and cannot survive in soil (28), prohibiting testing for B. cinerea resistance.
The establishment of MEKK1/MAPKKKα and MKK4/MKK5 as the upstream kinases in the MPK3/MPK6 cascade in regulation of camalexin biosynthesis is based mainly on biochemical and gain-of-function genetic evidence. The gain-of-function MAPKKs used in this study have only two Ser/Thr residues mutated to Asp. Such mutant proteins are likely to maintain their specificity in vivo because the kinase-interacting domain, which is involved in recognizing the downstream MAPKs, is still intact (26). The absence of activation of other MAPKs in GVG-NtMEK2 DD, GVG-MKK4 DD, and GVG-MKK5 DD plants supports this notion (12) (Fig. 1B). The gain-of-function MAPKKKs used in this and other reports lack the regulatory domain (13, 14), which may lead to nonspecific activation of downstream components in other MAPK cascades. Despite this concern, the correlation between MPK3/MPK6 activation and camalexin production in independent transgenic lines is strong (Fig. 2). The fact that MPK4 is activated in GVG_-Δ_MEKK1, but not _GVG_-Δ_MAPKKK_α, plants suggests that the active MAPKKK mutants maintain some specificity in vivo. Recent genetic analysis of mekk1 mutant demonstrated that MPK4 is indeed downstream of MEKK1 (27). The use of a conditional promoter in this study also reduces the potential secondary nonspecific effects of the gain-of-function mutants. More importantly, loss of MPK3 or/and MPK6 compromised camalexin induction after fungal infection (Fig. 3), providing loss-of-function evidence implicating this MAPK cascade in pathogen-induced camalexin induction.
The MPK3/MPK6 cascade is involved in regulating ethylene biosynthesis in plants under stress (39). Pathogen-induced production of camalexin exhibits no cross-talk with the ethylene pathway (18). In agreement with this, we found that camalexin is still highly induced in GVG-NtMEK2DD/etr1–1, GVG-NtMEK2DD/ein2, and GVG-NtMEK2DD/ein3 double mutants (Fig. S7), suggesting that camalexin induction after MPK3/MPK6 activation is independent of ethylene. Conversely, the loss of camalexin induction in the GVG-NtMEK2 DD/pad plants does not affect ethylene induction (data not shown). Induction of constitutively active NtMEK2 or MKK4/MKK5 also leads to cell death after 24 h (11, 12, 26). The activation of genes in the Trp and camalexin pathways occurs within 3 h (Figs. 4 and 5), long before cell death occurs. As a result, we conclude that camalexin production is not a secondary effect of cell death. We can also conclude that cell death is independent of camalexin production because cell death was not inhibited in camalexin-deficient GVG-NtMEK2 DD/pad2 and GVG-NtMEK2 DD/pad3 double mutants (Fig. S8).
Based on these analyses, we conclude that the MPK3/MPK6 cascade is capable of regulating multiple defense responses. In addition to the defense gene induction, ethylene biosynthesis, and HR-like cell death reported previously, we have now provided gain- and loss-of-function evidence supporting the regulatory role of this cascade in phytoalexin induction, another important defense response. The combined effect of these multiple downstream responses ultimately determines the role of this MAPK cascade in disease resistance.
Materials and Methods
Plant Growth and Treatments.
A. thaliana wild type (Col-0), mutants, and transgenics were maintained at 22°C in a growth chamber with a 14-hr light cycle (100 μE/m−2sec−1). For B. cinerea resistance tests, plants were grown under a 12-hr light cycle. Seedlings were grown in 50-ml GC vials with 6 ml of half-strength Murashige and Skoog medium in a growth chamber at 22°C under continuous light (70 μE/m−2sec−1). Unless indicated otherwise, 2-week-old seedlings were used for experiments. Seedlings collected at various times after the addition of DEX (1 μM) or inoculation of B. cinerea spores (4 × 105 spores per vial) were frozen in liquid nitrogen and stored at −80°C until use. B. cinerea maintenance and spore preparation were as described (20). Detailed procedures for seedling inoculation and visualization of fungal structures are given in SI Text.
At least two independent repetitions were performed for experiments with multiple time points. For single time-point experiments, at least three independent repetitions were done.
_Agrobacterium_-Mediated Transformation.
Constitutively active MEKK1 and MAPKKKα deletion mutants (13, 14) with an NcoI site added to introduce an ATG codon were first cloned in-frame into an intermediate vector with a double HA-epitope tag at the 3′ end. The inserts were then moved into binary vector pTA7002. Transgenic Arabidopsis plants were generated and selected as described (39).
Generation of Double Mutants.
mpk3 and mpk6 mutants were described (28, 39). Homozygous F3 or F4 plants from various crosses were used for experiments. Transgenes were followed by hygromycin resistance. CAPS markers were used to follow mutations in PAD3 (pad3–1) and PAD4 (pad4–1) (22, 30). Homozygous pad1 (pad1–1) was identified by a recessive leaf phenotype associated with the mutation (24). Homozygous pad2 (pad2–1) plants were identified based on reduced camalexin induction after Pseudomonas syringae infection (25, 33).
Camalexin Measurement.
Camalexin was extracted from Arabidopsis seedlings and quantified by using established procedures (23, 24). The excitation and emission spectra were determined by using a Model 8100 spectrofluorometer (SLM-AMINCO Instrument). Camalexin accumulation in the culture medium, which reflects its production and accumulation in the seedlings, was determined by fluorospectrometry with a standard curve established by using known concentrations of camalexin.
Protein Extraction, Immunoblot Analysis, and In-Gel Kinase Assay.
Protein was extracted from seedlings as described (39). The concentration of protein extracts was determined by using the Bio-Rad protein assay kit with BSA as the standard. Immunoblot detection of tagged transgene products was performed as described (39). Myelin basic protein (MBP) was used as the substrate for the in-gel kinase assay (39, 40).
RNA Blot and Quantitative (q)RT-PCR Analyses.
Total RNA was extracted by using TRIzol reagent (Invitrogen). RNA (5 μg per lane) was separated on 1.2% formaldehyde–agarose gels, transferred to an Immobilon-Ny+ membrane (Millipore), and hybridized with random primer-labeled cDNA insert as described (11). Relative abundance of PAD3 mRNA was determined by using the NIH Image program. Equal sample loading was confirmed by ethidium bromide staining of the gel.
After TURBO DNase treatment, 2 μg of total RNA were used for reverse transcription. Quantitative PCR analysis was performed by using an Optican 2 real-time PCR machine (MJ Research) as described (26, 39). After normalization to an _EF1_α control, the relative levels of gene expression were calculated. Primer pairs are listed in SI Text.
Acknowledgments.
We thank N.-H. Chua for providing the pTA7002 vector and the Arabidopsis Biological Resource Center for seed stocks. This work was supported by National Basic Research Program of China Grant 2003CB114304 and National Natural Science Foundation of China Grants 30421002 and 30770203 (to D.R.), National Science Foundation (NSF) Arabidopsis Grant 2010 IBN-0419648 (to J.G.), and NSF Grants IBN-0133220 and MCB-0543109 (to S.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Boller T. Peptide signalling in plant development and self/non-self perception. Cur Opin Cell Biol. 2005;17:116–122. doi: 10.1016/j.ceb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 3.Dangl JL, Jones JDG. Plant pathogens and integrated defense responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 4.Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JDG. Molecular genetics of plant disease resistance. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 5.Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 6.Zhang S, Klessig DF. MAPK cascades in plant defense signaling. Trends Plants Sci. 2001;6:520–527. doi: 10.1016/s1360-1385(01)02103-3. [DOI] [PubMed] [Google Scholar]
- 7.Tena G, Asai T, Chiu W-L, Sheen J. Plant mitogen-activated protein kinase signaling cascades. Cur Opin Plant Biol. 2001;4:392–400. doi: 10.1016/s1369-5266(00)00191-6. [DOI] [PubMed] [Google Scholar]
- 8.Pedley KF, Martin GB. Role of mitogen-activated protein kinases in plant immunity. Cur Opin Plant Biol. 2005;8:541–547. doi: 10.1016/j.pbi.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Nakagami H, Pitzschke A, Hirt H. Emerging map kinase pathways in plant stress signalling. Trends Plants Sci. 2005;10:339–346. doi: 10.1016/j.tplants.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Ichimura K, et al. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plants Sci. 2002;7:301–308. doi: 10.1016/s1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- 11.Yang K-Y, Liu Y, Zhang S. Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA. 2001;98:741–746. doi: 10.1073/pnas.98.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren D, Yang H, Zhang S. Cell death mediated by mitogen-activated protein kinase pathway is associated with the generation of hydrogen peroxide in Arabidopsis. J Biol Chem. 2002;277:559–565. doi: 10.1074/jbc.M109495200. [DOI] [PubMed] [Google Scholar]
- 13.Asai T, et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 14.del Pozo O, Pedley KF, Martin GB. MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 2004;23:3072–3082. doi: 10.1038/sj.emboj.7600283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon RA. Natural products and plant disease resistance. Nature. 2001;411:843–847. doi: 10.1038/35081178. [DOI] [PubMed] [Google Scholar]
- 16.Hammerschmidt R. Phytoalexins: What have we learned after 60 years? Ann Rev Phytopathol. 1999;37:285–306. doi: 10.1146/annurev.phyto.37.1.285. [DOI] [PubMed] [Google Scholar]
- 17.Morrissey JP, Osbourn AE. Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiol Mol Biol Rev. 1999;63:708–724. doi: 10.1128/mmbr.63.3.708-724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomma BPHJ, Nelissen I, Eggermont K, Broekaert WF. Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 1999;19:163–171. doi: 10.1046/j.1365-313x.1999.00513.x. [DOI] [PubMed] [Google Scholar]
- 19.Ferrari S, et al. Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol. 2007;144:367–379. doi: 10.1104/pp.107.095596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrari S, Plotnikova JM, De Lorenzo G, Ausubel FM. Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires eds4 and pad2, but not sid2, eds5 or pad4. Plant J. 2003;35:193–205. doi: 10.1046/j.1365-313x.2003.01794.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhao J, Last RL. Coordinate regulation of the tryptophan biosynthetic pathway and indolic phytoalexin accumulation in Arabidopsis. Plant Cell. 1996;8:2235–2244. doi: 10.1105/tpc.8.12.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou N, Tootle TL, Glazebrook J. Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome p450 monooxygenase. Plant Cell. 1999;11:2419–2428. doi: 10.1105/tpc.11.12.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuji J, Jackson EP, Gage DA, Hammerschmidt R, Somerville SC. Phytoalexin accumulation in Arabidopsis thaliana during the hypersensitive reaction to Pseudomonas syringae pv syringae. Plant Physiol. 1992;98:1304–1309. doi: 10.1104/pp.98.4.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glazebrook J, Ausubel FM. Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA. 1994;91:8955–8959. doi: 10.1073/pnas.91.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glazebrook J, et al. Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics. 1997;146:381–392. doi: 10.1093/genetics/146.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin H, et al. Function of a mitogen-activated protein kinase pathway in N-gene mediated resistance in tobacco. Plant J. 2003;33:719–731. doi: 10.1046/j.1365-313x.2003.01664.x. [DOI] [PubMed] [Google Scholar]
- 27.Suarez-Rodriguez MC, et al. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 2007;143:661–669. doi: 10.1104/pp.106.091389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. Stomatal development and patterning are regulated by environmentally responsive map kinases in Arabidopsis. Plant Cell. 2007;19:63–73. doi: 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen M, et al. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/s0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- 30.Jirage D, et al. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA. 1999;96:13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glazebrook J, et al. Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 2003;34:217–228. doi: 10.1046/j.1365-313x.2003.01717.x. [DOI] [PubMed] [Google Scholar]
- 32.Schuhegger R, et al. CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 2006;141:1248–1254. doi: 10.1104/pp.106.082024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parisy V, et al. Identification of PAD2 as a γ-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 2007;49:159–172. doi: 10.1111/j.1365-313X.2006.02938.x. [DOI] [PubMed] [Google Scholar]
- 34.Glawischnig E, Hansen BG, Olsen CE, Halkier BA. Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc Natl Acad Sci USA. 2004;101:8245–8250. doi: 10.1073/pnas.0305876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nafisi M, et al. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell. 2007;19:2039–2052. doi: 10.1105/tpc.107.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan J, Zhang S, Stacey G. Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol Plant Pathol. 2004;5:125–135. doi: 10.1111/j.1364-3703.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 37.Nühse T, Peck SC, Hirt H, Boller T. Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK6. J Biol Chem. 2000;275:7521–7526. doi: 10.1074/jbc.275.11.7521. [DOI] [PubMed] [Google Scholar]
- 38.Williamson B, Tudzynski B, Tudzynski P, Van Kan JAL. Botrytis cinerea: the cause of grey mould disease. Mol Plant Pathol. 2007;8:561–580. doi: 10.1111/j.1364-3703.2007.00417.x. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Zhang S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by mpk6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell. 2004;16:3386–3399. doi: 10.1105/tpc.104.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang S, Klessig DF. Salicylic acid activates a 48 kD MAP kinase in tobacco. Plant Cell. 1997;9:809–824. doi: 10.1105/tpc.9.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]