G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription (original) (raw)
Abstract
Methylation of DNA and lysine 9 of histone H3 (H3K9) are well-conserved epigenetic marks for transcriptional silencing. Although H3K9 methylation directs DNA methylation in filamentous fungi and plants, this pathway has not been corroborated in mammals. G9a and GLP/Eu-HMTase1 are two-related mammalian lysine methyltransferases and a G9a/GLP heteromeric complex regulates H3K9 methylation of euchromatin. To elucidate the function of G9a/GLP-mediated H3K9 methylation in the regulation of DNA methylation and transcriptional silencing, we characterized ES cells expressing catalytically inactive mutants of G9a and/or GLP. Interestingly, in ES cells expressing a G9a-mutant/GLP complex that does not rescue global H3K9 methylation, G9a/GLP-target genes remain silent. The CpG sites of the promoter regions of these genes were hypermethylated in such mutant ES cells, but hypomethylated in G9a- or _GLP_-KO ES cells. Treatment with a DNA methyltransferase inhibitor reactivates these G9a/GLP-target genes in ES cells expressing catalytically inactive G9a/GLP proteins, but not the wild-type proteins. This is the first clear evidence that G9a/GLP suppresses transcription by independently inducing both H3K9 and DNA methylation.
Keywords: DNA methylation, G9a, GLP, histone lysine methylation
Introduction
Genetic information of eukaryotes is stored as chromatin, which consists of genomic DNA, histones and a wide array of chromosomal proteins. Chromatin templates are subjected to various post-translational modifications to regulate many distinct biological functions (Strahl and Allis, 2000; Margueron et al, 2005). Among them, methylation of lysine 9 of histone H3 (H3K9) is a well-conserved epigenetic mark for heterochromatin formation and transcriptional silencing (Jenuwein, 2006). Methylation of H3K9 creates a binding site for the heterochromatin protein 1 (HP1) family molecules (Bannister et al, 2001; Lachner et al, 2001). Although recruitment of HP1 may be involved in heterochromatin formation, it is not well known how H3K9 methylation directs transcriptional silencing.
Methylation of genomic DNA is another crucial epigenetic mark for transcriptionally silent chromatin (Bird, 2002). Methylation of CpG sites regulates transcription through recruitment of the methyl CpG binding molecules or suppressing DNA binding of various transcriptional regulators such as CTCF (Wade, 2001). Furthermore, the hemimethylated CpG binding molecule(s) is important for the maintenance of DNA methylation though replication (Bostick et al, 2007; Sharif et al, 2007).
It is known that H3K9 methylation directs DNA methylation in filamentous fungi (Tamaru and Selker, 2001) and plants (Jackson et al, 2002). In Neurospora crassa, H3K9 methylation catalysed by HMTase DIM-5 directs all cytosine methylation. Similarly, in Arabidopsis thaliana, HMTase KRY/SUV4 catalyses H3K9 methylation and this epigenetic mark directs CMT3-mediated non-CpG methylation. However, the interplay between these epigentic marks in mammals is not well understood.
G9a and GLP/Eu-HMTase1 are two related mammalian lysine methyltransferases, both of which are crucial for H3K9 methylation (mainly mono- and di-methylation, me1 and 2) of euchromatin (Tachibana et al, 2002, 2005). Although G9a and GLP can form a homo- or heteromeric complex through their SET-domain interaction if they are transiently expressed, endogenous G9a and GLP mostly exist as G9a/GLP heteromer (Tachibana et al, 2005). Genetic analyses revealed that both G9a and GLP are essential for global H3K9me1 and 2 ((Tachibana et al, 2005) and Supplementary Figure S1). These findings indicate that the G9a/GLP heteromeric complex is the only functional form for global H3K9 methylation in vivo, even though recombinant G9a or GLP can independently methylate H3K9 in vitro (Tachibana et al, 2001, 2005).
RNA expression microarray analyses revealed that an overlapping set of genes are aberrantly expressed in G9a- and _GLP_-KO ES cells, indicating that G9a/GLP-mediated H3K9 methylation is involved in transcriptional silencing. Indeed, H3K9 methylation on the promoter region of one such G9a/GLP-target gene, Mage-a2, is significantly diminished in G9a- and _GLP_-KO ES cells (Tachibana et al, 2002, 2005). Furthermore, some studies suggest a functional interaction between G9a-mediated H3K9 methylation and DNA methylation (Xin et al, 2003; Estève et al, 2006; Feldman et al, 2006; Ikegami et al, 2007). We have also found that methylation of CpG sites of the Mage-a2 promoter region are regulated by G9a. These sites are hypomethylated in G9a- and _GLP-_KO ES cells, but hypermethylated in wild-type and transgene-complemented G9a- or _GLP_-KO ES cells (Figure 3B).
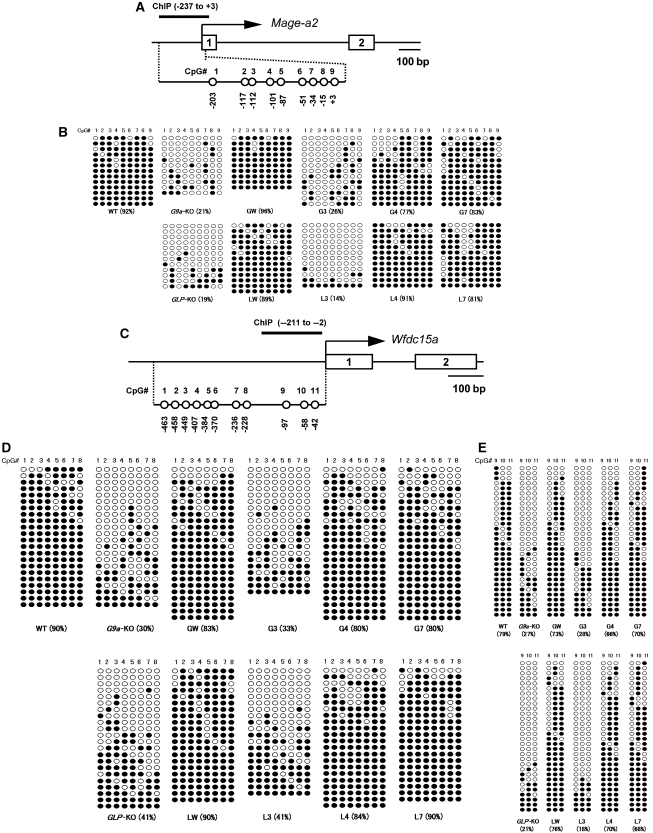
Figure 3.
G9a/GLP complex formation induces DNA methylation of the Mage-a2 promoter and the 5′-upstream region of the Wfdc15a gene. (A) A schematic diagram of CpG positions and amplicon for ChIP analysis within Mage-a2 promoter is shown. DNA methylation status of eight CpG sequences (no. 1-8) within the promoter (0 to −287) and a CpG (no. 9) located at the 5′ end of the transcribed region were examined in this study. (B) Methylation differences among serial ES lines expressing mutant G9a/GLP complex at the Mage-a2 promoter. DNA methylation status of CpG positions nos. 1–9 shown in (A) was measured by bisulphite sequencing. Open circles indicate unmethylated cytosines and filled circles indicate methylated cytosines. (C) Schematic diagram of CpG positions and amplicon for ChIP analysis within the 5′-upstream region of Wfdc15a. (D, E) Methylation differences among serial ES lines expressing mutant G9a/GLP complex at the position no. 1–8 (D) and no. 9–11 (E) of CpG sequences on the 5′-upstream region of Wfdc15a.
To further elucidate how G9a/GLP-mediated H3K9 methylation is crucial for the regulation of transcriptional silencing and DNA methylation, we established several ES cell lines that express only catalytically inactive mutants of G9a or/and GLP.
Results
Establishment of ES cells expressing G9a-mutant/GLP complexes that cannot catalyse H3K9 methylation in vivo
To elucidate whether G9a/GLP-mediated H3K9 methylation is crucial for transcriptional silencing and DNA methylation of target genes, we wished to establish ES cell lines that express only catalytically inactive mutants of G9a or/and GLP. To establish such ES cell lines, we screened amino-acid substitution or deletion mutants of G9a and GLP SET-domains for their histone methyltransferase (HMTase) activities in vitro. On the basis of the crystal structure of another H3K9 HMTase, DIM-5 (Zhang et al, 2002), we introduced four different mutations in both G9a and GLP SET-domains (Figure 1A and B). In vitro methyltransferase assays using recombinant histone H3/H4 and glutathione _S_-transferase fused to the SET-domains of G9a or GLP showed that each of the G9a and GLP SET-domain mutants were severely impaired for their HMTase activities (<0.1–0.5% catalytic activities of wild-type G9a or GLP SET-domains)(Figure 1C).
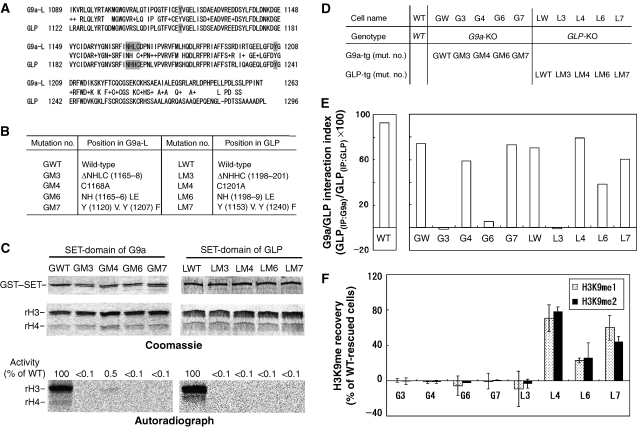
Figure 1.
Reconstitution of G9a/GLP complex carrying mutations in their SET-domains. (A) Amino-acid alignment of the SET-domains of mG9a-L (Genbank, 21832048) and mGLP (Genbank, 60115440). Conserved and similar residues (shown as +) are shown between the two sequences. Mutagenized amino acids are shown in grey boxes. (B) Table showing serial mutants of G9a and GLP. (C) In vitro histone methyltransferase activity of mutant SET-domains of G9a (left) and GLP (right). GST-fused carboxy terminus of mouse G9a (969-1263 of G9a-L) and that of mouse GLP (1002–1296) were incubated with recombinant histone H3/H4 and _S_-adenosyl-[_methyl_-14C]-L-methionine, and then separated by SDS–PAGE. Coomassie staining shows enzymes and substrates. Autoradiography shows 14C-methyl group incorporated into recombinant H3. Activities of mutant enzymes were quantified using image J (http://rsb.info.nih.gov/ij/) and those of GWT or LWT are normalized to 100%. (D) List of ES cell lines expressing mutant G9a/GLP complexes. Serial cell lines were established by stable introduction of the expression plasmids for full-length of G9a-L or GLP into the corresponding KO cells shown in Supplementary Figure S1. Expression levels of mutant G9a or GLP proteins are shown in Supplementary Figure S2. (E) The ability of serial mutant proteins to form heterodimers are numerically represented as G9a/GLP interaction index. Protein levels within anti-G9a or anti-GLP immune complexes shown in Supplementary Figure S3 were measured by image J software, and the ratio of GLP protein immunoprecipitated with anti-G9a to those immunoprecipitated with anti-GLP were calculated. The interaction index of wild-type (WT) ES cells (top left panel) is ∼100%, indicating that endogenous G9a and GLP form a stoichiometric (1:1) heteromer in wild-type cells. (F) H3K9 methylation status of serial mutant ES lines were measured by immunoblot analysis and numerically represented as H3K9me recovery (Raw data are presented in Supplementary Figure S4). The values of signal intensities for methylated H3K9 of GW and LW are normalized to 100%, and those of the corresponding KO cells are normalized to 0%. Data represents an average value of the H3K9 recovery of two clones established independently.
We introduced full-length G9a or GLP mutants harbouring one of these mutations (GM3, 4, 6, 7 and LM3, 4, 6, 7) into _G9a_-KO or _GLP_-KO ES cells, respectively (listed in Figure 1D). Whereas the LM3 or LM6 GLP mutants were poorly expressed (∼20% of wt GLP level), the remaining mutants were expressed at levels similar to endogenous G9a or GLP (Supplementary Figure S2). As G9a/GLP heteromeric formation is essential for H3K9 methyltransferase activities in vivo, we examined the ability of these mutants to form such heteromeric complexes. As shown in Figure 1E and Supplementary Figure S3, G9a and GLP mutants that possess mutation GM3, GM6 or LM3 do not form heteromers. In contrast, the G9a or GLP mutants GM4, GM7, LM4, LM6 or LM7 do form heteromers with their partner molecules. These findings indicate that the NHXC motifs of G9a and GLP SET-domains are essential for both HMTase activity and complex formation, but these two functions are separable.
We next examined the effects of these mutations on H3K9 methyltransferase activity in vivo (summarized in Figure 1F). As described previously and shown in Supplementary Figure S4, global H3K9me2 and me1 are dramatically impaired in _G9a_- and _GLP-_KO ES cells and completely restored by stable expression of exogenous wild-type proteins (Tachibana et al, 2002, 2005). Among the ES cell lines expressing each G9a or GLP mutant, H3K9 methylation was not recovered in G3, G6 and L3 ES cells, in which mutant G9a or GLP could not form a heteromer (see Figure 1E), confirming that the formation of the G9a/GLP heteromeric complex is essential for their in vivo H3K9 methylation. Interestingly, while L4 and L7 ES cells, which express GLP mutants LM4 and LM7, respectively, recovered global H3K9me1 and 2, such methylation was not rescued in G4 and G7 ES cells in which G9a mutants containing GM4 and GM7, respectively, were expressed. As shown in Figure 1E, these G9a and GLP mutants did form heteromeric complexes. Therefore, these data suggest that catalytic activity of GLP but not G9a is dispensable for their H3K9 methyltransferase activities in vivo.
Silencing of G9a/GLP-target genes is maintained in ES cells expressing catalytically inactive G9a/GLP complexes
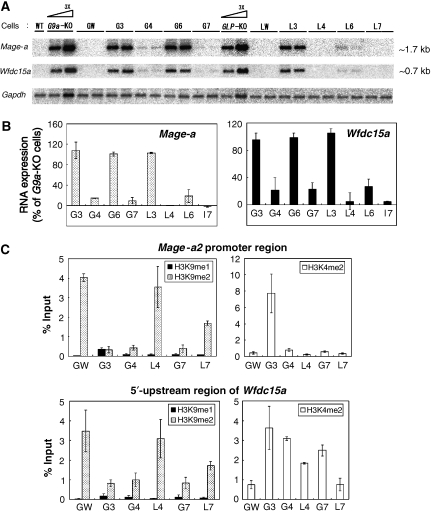
Although it is not clear why catalytic activity of GLP is dispensable for H3K9 methylation in vivo, we generated ES cells that express catalytically inactive G9a/GLP heteromeric complexes that do not rescue global H3K9me1 and 2 to elucidate the impact of such mutant complexes on transcriptional silencing. We examined the expression of two of G9a-target genes, Mage-a and Wfdc15a, shown previously to be targets of G9a and GLP. The Mage-a gene, a homologue of one of the cancer-testis antigen genes, is located on the X chromosome (De Plaen et al, 1999). The Wfdc15a gene is located on mouse chromosome 2 (2 H3). As shown in Figure 2A and B and Supplementary Figure S1, both genes are reactivated in G9a- or _GLP_-KO ES cells. Moreover, stable expression of exogenous WT genes in the corresponding KO ES cells reestablished transcriptional silencing of these genes (Figure 2A, lanes GW and LW). These observations confirm that transcripts of Mage-a and Wfdc15a are negatively regulated by the G9a/GLP heteromeric complex. Neither transcript was suppressed in the newly established ES cell lines G3, G6 and L3, indicating again that the formation of G9a/GLP heteromeric complex is necessary for their transcriptional silencing function. However, unexpectedly, G9a/GLP complex-mediated gene suppression was maintained in G4 and G7 ES cells that do not rescue global H3K9me1 and 2 (compare Figures 1F and 2B). To determine whether H3K9 methylation is affected in the promoter region of Mage-a2 and the 5′-upstream region of Wfdc15a in these cells, we carried out chromatin immunoprecipitation (ChIP) analysis in GW, G3, G4, G7, L4 and L7 cells (Figure 2C). Although H3K9 mono- and di-methylation levels at both loci were depleted in the G4 and G7 lines, to a level similar to that observed for the G3 line, these marks were still present in L4 and L7. Profiles of H3K4me2 on the Mage-a2 promoter in these mutant lines were also examined by ChIP analysis (Figure 2C, top). Among them, only G3 cells showed increased H3K4me2 levels, consistent with the transcriptional activity of the Mage-a gene. In contrast, the levels of H3K4me2 on the 5′-upstream region of Wfdc15a have less correlation with transcriptional competency compared with those of Mage-a2 (Figure 2C, bottom). Collectively, these data clearly suggest the existence of a new mechanism of G9a/GLP complex-mediated transcriptional silencing, which occurs independent of H3K9 methylation or HMTase activity. Consistent with our findings, treatment of the G9a-specific inhibitor, BIX-01294, only very poorly reactivated G9a-target genes in murine ES cells (Kubicek et al, 2007).
Figure 2.
Effect of G9a or GLP mutations on transcriptional suppression. (A) Northern blot analyses of Mage-a and Wfdc15a genes in ES cells expressing mutant G9a/GLP are shown. Total RNA collected from two independent clones were separated and probed with radiolabelled cDNAs. (B) Signal intensities for the transcripts of Mage-a (left) and Wfdc15a (right) were calculated by image J software and numerically represented. The value for _G9a-_KO cells was set at 100%. Data represents an average value between two independent clones. (C) Differences in the histone modification patterns on the Mage-a2 promoter region (top) and 5′-upstream region of Wfdc15a (bottom) in the serial ES cell lines were measured by chromatin immunoprecipitation (ChIP) analysis using anti-H3K9me1 and me2 (left) and H3K4me2 (right). Each experiment was performed twice.
DNA methylation of the G9a/GLP-target promoter regions is maintained in ES cells expressing catalytically inactive G9a/GLP complexes
We next examined the status of CpG methylation on the promoter and 5′-upstream region of G9a/GLP-target genes. We analysed the status of eight CpG sites within the promoter region (0 to −287) and the first CpG site within exon one of the Mage-a2 gene (Figure 3A) and eleven CpG sites in the 5′-upstream of Wfdc15a (Figure 3C) by bisulfite sequencing analysis. DNA methylation status of the Mage-a2 promoter region in the ES lines analysed is summarized in Figure 3B. As described above, all nine CpG sites are heavily methylated in WT cells, whereas those of _G9a_- or _GLP_-KO cells are hypomethylated. Furthermore, those hypomethylation phenotypes can be rescued by introduction of the exogenous corresponding HMTases (see panels _G9a-_KO, _GLP-_KO, GW and LW). Such restoration of DNA methylation was not induced in G3 and L3 cells in which the mutant G9a or GLP does not form a heteromeric complex with endogenous partner. Most importantly, these CpG sites on the _Mage-a_2 promoter region were significantly methylated in G4 and G7 cells in which both global and specific H3K9 methylation were not restored, but the G9a/GLP-target genes were nevertheless silent. The pattern of DNA methylation in the 5′-upstream region of Wfdc15a was similar to that of the Mage-a2 promoter region in the ES lines examined—that is, hypermethylated in WT, GW, LW, G4, L4, G7 and L7 cells, but hypomethylated in _G9a_-KO, _GLP_-KO, G3 and L3 ES cells (Figure 3D and E). Protein amounts of Dnmt1, 3a and 3b were not reduced in _G9a_- or _GLP_-KO and several ES cell lines expressing mutant G9a/GLP (Supplementary Figure S2), suggesting that the DNA hypomethylation phenotype was not induced by an intrinsic reduction of DNA methylation machineries. To summarize, these data strongly suggest that the G9a/GLP complex regulates DNA methylation independent of the HMTase activities of these enzymes.
Inhibition of DNA methylation reactivates the G9a/GLP-target genes specifically in the ES cells expressing catalytically inactive G9a/GLP complexes
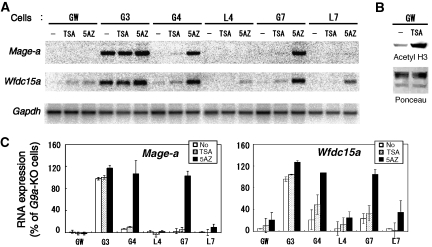
To further elucidate whether the G9a/GLP complex suppresses target genes through the combination of H3K9 and DNA methylation, we cultured ES cells expressing wt or mutant G9a or GLP in the presence of the DNA methyltransferase inhibitor, 5-Aza-2′-deoxycytidine (5-Aza-dC) or the histone deacetylase inhibitor, tricostatin-A (TSA). As shown in Figure 4A (and summarized in Figure 4C), TSA treatment had no or very small effects on transcriptional activation of Mage-a and Wfdc15a genes in the examined ES cell lines, including GW, G3, G4, L4, G7 and L7, although global H3 acetylation was drastically increased (Figure 4B). On the other hand, 5′Aza-dC treatment significantly reactivated both genes in G4 and G7 cells (Figure 4A and C) to levels similar to those seen in G9a- or _GLP-_KO ES cells without 5′Aza-dC treatment (Figure 2A and data not shown). In contrast, these G9a/GLP-negative target genes were still silent or very mildly reactivated in GW, L4 and L7 cells after the same 5′Aza-dC treatment. We examined the CpG methylation status of G9a-target genes in the 5′Aza-dC-treated cell lines using bisulphite sequencing. As shown in Supplementary Figure S5 (and in comparison with Figure 3B–E), CpG methylation was significantly reduced in all 5′Aza-dC-treated cells, indicating that transcriptional reactivation in G4 and G7 cells was induced by the reduction of DNA methylation. These serial experiments strongly support that the G9a/GLP heteromeric complex induces both H3K9 and DNA methylation on their target gene loci and that these two epigenetic marks cooperatively suppress their targets transcription.
Figure 4.
Inhibition of DNA methyltransferase activity reactivates G9a/GLP-target gene expression independent of H3K9 methylation. (A) Serial ES lines were cultured with histone deacetylase inhibitor, tricostatin-A (TSA) or the DNA methyltransferase inhibitor, 5-Aza-2′-deoxycytidine (5AZ). Total RNA from treated cells was isolated and analysed through northern blotting using radiolabelled cDNA. (B) The effect of TSA on H3 acetylation was confirmed by immunoblot analysis with specific antibodies directed against acetyl histone H3. Ponceau staining represents equal loading of histones. (C) Reactivation of the transcripts for Mage-a (left) and Wfdc15a (right) is represented numerically. The values for _G9a-_KO cells without drug treatment are normalized to 100%. Experiments were performed twice.
DNA methylation does not direct H3K9 methylation of the G9a/GLP-target genes
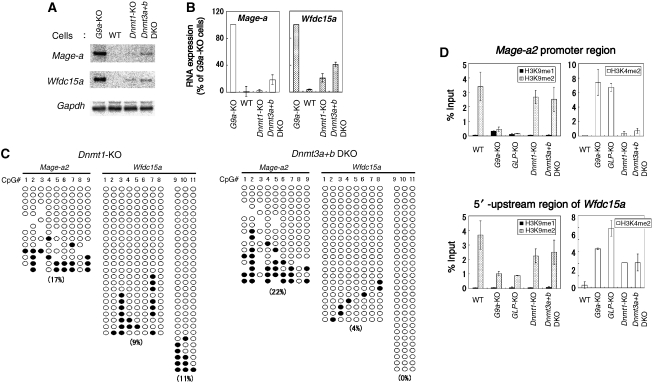
Bisulphite sequencing analysis clearly demonstrated that G9a/GLP-regulated DNA methylation is not directed by their HMTase activities or deposited H3K9 methylation, at least on some target gene loci. However, whether DNA methylation or the DNA methylation machinery directs G9a/GLP-mediated transcriptional silencing or H3K9 methylation remains controversial. In humans, DNA methyltransferase 1 (DNMT1) knockdown in HeLa cells or _DNMT1_-null HCT116 cells shows a reduction of G9a-mediated H3K9 methylation (Estève et al, 2006). On the other hand, no clear reduction of global H3K9 methylation was seen in the Dnmt1, 3a, 3b triple mouse KO ES cells (Tsumura et al, 2006). To clarify this issue, we analysed the status of transcription of Mage-a and Wfdc15a genes, DNA methylation and H3K9 methylation of the Mage-a2 promoter and 5′-upstream of Wfdc15a in _Dnmt_-KO ES cells. We utilized Dnmt1- and Dnmt3a+b double KO (DKO) cells, both of which show a dramatic reduction of DNA methylation (Li et al, 1992; Okano et al, 1999). As shown in Figure 5A and 5B, Mage-a and Wfdc15a genes were mostly silent or very poorly transcribed in the _Dnmt_-KO ES cells. However, the CpG sites on the Mage-a2 promoter region and 5′-upstream of Wfdc15a were hypomethylated in both _Dnmt1_-KO and _Dnmt3a+b_-DKO ES cells to levels similar or lower than those of G9a- or _GLP-_KO ES cells, respectively (Figure 5C and in comparison with Figure 3B–E). These observations indicate that both Dnmt1 and Dnmt3a+b are involved in DNA methylation on those G9a/GLP-target gene loci. Importantly, the status of H3K9me2 on the Mage-a2 promoter region and 5′-upstream region of Wfdc15a were maintained in both _Dnmt1_-KO and _Dnmt3a+b_-DKO ES cells at levels similar to those of wild-type ES cells (Figure 5D). These facts clearly suggest that G9a/GLP-mediated H3K9 methylation at G9a/GLP-target gene loci are not directed by the DNA methyltransferase machinery or deposited DNA methylation. Furthermore, the phenotypes of _Dnmts_-KO ES cells indicate that G9a/GLP-mediated H3K9 methylation can suppress transcription even in the absence of DNA methylation. To identify the de novo DNA methylation enzyme(s) responsible for the regulation of G9a/GLP-target gene loci, we analysed CpG methylation status in several Dnmt3a+b DKO ES lines that stably expressed exogenous Dnmt3a1, 3a2 or 3b1 ((Oda et al, 2006), Figure 6A and B). Methylation of the CpG sites on Mage-a2 and Wfdc15a was significantly recovered in all the rescued Dnmt3a+b DKO ES cell lines expressing Dnmt3a1, 3a2 or 3b1 (in comparison of Figure 6B with 5C). The relatively poor expression of exogenous Dnmt3b1 may account for the ‘partially rescued' phenotype (Figure 6A). Collectively, these genetic data suggest that both Dnmt3a and 3b redundantly regulate DNA methylation at G9a/GLP-target loci.
Figure 5.
H3K9 methylation suppresses G9a/GLP-target genes in DNA methyltransferase-deficient ES cells. (A) Northern blot analyses of _Dnmts-_KO ES cells are shown. Total RNA was separated and probed with radiolabelled cDNAs. (B) Expression levels for Mage-a (left) and Wfdc15a (right) are numerically represented. The values for _G9a-_KO cells are normalized to 100%. Each experiment was carried out twice. (C) Cytosine methylation status of Mage-a2 promoter and 5′-upstream region of Wfdc15a in _Dnmt1-_KO (left) and Dnmt3a+b DKO (right) ES cells. CpG positions are as described in Figure 3B and D. (D) Histone methylation patterns of the Mage-a2 promoter region (top) and 5′-upstream region of Wfdc15a (bottom) in _Dnmts-_KO ES cells were determined by ChIP analyses. H3K9me2 around the promoter is still maintained in _Dnmt1-_KO and Dnmt3a+b DKO ES cells. Experiments were carried out twice.
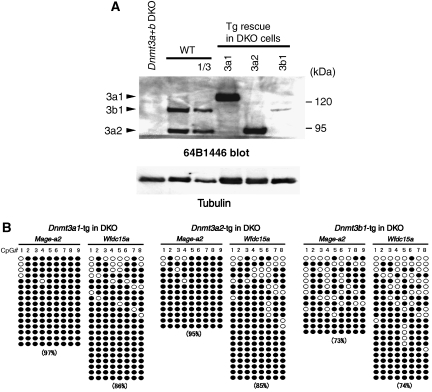
Figure 6.
Dnmt3a and 3b redundantly methylate CpG sequences on G9a/GLP-target genes. (A) Dnmt3 protein expression of several rescued ES lines on Dnmt3a+b DKO background, were analysed with mouse monoclonal antibody 64B1446, which recognizes carboxy terminus of both Dnmt3a and 3b (Chen et al, 2002). (B) Cytosine methylation status of Mage-a2 promoter and 5′-upstream region of Wfdc15a in Dnmt3-rescued line. CpG positions are as described in Figure 3B and D.
Discussion
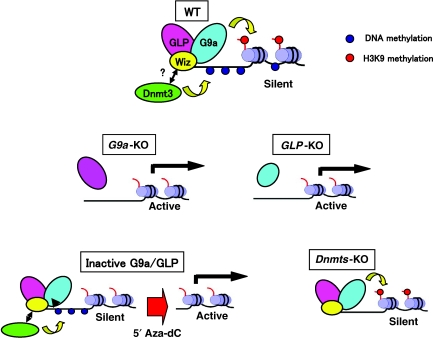
In this report, we demonstrate that the G9a/GLP heteromeric complex regulates both H3K9 and DNA methylation and that these two epigenetic marks cooperatively silence transcription. However, these two epigenetic marks are deposited independent of one another (see model—Figure 7).
Figure 7.
Schematic representation of G9a/GLP complex-mediated gene suppression. One candidate molecule links G9a/GLP to DNA methyltransferase is Wiz, as it binds preferentially G9a/GLP heteromer (Ueda et al, 2006). Genetic analyses indicate that orphan G9a (right) or GLP (left) is both incapable of catalysing H3K9 methylation and recruiting DNA methylation, accompanied with the reactivation of the target genes (see panels of _G9a-_KO and _GLP_-KO). Mutant complex incapable of H3K9 methylation, such as GM4/LWT or GM7/LWT, can still suppress transcription via recruiting DNA methyltransferase(s) (see panels of inactive G9a/GLP). This suppression is cancelled by 5′Aza-dC treatment. G9a/GLP complex can also suppress transcription even without DNA methylation, solely by depositing H3K9 methylation (see panel of _Dnmts-_KO).
How does the G9a/GLP complex regulate DNA methylation at the molecular level? G9a/GLP-target genes were fully reactivated in G4 or G7 cells in the presence of 5′Aza-dC, suggesting that induction of DNA methylation on those G9a/GLP-target genes is directed by the G9a/GLP complex rather than a lack of transcriptional competence. One simple mechanism is that the G9a/GLP heteromeric complex directly recruits the DNA methyltransferase(s)/DNA methyltransferase machinery. Indeed, it has been reported that human G9a interacts with DNMT1 (Estève et al, 2006) and HP1 interacts with DNMT1 (Smallwood et al, 2007). Furthermore, our proteomics analysis shows that HP1 (mainly HP1γ) is in the G9a/GLP complex in mouse ES cells (data not shown). However, we have been unable to detect an association between G9a(/GLP) and Dnmt1 in ES cells (data not shown). The DNA methylation phenotypes of Mage-a2 and Wfdc15a also suggest that the G9a/GLP complex induces de novo DNA methylation. Therefore, G9a/GLP may associate with de novo DNA methyltransferases. Although genetic evidence indicates that the de novo DNA methyltransferases Dnmt3a and 3b are responsible for DNA methylation at the G9a/GLP-target loci (Figure 6B), our anti-G9a/GLP or anti-Dnmt3s co-IP experiments fail to detect clear G9a/GLP-Dnmt3 s association in ES cells (data not shown). We have also performed ChIP analysis using antibodies against G9a, GLP and Dnmts and confirmed immunoprecipitation of those molecules from the formalin-fixed samples, but so far we are unable to obtain reliable data exhibiting enrichment of G9a/GLP and Dnmts at specific loci including Mage-a2 and Wfdc15a or showing a good correlation between the DNA methylation phenotypes and enrichment of specific Dnmts (data not shown). Targeting of G9a/GLP or Dnmts may be quite transient or enrichment of ChIPed G9a/GLP- or Dnmts-nucleosomes may be too low to show significant data.
How is the G9a/GLP-mediated DNA methylation phenotype relevant to global DNA methylation? Although we do not detect as dramatic reduction of CpG methylation in _G9a_-KO ES cells as shown in _Dnmt1_- or _Dnmt3a+b_-DKO ES cells (not shown), Dong et al. describe that _G9a_-KO ES cells show a drastic reduction in DNA methylation of endogenous retroelements, which represent 5–10% of the mammalian genome (Dong et al, 2008). They also show that G9a-mediated DNA methylation of the retroelements is independent of its catalytic activity, which is consistent with our findings. If G9a/GLP is required only for de novo DNA methylation, depletion of DNA methylation in the _G9a_-KO or _GLP_-KO lines may be minimized by maintenance of methyltransferase activity. To clarify how this G9a/GLP-mediated DNA methylation system is maintained or effective in different types of cells, we examined CpG methylation status of the G9a-target loci in _G9a-_KO embryos (E9.5). Interestingly, but in contrast to G9a- or _GLP-_KO ES cells, _G9a-_KO embryos (E9.5) did not exhibit a DNA hypomethylation phenotype at G9a-target gene loci (Supplementary Figure S6). As global H3K9 methylation is severely affected in E9.5 _G9a-_KO embryos in the same way as in _G9a-_KO ES cells (Tachibana et al, 2002), these data suggest that the G9a/GLP-independent regulation mechanism(s) also controls CpG methylation of the G9a-taget loci in cells of E9.5 embryos.
What is the biological significance of the dual silencing mechanism of G9a/GLP seen in ES cells? Compared with differentiated somatic cells, pluripotent ES cells maintain a chromatin structure that is generally permissive for transcriptional activation including relatively low overall levels of DNA methylation (Kimura et al, 2004; Meshorer and Misteli, 2006). In this regard, ES cells may effectively utilize the dual methylation system observed here to secure the silencing state of G9a/GLP-target gene loci upon cellular differentiation. Another possible benefit is that such dual silencing mechanism can also stably maintain the silent status throughout differentiation of specific lineage of cells. DNA methylation is quite dynamically regulated during development of specific lineage of cells, such as germ-lineage cells (Reik et al, 2001; Surani, 2001). During temporal loss of DNA methylation in certain developmental stages, the silent states can be maintained if the G9a/GLP dual silencing mechanism regulates gene transcription (similar to the case of _Dnmts_-KO in Figure 7). This mechanism also applies to the dynamics of H3K9 methylation regulated by HMTases and histone lysine demethylases or other mechanisms (Klose and Zhang, 2007; Shi, 2007). Lastly, our data suggest that other epigenetic marks also function as one of multiple layers of regulatory mechanisms.
Materials and methods
In vitro HMTase assay
Methyltransferase assays were performed as described previously (Tachibana et al, 2001). Briefly, 15 μl of reaction mixture containing 1 μg of recombinant H3/H4, 1 μg of recombinant enzymes and 50 nCi of _S_-adenosyl-[_methyl_-14C]-L-methionine in methylase activity buffer (50 mM Tris, pH 8.5, 20 mM KCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, 250 mM sucrose) was incubated for 120 min at room temperature. The reaction products were separated by 15% SDS–PAGE. Detection of _methyl_-14C was performed using a BAS-5000 imaging analyser (Fuji Film).
Cell culture
Embryonic stem cells were maintained in Dulbecco's modified Eagle's medium (Sigma) containing 10% Knockout SR (Invitrogen) and 1% fetal calf serum (described as ES medium hereafter). To generate ES cells stably expressing mutant G9a or GLP, the expression vectors designated as pCAG-G9a-L-IRESpuro or pCAG-GLP-IRESbsd were introduced into the corresponding KO ES cells by using Lipofectamine 2000 reagent (Invitrogen). The stable transfectant clones expressing mutant G9a or GLP were selected in the ES medium containing puromycin (1 μg/ml) or blasticidin (10 μg/ml), respectively. For northern blot analysis, ES cells were cultured in the ES medium with tricostatin-A (10 ng/ml) for 24 h or 0.5 μM 5-Aza-2′-Deoxycytidine for 96 h.
G9a/GLP complex formation analysis
Serial ES lines were harvested with PBS containing 0.05% trypsin and 0.2 mM EDTA. The cellular pellets were washed once in ice-cold PBS, snap-frozen in liquid nitrogen and stored at −80 °C before analysis. For G9a/GLP interaction analysis, the cellular pellets were lysed with RIPA buffer (PBS containing 1% NP40, 0.5% deoxycholate, 0.1% SDS and protease inhibitor cocktail (Nakalai)). The cellular extracts were prepared by removing unresolved pellets after centrifugation, and then incubated with 2 μg of either anti-G9a (A8620A, Perseus Proteomix) or anti-GLP (B0422, Perseus Proteomix) overnight. The immune complexes were collected with 15 μl of protein G slurry (1:1 ratio, GE Healthcare) for 3 h, washed three times with PBS containing 0.1% NP40 and then separated by SDS–PAGE.
Histone modification analysis
Histones corresponding to 5 × 104 nuclei were purified by acid extraction (Tachibana et al, 2002), separated by SDS–PAGE, transferred to nitrocellulose membranes, blocked with 5% milk and probed with the specific antibodies against H4 (Millipore, 07-108), H3K9me1 (Millipore, 07-450), H3K9me2 (Millipore, 07-441) and H3K9me3 (Millipore, 07-523).
For ChIP analysis, we slightly modified the previous protocols for native ChIP analysis (O'Neill and Turner, 1995; Gregory et al, 2001). ES cells (∼5 × 106) suspended in 50 μl of 0.3 M sucrose-containing buffer 1 (60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.1 mM EGTA, 0.5 mM dithiothreitol, 0.1 mM PMSF, 3.6 ng/ml aprotinin, 15 mM Tris–HCl pH 7.5) were lysed by addition of 50 μl of buffer 1 containing 0.8% NP40 and 0.3 M sucrose and put on ice for 10 min. After addition of 0.8 ml of buffer 1 containing 1.2 M sucrose to the suspension, chromatin fraction was collected as pellets by centrifugation (8500 r.p.m., 10 min). The pellets were resuspended in 200 μl of micrococcal nuclease (MNase) digestion buffer (0.32 M sucrose, 4 mM MgCl2, 1 mM CaCl2, 0.1 mM PMSF, 50 mM Tris–HCl pH 7.5), incubated at 37 °C for 15 min with MNase (0.3–0.6 U, Takara). Digestion was stopped by addition of EDTA to a final concentration of 20 mM. Approximately 200 μl of the supernatant was obtained by centrifugation (13000 r.p.m., 10 min), combined with 1.8 ml of incubation buffer (50 mM NaCl, 5 mM EDTA, 0.1% NP40, 0.1 mM PMSF, 20 mM Tris–HCl pH 7.5), incubated overnight with antibodies against methylated H3 and then incubated for further 4 h after addition of 15 μl of protein G slurry (1:1 ratio, GE Healthcare). Antibodies used were mouse monoclonal antibodies against H3K9me1 (CMA306), H3K9me2 (ab1220, Abcam) and H3K4me2 (CMA303) (Kimura et al, 2008). The immune complexes were sequentially washed in 75 mM NaCl-containing buffer A (75 mM NaCl, 10 mM EDTA, 50 mM Tris–HCl pH 7.5), 125 mM NaCl-containing buffer A and 175 mM NaCl-containing buffer A. DNA was obtained by phenol/chloroform extraction following proteinase K treatment (0.1 mg/ml) of the immune complex in digestion buffer (400 mM NaCl, 10 mM EDTA, 0.5% SDS, 20 mM Tris–HCl pH 8.0, 0.1 mg/ml proteinase K) for 60 min at 56 °C, and then analysed by real-time PCR using primers specific for Mage-a2 promoter (Mage-a2A, 5′-TTGGTGGACAGGGAAGCTAGGGGA-3′; Mage-a2B, 5′-CGCTCCAGAACAAAATGGCGCAGA-3′), and 5′-upstream region of Wfdc15a (AV2P.4, 5′-GGATAACCAGCAGGGAAGCTGAGG-3′; AV2P.5, 5′-TCTCTAGGTGCCTGAACCTACAGC-3′).
DNA methylation analysis
Bisulphite treatment of the genomic DNA isolated from serial ES lines was carried out with EpiTect bisulfite Kit (Qiagen). Nested PCR was performed to amplify the sequences around the Mage-a2 promoter and 5′-upstream region of Wfdc15a. The first-round PCR (35 cycles) and the second-round PCR (20 cycles) were carried out using specific primer sets as follows. For CpG position no. 1–9 of Mage-a2 (see Figure 3A), Magea2.1bi (5′-GGATTTTGAAAGAAATGGTGATTT-3′) and Magea2.2bi (5′-AATTCTTACCTCAACTAACACAAC- 3′) were used for the first-round PCR and Magea2-NesF-bi-Bam (5′-ATAGGATCCTATATTGGTGGATAGGGAAGTTAGGG-3′) and Magea2-R-bi-RI (5′-ATAGAATTCTCAACTAACACAACATAAAAACCTCC-3′) were used for the second-round PCR. For CpG position no. 1–8 of Wfdc15a (see Figure 3C), SAV-bi.b (5′-TTGGTGTTTAGGTAGATATAGTTAATAAGA-3′) and SAV2bi.d (5′-CTTCTTAAAAACTAACAAAACTAATCTAACATTTC-3′) were used for the first-round PCR and SAV-nes-BF-2 (5′-ATAGGATCCAAGGAAAATGTATTTATAAATTAAAG-3′) and SAV-nes-RR-3 (5′-TATGAATTCAAAACTAATCTAACATTTCCTAATAC-3′) were used for the second-round PCR. For CpG position no. 9–11 of Wfdc15a, AV2bi.1 (5′-GGATAATTAGTAGGGAAGTTGAGG-3′) and AV2bi.4 (5′-ACCAAACTAACCCTACTAATCCCTA-3′) were used for the first-round PCR and AVbi.1-RI (5′-ATAGAATTCAAGTTGAGGTTATAGGTTAGG-3′) and AV2bi.4-Bam (5′-TAAGGATCCAAAACATTATCTTCCTTCCTC-3′) were used for the second-round PCR. The second-round PCR products were subcloned into pBluescript vectors digested with _Eco_RI and _Bam_HI.
RNA expression analysis
Total RNAs were isolated with Sepasol-RNA I (Nakarai). Three micrograms of them were separated by 1% agarose-formaldehyde gel electrophoresis, transferred to a nylon membrane, and probed with 32P-labelled cDNAs as follows. The subcloned RT–PCR products corresponding to nucleotides 321–780 of GenBank entry NM_020020 (Mage-a8), 28–475 of AV206552 (Wfdc15a) and 247-1120 of BC083080 (Gapdh) were used as probes.
Supplementary Material
Supplementary Figures S1–6
Acknowledgments
We are grateful to Dr E Li (Novartis) and Dr M Okano (RIKEN) for providing _Dnmts_-KO ES cells, the rescued ES lines and specific antibody against Dnmts, and Drs R Hirasawa and H Sasaki (NIG) for technical advices. We thank Dr M Lorincz (The University of British Columbia) for valuable scientific comments and editorial help and our laboratory members for assistance. We also thank Dr EM Oltz for critically reading the manuscript. This work was supported by a Grant-in-Aid from the Ministry of Education, Science, Technology and Culture of Japan.
References
- Bannister A, Zegerman P, Partridge J, Miska E, Thomas J, Allshire R, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim J, Estève P, Clark A, Pradhan S, Jacobsen S (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764 [DOI] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Xie S, Li E (2002) A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem 277: 38746–38754 [DOI] [PubMed] [Google Scholar]
- De Plaen E, De Backer O, Arnaud D, Bonjean B, Chomez P, Martelange V, Avner P, Baldacci P, Babinet C, Hwang S, Knowles B, Boon T (1999) A new family of mouse genes homologous to the human MAGE genes. Genomics 55: 176–184 [DOI] [PubMed] [Google Scholar]
- Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schübeler D, Tachibana M, Shinkai Y, Lorincz MC (2008) DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J; E-pub ahead of print 25 September 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estève P, Chin H, Smallwood A, Feehery G, Gangisetty O, Karpf A, Carey M, Pradhan S (2006) Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev 20: 3089–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y (2006) G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol 8: 188–194 [DOI] [PubMed] [Google Scholar]
- Gregory R, Randall T, Johnson C, Khosla S, Hatada I, O'Neill L, Turner B, Feil R (2001) DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs1. Mol Cell Biol 21: 5426–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Iwatani M, Suzuki M, Tachibana M, Shinkai Y, Tanaka S, Greally J, Yagi S, Hattori N, Shiota K (2007) Genome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cells. Genes Cells 12: 1–11 [DOI] [PubMed] [Google Scholar]
- Jackson J, Lindroth A, Cao X, Jacobsen S (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Jenuwein T (2006) The epigenetic magic of histone lysine methylation. FEBS J 273: 3121–3135 [DOI] [PubMed] [Google Scholar]
- Kimura H, Hayashi-Takanaka Y, Goto Y, Takizawa N, Nozaki N (2008) Organization of histone H3 modifications revealed by a panel of specific monoclonal antibodies. Cell Struct Funct 33: 61–73 [DOI] [PubMed] [Google Scholar]
- Kimura H, Tada M, Nakatsuji N, Tada T (2004) Histone code modifications on pluripotential nuclei of reprogrammed somatic cells. Mol Cell Biol 24: 5710–5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R, Zhang Y (2007) Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8: 307–318 [DOI] [PubMed] [Google Scholar]
- Kubicek S, O'Sullivan R, August E, Hickey E, Zhang Q, Teodoro M, Rea S, Mechtler K, Kowalski J, Homon C, Kelly T, Jenuwein T (2007) Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 25: 473–481 [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Li E, Bestor T, Jaenisch R (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Margueron R, Trojer P, Reinberg D (2005) The key to development: interpreting the histone code? Curr Opin Genet Dev 15: 163–176 [DOI] [PubMed] [Google Scholar]
- Meshorer E, Misteli T (2006) Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol 7: 540–546 [DOI] [PubMed] [Google Scholar]
- O'Neill L, Turner B (1995) Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J 14: 3946–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M, Yamagiwa A, Yamamoto S, Nakayama T, Tsumura A, Sasaki H, Nakao K, Li E, Okano M (2006) DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes Dev 20: 3382–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell D, Haber D, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257 [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J (2001) Epigenetic reprogramming in mammalian development. Science 293: 1089–1093 [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo T, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912 [DOI] [PubMed] [Google Scholar]
- Shi Y (2007) Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet 8: 829–833 [DOI] [PubMed] [Google Scholar]
- Smallwood A, Estève P, Pradhan S, Carey M (2007) Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev 21: 1169–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B, Allis C (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Surani M (2001) Reprogramming of genome function through epigenetic inheritance. Nature 414: 122–128 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y (2001) Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem 276: 25309–25317 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y (2002) G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y (2005) Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 19: 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Selker E (2001) A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414: 277–283 [DOI] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda H, Nakayama J, Okano M (2006) Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11: 805–814 [DOI] [PubMed] [Google Scholar]
- Ueda J, Tachibana M, Ikura T, Shinkai Y (2006) Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J Biol Chem 281: 20120–20128 [DOI] [PubMed] [Google Scholar]
- Wade P (2001) Methyl CpG-binding proteins and transcriptional repression. Bioessays 23: 1131–1137 [DOI] [PubMed] [Google Scholar]
- Xin Z, Tachibana M, Guggiari M, Heard E, Shinkai Y, Wagstaff J (2003) Role of histone methyltransferase G9a in CpG methylation of the Prader–Willi syndrome imprinting center. J Biol Chem 278: 14996–15000 [DOI] [PubMed] [Google Scholar]
- Zhang X, Tamaru H, Khan S, Horton J, Keefe L, Selker E, Cheng X (2002) Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell 111: 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–6