MicroRNA-target pairs in human renal epithelial cells treated with transforming growth factor β1: a novel role of miR-382 (original) (raw)
Abstract
We reported previously an approach for identifying microRNA (miRNA)-target pairs by combining miRNA and proteomic analyses. The approach was applied in the present study to examine human renal epithelial cells treated with transforming growth factor β1 (TGFβ1), a model of epithelial–mesenchymal transition important for the development of renal interstitial fibrosis. Treatment of human renal epithelial cells with TGFβ1 resulted in upregulation of 16 miRNAs and 18 proteins and downregulation of 17 miRNAs and 16 proteins. Of the miRNAs and proteins that exhibited reciprocal changes in expression, 77 pairs met the sequence criteria for miRNA–target interactions. Knockdown of miR-382, which was up-regulated by TGFβ1, attenuated TGFβ1-induced loss of the epithelial marker E-cadherin. miR-382 was confirmed by 3′-untranslated region reporter assay to target five genes that were downregulated at the protein level by TGFβ1, including superoxide dismutase 2 (SOD2). Knockdown of miR-382 attenuated TGFβ1-induced downregulation of SOD2. Overexpression of SOD2 ameliorated TGFβ1-induced loss of the epithelial marker. The study provided experimental evidence in the form of reciprocal expression at the protein level for a large number of predicted miRNA-target pairs and discovered a novel role of miR-382 and SOD2 in the loss of epithelial characteristics induced by TGFβ1.
INTRODUCTION
MicroRNAs (miRNAs) are small regulatory RNA molecules encoded by specific genes in the genomes (1–5). Typically, miRNAs bind to the 3′-untranslated region (UTR) of target mRNAs and decrease the abundance of target proteins. The effect is mediated mainly by translational repression (6,7) and, in some cases, decreases in mRNA abundance (8–11). Numerous studies have established miRNAs as broad and powerful regulators of protein expression in physiology and disease (11–15).
A major challenge in miRNA research is identification of specific target proteins (14,16). An miRNA could potentially target multiple genes since its binding to 3′-UTR requires only partial complementarities. In silico sequence analyses predicted several thousand proteins as potential targets of miRNAs, providing a valuable starting point (17–19). However, of the several hundred thousand predicted miRNA-target pairs in human, rat and mouse, only a small fraction has been validated experimentally. Experimental validation of miRNA-target pairs is laborious and time consuming (16), considering the high rate of ‘hit and miss’ that many investigators have experienced. The scarcity of miRNA-target pairs that are supported by experimental evidence significantly limits the field of miRNA research.
We recently developed a new approach for identifying miRNA-target pairs by combining miRNA, proteomic and bioinformatic analyses (20). A miRNA-target pair is considered a high-probability pair if the miRNA and the target protein are reciprocally expressed.
Treatment of renal epithelial cells with transforming growth factor β1 (TGFβ1) is a model commonly used to study renal interstitial fibrosis (21,22). Renal epithelial cells treated with TGFβ1 undergo epithelial–mesenchymal transition, a process that may contribute to the accumulation of extracellular matrix proteins and the development of interstitial fibrosis in vivo. Interstitial fibrosis is a prominent feature of chronic renal injury in humans and animal models and a strong predictor of the progression of chronic renal disease in humans. Signs of epithelial–mesenchymal transition have been detected in several models of chronic renal injury including the Dahl salt-sensitive rat (23).
Using the proteomic approach (20), we identified 77 predicted miRNA-target pairs that exhibited reciprocal changes of expression in human renal epithelial cells in response to TGFβ1. Further experiments revealed a novel role of miR-382 and its target superoxide dismutase 2 (SOD2) in TGFβ1-induced loss of epithelial characteristics in human kidney cells.
MATERIALS AND METHODS
Cell culture and TGFβ1 treatment
HK-2, a human kidney epithelial cell line, was obtained from and cultured as suggested by ATCC (20,24,25). Cells at ∼40% confluency were treated with recombinant human TGFβ1 (3 ng/ml) (R&D Systems) or the vehicle control in DMEM for the indicated time period.
MiRNA expression profiling
Expression levels of 376 human miRNAs were measured by real-time reverse transcription polymerase chain reaction (RT–PCR). Total RNA was extracted using TRIzol (Invitrogen) from cells treated with TGFβ1 or vehicle for 24 h. Small RNA was isolated using a miRNA isolation kit and reverse-transcribed (200 ng) using a miRNA first strand kit (SABiosciences). Real-time PCR was performed in 384-well, human genome miRNA PCR plates (SABiosciences) using a SYBR Green/ROX qPCR master mix (SABiosciences) and the ABI 7900HT instrument (Applied Biosystems). The human genome miRNA PCR plate contained real-time PCR primers for 376 human miRNAs, four normalizer small RNAs and several quality controls. Three independent replicates, representing cells at two different passage numbers, were analyzed for each treatment condition. Each replicate sample was a pool of two dishes of cells. Differences in miRNA levels were calculated using the ΔΔCt method (25,26). The criteria of differential expression were P < 0.02 and fold changes >20%, which had a false positive rate of ∼10% calculated as described previously (20,23).
Taqman real-time PCR
Expression levels of several miRNAs and protein-coding genes were quantified using real-time PCR with the Taqman chemistry (Applied Biosystems) as described previously (11,20). To avoid any bias introduced during the isolation of small RNA, total RNA samples were used in these analyses. The 5S and 18S rRNA was used as internal normalizer for miRNA and protein-coding genes, respectively.
Differential in-gel electrophoresis and mass spectrometry analysis
Proteomic analysis was performed using the differential in-gel electrophoresis (DIGE) and mass spectrometry method as described previously (20,24). Briefly, protein samples from vehicle or TGFβ1 treated cells (n = 4) were labeled with CyDye DIGE Fluor minimal dyes Cy3 or Cy5 (GE Healthcare). The dyes were switched between vehicle- and TGFβ1-treated cells for every other gel. A pool of all of the samples was labeled with Cy2. Three labeled samples, including a vehicle-treated sample, a TGFβ1-treated sample and the pool, were combined and separated on one gel by two-dimensional electrophoresis. A total of four analytical gels were run to analyze four pairs of control and TGFβ1-treated samples. The protein abundance was compared between vehicle- and TGFβ1-treated samples according to the intensity of each fluorescent dye within a spot on the gels. Criteria for differential expression were P < 0.05, a difference >1.2-fold, and the appearance of the spot in at least three of the four gels. Differentially expressed protein spots were picked from preparative gels. The identities of the proteins were obtained using MALDI-TOF mass spectrometry followed by Mascot searches with the settings described previously (20,24).
Western blot
The relative abundance of several proteins was analyzed using western blot as we described previously (11,25). Coomassie blue staining of the entire membrane was used to confirm equal loading on the gel. Antibodies were from Santa Cruz Biotechnology.
Bioinformatic analysis of MiRNA-target pairs
Bioinformatic analysis was performed as described previously (20). Reciprocal expression was defined as a miRNA being upregulated and a protein being downregulated in the TGFβ1-treated cells, or vice versa. Sequence-based predictions of miRNA targets were retrieved from two algorithms, TargetScan v5.1 (http://www.targetscan.org/) and MicroCosm Targets v5 (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/).
3′-UTR reporter analysis
A 3′-UTR reporter analysis was performed to examine interactions between specific miRNAs and target sequences, as we described previously (11). Briefly, a segment of the 3′-UTR region of a miRNA target gene was cloned into pMIR-REPORT luciferase plasmid (Ambion). Each 3′-UTR reporter construct was transfected into HeLa cells. Activities of luciferase and the internal normalizer β-galactosidase, in the presence of a specific or control anti-miR or pre-miR (Ambion), were measured using the Dual-Light system (ABI) in a microplate format.
Site-directed mutagenesis
Mutated or partially deleted 3′-UTR reporter constructs were generated using site-directed mutagenesis as we described previously (27). In the mutated construct, CAA in the miR-382 seed region-binding site within the 3′-UTR segment of SOD2 were mutated to TGG. ACAA in the seed region binding site were deleted in the partial deletion construct.
Oligonucleotides
Locked nucleic acid (LNA)-modified anti-miR oligonucleotides were obtained from Exiqon. Additional anti-miR and pre-miR were obtained from Ambion. HK2 cells were transfected with the oligonucleotides (100 nM) using Oligofectamine (Invitrogen).
Plasmid transfection
Superoxide dismutase 2 (SOD2) human cDNA ORF clone was obtained from OriGene. The clone expresses SOD2 driven by a CMV promoter and tagged with C-terminal Myc-DDK. The clone was amplified and purified using techniques described previously (11,27). HK2 cells were transfected with SOD2 plasmid or an empty vector (2 µg for each 3.5-cm dish) using Lipofectamine 2000.
Statistics
Data were analyzed using the Student’s _t_-test and multiple-group analysis of variance (ANOVA). A P < 0.05 was considered significant except in those special cases described above. Data are shown as mean ± SEM.
RESULTS
TGFβ1 substantially altered the MiRNA expression profile in HK2 cells
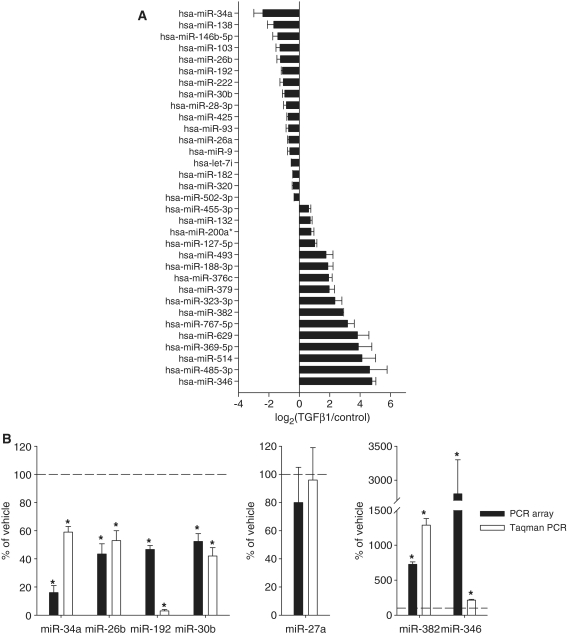
Treatment with TGFβ1 (3 ng/ml) for 24 h resulted in substantial alterations in the miRNA expression profile in HK2 cells, a human renal epithelial cell line. At a permutation-based false positive rate of 10%, 16 of the 376 miRNAs examined were found upregulated and 17 downregulated (Figure 1A and Supplementary Table S1). Nine miRNAs were altered by >4-fold, including five that were altered by >10-fold. Our laboratory has examined changes in miRNA expression profiles under various pathophysiological conditions in separate studies. The changes in most cases were much more modest than those observed in the present study.
Figure 1.
TGFβ1 had widespread effects on miRNA expression in human renal epithelial cells. HK2 cells were treated with vehicle or 3 ng/ml of TGFβ1 for 24 h. (A) 33 of the 376 miRNAs examined with real-time qPCR array were differentially expressed. n = 3. See ‘Materials and Methods’ section for the criteria of differential expression. (B) Verification of the result of miRNA expression profiling. RNA samples used for the individual Taqman qPCR analysis were independent of those used in the high-throughput profiling with the SYBR Green qPCR array. n = 3 for qPCR array, n = 4 for Taqman qPCR; *significantly different from vehicle (P < 0.05 for Taqman qPCR; see ‘Materials and Methods’ section for the criteria of differential expression for qPCR array).
The expression profiling was performed with high-throughput real-time PCR using the SYBR Green chemistry. Seven individual miRNAs, including four downregulated, two upregulated and one not differentially expressed, were further examined in an independent set of RNA samples using the Taqman chemistry. As shown in Figure 1B, individual real-time PCR analyses using the Taqman chemistry confirmed the result of the PCR array analysis for all seven miRNAs.
TGFβ1 altered the proteomic profile in HK2 cells
Treatment with TGFβ1 (3 ng/ml) for 48 h altered the abundance of 34 proteins in HK2 cells. Approximately 2000 protein spots were detectable on each of the four analytical DIGE gels. The intensities of 148 spots were found to be significantly different between vehicle- and TGFβ1-treated samples (n = 4). MALDI-TOF analysis successfully assigned identities to 97 of the 148 spots. The 97 spots represented 34 distinct proteins, 18 of which were upregulated by TGFβ1 and 16 downregulated (Table 1; see Supplementary Table S2 for additional details of protein identification). A recent study reported that treatment of HK2 cells with TGFβ1 at 2 ng/ml for 48 h resulted in the differential expression of 1207 genes at the mRNA level (P < 0.05 and fold change >2) (28). None of the 34 proteins we identified was considered differentially expressed at the mRNA level in that study.
Table 1.
Proteins differentially expressed in HK2 cells in response to TGFβ1
| Common protein name | Uniprot | Gene symbol | log2(T/C)a | _P_-value |
|---|---|---|---|---|
| Superoxide dismutase, mitochondrial | P04179 | SOD2 | −1.16 | 4.00E-05 |
| 60 kDa heat shock protein, mitochondrial | P10809 | HSPD1 | −0.58 | 0.0095 |
| T-complex protein 1 subunit gamma | P49368 | CCT3 | −0.44 | 0.012 |
| Cathepsin D | P07339 | CTSD | −0.41 | 0.0018 |
| Endoplasmic reticulum protein ERp29 | P30040 | ERP29 | −0.41 | 0.0041 |
| Lamin-A/C | P02545 | LMNA | −0.39 | 0.0016 |
| Heterogeneous nuclear ribonucleoprotein K | P61978 | HNRPK | −0.38 | 0.0031 |
| Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial | Q13011 | ECH1 | −0.36 | 2.80_E_–05 |
| Thioredoxin-dependent peroxide reductase, mitochondrial | P30048 | PRDX3 | −0.36 | 0.00036 |
| ATP synthase D chain, mitochondrial | O75947 | ATP5H | −0.34 | 0.0011 |
| Nucleophosmin | P06748 | NPM1 | −0.32 | 0.0041 |
| Heterogeneous nuclear ribonucleoprotein L | P14866 | HNRPL | −0.30 | 0.0015 |
| Paraspeckle component 1 | Q8WXF1 | PSPC1 | −0.29 | 0.0028 |
| RNA-binding protein 8A | Q9Y5S9 | RBM8A | −0.29 | 0.0059 |
| Enoyl-CoA hydratase, mitochondrial | P30084 | ECHS1 | −0.28 | 0.016 |
| Zinc finger protein 160 | Q9HCG1 | ZNF160 | −0.26 | 0.0012 |
| Heterogeneous nuclear ribonucleoproteins A2/B1 | P22626 | HNRPA2B1 | 0.26 | 0.013 |
| 78-kDa glucose-regulated protein precursor | P11021 | HSPA5 | 0.26 | 0.029 |
| Endoplasmic reticulum lipid raft-associated protein 2 | O94905 | ERLN2 | 0.28 | 0.0029 |
| Protein disulfide-isomerase A3 | P30101 | PDIA3 | 0.30 | 0.018 |
| Alpha-actinin-4 | O43707 | ACTN4 | 0.34 | 0.042 |
| Tropomodulin-3 | Q9NYL9 | TMOD3 | 0.34 | 0.0044 |
| Ezrin | P15311 | EZR | 0.38 | 0.018 |
| Heat shock protein HSP 90-beta | P08238 | HSP90AB1 | 0.41 | 0.02 |
| Heterogeneous nuclear ribonucleoprotein D0 | Q14103 | HNRPD | 0.42 | 0.031 |
| Eukaryotic translation initiation factor 3 subunit I | Q13347 | EIF3I | 0.43 | 0.017 |
| Proliferation-associated protein 2G4 | Q9UQ80 | PA2G4 | 0.45 | 0.024 |
| Actin, cytoplasmic 1 | P60709 | ACTB | 0.46 | 0.0015 |
| Eukaryotic initiation factor 4A-I | P60842 | EIF4A1 | 0.50 | 0.0084 |
| Tubulin beta chain | P07437 | TUBB | 0.55 | 0.025 |
| Membrane-organizing extension spike protein | P26038 | MSN | 0.58 | 0.016 |
| Keratin, type I cytoskeletal 18 | P05783 | KRT18 | 0.59 | 0.0021 |
| Tubulin alpha-1B chain | P68363 | TUBA1B | 0.59 | 0.019 |
| Tropomyosin alpha-1 chain | P09493 | TPM1 | 1.87 | 0.00088 |
The differentially expressed proteins shared several functional themes including regulation of apoptosis, RNA binding and gene transcription (Supplementary Table S3). Many differentially expressed proteins are involved in the cytoskeleton, mitochondria and endoplasmic reticulum. For example, seven mitochondrial proteins were differentially expressed, including ATP5H, CTSD, ECH1, ECHS1, HSPD1, PRDX3 and SOD2.
A protein was represented in multiple spots in several cases. We have found apparent shifts of a protein on the gel between experimental conditions in previous studies, indicating changes in protein processing or modification (20,24). In the present study, however, spots representing the same protein were invariably up- or downregulated in the same direction by TGFβ1. Data for only one representative spot was shown in Table 1 and Supplementary Table S1 in those cases.
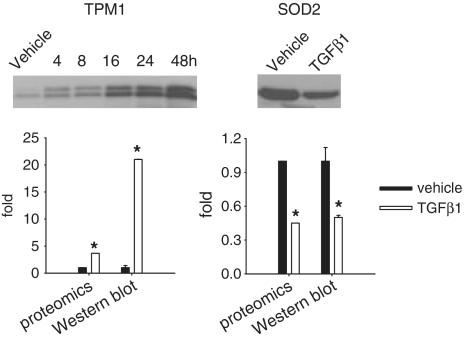
Western blot analysis of tropomyosin 1 (TPM1) and SOD2 was performed in protein samples independent of those used in the proteomic analysis. The result (Figure 2) confirmed the upregulation of TPM1 and downregulation of SOD2 in response to TGFβ1. mRNA levels of TPM1 tended to be increased by TGFβ1 (to 1.8 ± 0.3-fold of vehicle, n = 6, P = 0.07). SOD2 mRNA levels were significantly decreased by TGFβ1 (to 24 ± 3% of vehicle, n = 6, P < 0.05). In the microarray study mentioned above (28), TPM1 and SOD2 exhibited statistically significant up- and downregulation, respectively, but did not reach the fold-change threshold of 2 used in that study.
Figure 2.
Verification of proteomic results. HK2 cells were treated as described in Table 1. Protein samples used for the western blot analysis were independent of those used for the proteomic analysis. TPM1, tropomyosin 1; SOD2, superoxide dismutase 2. The gel shown for TPM1 was a time course analysis. Only the 48-h time point was plotted in the bar graph for comparison with the proteomic data. n = 4 for proteomics, n = 3 for western blot; *significantly different from vehicle (P < 0.05 for western blot; see ‘Materials and Methods’ section for the criteria of differential expression for the proteomic analysis).
MiRNA-target pairs supported by reciprocal expression
The differentially expressed miRNAs and proteins shown in Figure 1 and Table 1 formed 578 pairs of reciprocally expressed miRNAs and proteins. Of the 578 pairs, 77 were predicted to be miRNA-target pairs according to TargetScan v5.0 or MicroCosm Targets v5 based on sequence characteristics (Table 2). The 77 pairs involved 31 miRNAs and 28 proteins.
Table 2.
MiRNA-target pairs supported by sequence characteristics and reciprocal expression in human renal epithelial cells treated with TGFβ1
| miRNA | miRNA log2(T/C)a | Protein target | Protein log2(T/C) | Target Scan | Micro Cosm |
|---|---|---|---|---|---|
| hsa-miR-346 | 4.78 | LMNA | −0.39 | Yes | |
| hsa-miR-485-3p | 4.63 | SOD2 | −1.16 | Yes | |
| hsa-miR-514 | 4.12 | PSPC1 | −0.29 | Yes | |
| hsa-miR-514 | 4.12 | RBM8A | −0.29 | Yes | |
| hsa-miR-369-5p | 3.88 | ZNF160 | −0.26 | Yes | |
| hsa-miR-369-5p | 3.88 | CCT3 | −0.44 | Yes | |
| hsa-miR-629 | 3.82 | CCT3 | −0.44 | Yes | |
| hsa-miR-629 | 3.82 | CTSD | −0.41 | Yes | |
| hsa-miR-767-5p | 3.17 | ZNF160 | −0.26 | Yes | Yes |
| hsa-miR-767-5p | 3.17 | RBM8A | −0.29 | Yes | |
| hsa-miR-767-5p | 3.17 | HNRNPL | −0.3 | Yes | |
| hsa-miR-767-5p | 3.17 | NPM1 | −0.32 | Yes | |
| hsa-miR-767-5p | 3.17 | LMNA | −0.39 | Yes | |
| hsa-miR-382 | 2.86 | PSPC1 | −0.29 | Yes | |
| hsa-miR-382 | 2.86 | NPM1 | −0.32 | Yes | |
| hsa-miR-382 | 2.86 | ECH1 | −0.36 | Yes | |
| hsa-miR-382 | 2.86 | HSPD1 | −0.58 | Yes | Yes |
| hsa-miR-382 | 2.86 | SOD2 | −1.16 | Yes | Yes |
| hsa-miR-323-3p | 2.34 | PSPC1 | −0.29 | Yes | |
| hsa-miR-323-3p | 2.34 | HNRNPK | −0.38 | Yes | Yes |
| hsa-miR-379 | 1.97 | RBM8A | −0.29 | Yes | |
| hsa-miR-376c | 1.93 | PSPC1 | −0.29 | Yes | |
| hsa-miR-376c | 1.93 | RBM8A | −0.29 | Yes | |
| hsa-miR-376c | 1.93 | ECH1 | −0.36 | Yes | |
| hsa-miR-376c | 1.93 | PRDX3 | −0.36 | Yes | Yes |
| hsa-miR-376c | 1.93 | HNRNPK | −0.38 | Yes | |
| hsa-miR-188-3p | 1.87 | ZNF160 | −0.26 | Yes | |
| hsa-miR-188-3p | 1.87 | PSPC1 | −0.29 | Yes | |
| hsa-miR-188-3p | 1.87 | ERP29 | −0.41 | Yes | |
| hsa-miR-493 | 1.75 | ZNF160 | −0.26 | Yes | |
| hsa-miR-493 | 1.75 | HNRNPL | −0.3 | Yes | |
| hsa-miR-127-5p | 1.01 | ZNF160 | −0.26 | Yes | |
| hsa-miR-127-5p | 1.01 | RBM8A | −0.29 | Yes | |
| hsa-miR-200a* | 0.77 | HNRNPK | −0.38 | Yes | |
| hsa-miR-200a* | 0.77 | SOD2 | −1.16 | Yes | |
| hsa-miR-132 | 0.73 | HNRNPL | −0.3 | Yes | |
| hsa-miR-132 | 0.73 | SOD2 | −1.16 | Yes | Yes |
| hsa-miR-455-3p | 0.61 | RBM8A | −0.29 | Yes | |
| hsa-miR-455-3p | 0.61 | CTSD | −0.41 | Yes | |
| hsa-miR-182 | −0.43 | HSPA5 | 0.26 | Yes | |
| hsa-miR-182 | −0.43 | ERLIN2 | 0.28 | Yes | |
| hsa-miR-182 | −0.43 | MSN | 0.58 | Yes | |
| hsa-let-7i | −0.51 | HSPA5 | 0.26 | Yes | |
| hsa-let-7i | −0.51 | PDIA3 | 0.3 | Yes | |
| hsa-let-7i | −0.51 | TUBB | 0.55 | Yes | |
| hsa-let-7i | −0.51 | MSN | 0.58 | Yes | |
| hsa-miR-9 | −0.62 | ERLIN2 | 0.28 | Yes | |
| hsa-miR-9 | −0.62 | MSN | 0.58 | Yes | |
| hsa-miR-26a | −0.69 | HNRNPA2B1 | 0.26 | Yes | |
| hsa-miR-26a | −0.69 | EIF3I | 0.43 | Yes | |
| hsa-miR-93 | −0.7 | HSPA5 | 0.26 | Yes | |
| hsa-miR-425 | −0.73 | HNRNPD | 0.42 | Yes | Yes |
| hsa-miR-28-3p | −0.85 | HNRNPA2B1 | 0.26 | Yes | |
| hsa-miR-28-3p | −0.85 | TMOD3 | 0.34 | Yes | Yes |
| hsa-miR-28-3p | −0.85 | HSP90AB1 | 0.41 | Yes | |
| hsa-miR-30b | −0.95 | HNRNPA2B1 | 0.26 | Yes | |
| hsa-miR-30b | −0.95 | HSPA5 | 0.26 | Yes | |
| hsa-miR-222 | −1.05 | HNRNPD | 0.42 | Yes | Yes |
| hsa-miR-222 | −1.05 | MSN | 0.58 | Yes | |
| hsa-miR-192 | −1.05 | HSPA5 | 0.26 | Yes | |
| hsa-miR-192 | −1.05 | ERLIN2 | 0.28 | Yes | |
| hsa-miR-192 | −1.05 | MSN | 0.58 | Yes | |
| hsa-miR-26b | −1.24 | HNRNPA2B1 | 0.26 | Yes | |
| hsa-miR-26b | −1.24 | EIF3I | 0.43 | Yes | |
| hsa-miR-103 | −1.27 | HNRNPA2B1 | 0.26 | Yes | |
| hsa-miR-103 | −1.27 | ERLIN2 | 0.28 | Yes | |
| hsa-miR-103 | −1.27 | EZR | 0.38 | Yes | |
| hsa-miR-103 | −1.27 | TPM1 | 1.87 | Yes | Yes |
| hsa-miR-146b-5p | −1.41 | ERLIN2 | 0.28 | Yes | |
| hsa-miR-146b-5p | −1.41 | HNRNPD | 0.42 | Yes | Yes |
| hsa-miR-146b-5p | −1.41 | PA2GA | 0.45 | Yes | |
| hsa-miR-138 | −1.68 | ERLIN2 | 0.28 | Yes | |
| hsa-miR-138 | −1.68 | ACTN4 | 0.34 | Yes | |
| hsa-miR-138 | −1.68 | EZR | 0.38 | Yes | |
| hsa-miR-138 | −1.68 | MSN | 0.58 | Yes | |
| hsa-miR-34a | −2.39 | HSPA5 | 0.26 | Yes | |
| hsa-miR-34a | −2.39 | TUBB | 0.55 | Yes |
Of the 28 proteins in Table 2, 11 were predicted targets of at least three miRNAs. No significant differences were observed between the magnitude of expression changes of these 11 proteins and the proteins that were predicted targets of one or two miRNAs each.
In addition to translational inhibition, miRNA can accelerate mRNA degradation, leading to reciprocal expression of miRNAs and target mRNAs. Moreover, miRNAs can sometimes be co-regulated with target mRNAs possibly due to compensatory changes in the transcription of target mRNAs. The differentially expressed miRNAs we identified were compared with mRNA abundance data from the study by Hills, et al. (28). The comparison indicated several pairs of miRNAs and predicted target mRNAs that were reciprocally regulated or co-regulated by TGFβ1 (Supplementary Table S4).
miR-382 contributed to TGFβ1-induced loss of epithelial characteristics of human renal cells
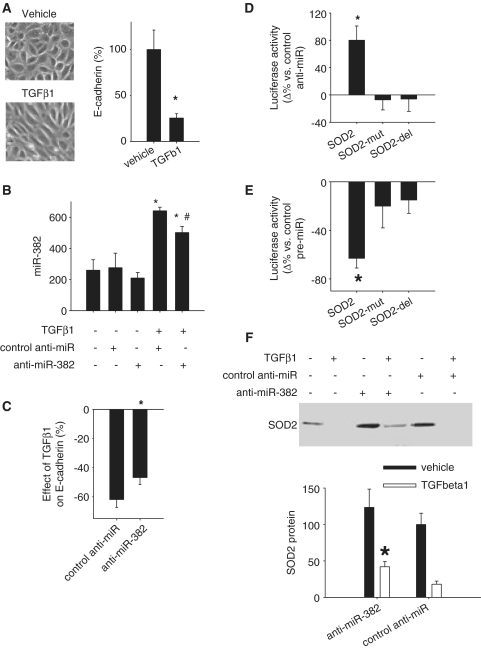
Treatment of HK2 cells with TGFβ1 induced elongation of the shape of the cells and a substantial loss of E-cadherin expression, a marker of epithelial cells (Figure 3A). The loss of epithelial characteristics is typical of the early phase of epithelial–mesenchymal transition, a process importantly involved in the development of renal interstitial fibrosis (21,22).
Figure 3.
miR-382 targeted SOD2 and contributed to TGFβ1-induced loss of epithelial characteristics in human renal epithelial cells. (A) TGFβ1 induced loss of epithelial characteristics in HK2 cells as indicated by typical changes in cell morphology and a suppression of E-cadherin mRNA, an epithelial marker. TGFβ1 was used at 3 ng/ml for 48 h. N = 6, *P < 0.05. (B) Knockdown of miR-382. HK2 cells were treated with vehicle or TGFβ1 in the presence of LNA anti-miR-382 or LNA scrambled control anti-miR (100 nM). n = 6, *P < 0.05 vs. no treatment; #P < 0.05 versus TGFβ1 plus control anti-miR. (C) Knockdown of miR-382 attenuated the suppression of E-cadherin mRNA by TGFβ1. n = 6–9, *P < 0.05 versus control anti-miR. (D) miR-382 interacted with the 3′-UTR of SOD2 in Hela cells. Hela cells were transfected with luciferase reporter constructs containing a 3′-UTR segment of SOD2 or a mutated (SOD2-mut) or partially deleted (SOD2-del) segment. The mutations and deletion were described in the text. The effect of anti-miR-382 (100 nM) compared to control, scramble anti-miR was shown. n = 4, *P < 0.05 versus control anti-miR. (E) miR-382 interacted with 3′-UTR of SOD2 in HK2 cells. HK2 cells were transfected with luciferase reporter constructs as described above. The effect of pre-miR-382 (100 nM), a miR-382 mimic, compared with control, scramble pre-miR was shown. n = 4, *P < 0.05 versus control pre-miR. (F) SOD2 expression was suppressed by TGFβ1, but partially restored by knockdown of miR-382. miR-382 in TGFβ1-treated HK2 cells was knocked down with LNA anti-miR (see Figure 3B). n = 6, *P < 0.05 vs. control anti-miR.
miR-382 was substantially upregulated by TGFβ1 in HK2 cells (Figure 1). LNA anti-miR-382, compared with LNA scrambled anti-miR, modestly but significantly knocked down the expression level of miR-382 in HK2 cells treated with TGFβ1 (Figure 3B). The modesty of the knockdown could be in part due to low transfection efficiency as HK2 cells are difficult to transfect. Importantly, the partial knockdown of miR-382 resulted in significant attenuation of TGFβ1-induced loss of E-cadherin (Figure 3C), indicating that miR-382 upregulation contributed to TGFβ1-induced loss of epithelial characteristics.
miR-382 targeted SOD2
The protein abundance of five predicted miR-382 targets, SOD2, NPM1, PSPC1, HSPD1 and ECH1, was reduced by TGFβ1 (Tables 1 and 2). We cloned a segment of the 3′-UTR of SOD2. The segment included the predicted miR-382 binding site (Table 3). The 3′-UTR segment was inserted into an expression construct downstream from a luciferase reporter gene and transfected into Hela cells. Luciferase activities were significantly increased when endogenous miR-382 was inhibited by an anti-miR, compared with a control anti-miR (Figure 3D). Anti-miR-382 did not have significant effects on luciferase activity when the seed region binding site was mutated or partially deleted (Figure 3D). Anti-miR-382 also increased luciferase activities when the luciferase gene was linked to a 3′-UTR segment cloned from NPM1, PSPC1, HSPD1 or ECH1 (Table 3). The increase ranged from 1.33- to 3.01-fold at 24 or 48 h after transfection (n = 4, two-way ANOVA P < 0.05 for each target gene). It suggested that endogenous miR-382 in HeLa cells suppressed the expression of luciferase gene by interacting with the 3′-UTR of each of the five predicted targets.
Table 3.
3′-UTR reporter constructs
| Target | miR-382 target site | _P_-value | 3′-UTR segment | primer sequences |
|---|---|---|---|---|
| SOD2 | 120 to 136 | 0.015 | 27 to 289 | Forward: 5-AAGCTCTTTATGACTGTTTTTGTAG-3; |
| Reverse: 5-ATGGATGGAATATTTTTGATGG-3 | ||||
| NPM1 | 1 to 20 | 0.048 | −66 to 144 | Forward: 5-AATTGCTTCCGGATGACTGAC-3; |
| Reverse: 5-CTTGGCAATAGAACCTGGACA-3 | ||||
| PSPC1 | 284 to 303 | 0.011 | 58 to 353 | Forward: 5-TTTACCTGTTATCTGGAAGAAATG-3; |
| Reverse: 5-TACAGCAAAGTTTAGTAAGACCGT-3 | ||||
| HSPD1 | 56 to 76 | 0.010 | 18 to 424 | Forward: 5-TACCTTTATTAATGAACTGTGAC-3; |
| Reverse: 5-CATAATTGGATACTTCTCTACTTTGT-3 | ||||
| ECH1 | 102 to 124 | 0.002 | 36 to 185 | Forward: 5-GCCTTGTCCCGCCTCAT-3; |
| Reverse: 5-ACTTTGCTTTATTGTTTGTGG-3 |
In HK2 cells, a miR-382 mimic significantly decreased luciferase activity when the cells were transfected with the reporter construct containing the 3′-UTR segment of SOD2. Again, mutations or partial deletion in the seed region-binding site abrogated the effect of the miR-382 mimic (Figure 3E). A miR-382 mimic, instead of an anti-miR, was used in these experiments because basal levels of miR-382 in HK2 cells that were not treated with TGFβ1 were not further reduced by the anti-miR (Figure 3B).
miR-485-3p and miR-200a* were two other miRNAs predicted to regulate SOD2 and showing reciprocal regulation by TGFβ1 compared with SOD2 (Table 2). SOD2 3′-UTR reporter analysis, however, showed that anti-miR-485-3p, compared with control anti-miR, did not significantly change luciferase activities in Hela cells [7 ± 27%, n = 4, non-significant (NS)] or HK2 cells (5 ± 20%, n = 4, NS). Similarly, anti-miR-200a* did not significantly change luciferase activities (−2 ± 7% in Hela cells, −18 ± 13% in HK2 cells, n = 5, NS). The 3′-UTR construct used for the miR-382 experiment (Table 3) contained a predicted miR-485-3p binding site in SOD2 transcript variant 1 (NM_000636) and was used for the miR-485-3p experiment. A separate 3′-UTR construct containing a predicted miR-200a* binding site was used for the miR-200a* experiment. The construct spanned nucleotides 20–199 of SOD2 transcript variant 3 (NM_001024466).
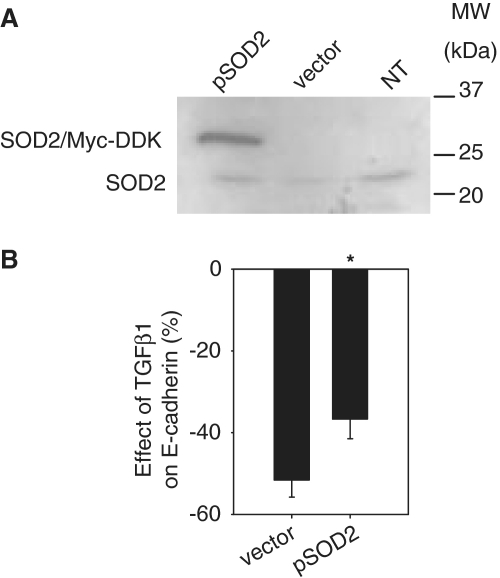
Knockdown of miR-382 significantly attenuated TGFβ1-induced downregulation of SOD2 (Figure 3F). A myc-DDK-tagged construct, driven by a CMV promoter, was used to overexpress SOD2 in HK2 cells (Figure 4A). Overexpression of SOD2 attenuated TGFβ1-induced loss of E-cadherin (Figure 4B). The data suggested that SOD2 was protective against TGFβ1-induced loss of epithelial characteristics. In TGFβ1-treated cells, endogenous SOD2 was suppressed in part by upregulated miR-382.
Figure 4.
SOD2 was protective against TGFβ1-induced loss of epithelial characteristics. (A) Overexpression of SOD2 with a Myc-DDK-tagged plasmid (pSOD2). HK2 cells were not transfected (NT) or were transfected with pSOD2 or an empty vector (2 µg for each 3.5-cm dish). Western blot was performed 24 h later using an SOD2 antibody. (B) Overexpression of SOD2 attenuated TGFβ1-induced downregulation of E-cadherin. n = 5, *P < 0.05.
DISCUSSION
We have provided experimental evidence in the form of reciprocal expression at the protein level for 77 predicted miRNA-target pairs, five of which were further validated with 3′-UTR reporter analyses. Moreover, we demonstrated a novel role of miR-382 and SOD2 in TGFβ1-induced loss of epithelial characteristics in human kidney cells.
The 77 pairs represent a substantial expansion of human miRNA-target pairs that are supported by reciprocal expression with the target examined at the protein level. Many studies of miRNA function would begin by identifying miRNAs that are differentially expressed under an experimental or disease condition. The next step is to identify target genes for the differentially expressed miRNAs. Definitive evidence for a miRNA-target pair consists of sequence characteristics, reciprocal expression, and 3′-UTR response. MiRNA-target pairs such as those reported in the present study would help to substantially narrow down the long list of sequence-based, predicted targets. It could accelerate miRNA studies by enabling investigators to focus on high-probability targets when performing laborious 3′-UTR and functional analysis. In addition, substantial expansion of experimentally supported miRNA-target pairs will provide an increasingly larger basis upon which algorithms for miRNA target prediction could be improved (29).
Our findings highlight the importance of considering multiple factors when studying the effect of an miRNA. We were able to confirm 5 of the 77 predicted pairs, all of which involved miR-382. However, two predicted pairs involving miR-485-3p and miR-200a* regulation of SOD2 were not confirmed. miR-485-3p had a large fold change following the TGFβ1 treatment, but its abundance, estimated from the PCR data, was approximately one order of magnitude lower than miR-382. miR-200a* appeared more abundant than miR-382, yet its change in response to TGFβ1 was much smaller. In addition, transcriptional regulation cannot be ruled out for some of the target genes. Many factors, including abundance, relative change, binding kinetics, sub-cellular localization, characteristics of specific target sequences and the cellular milieu, could conceivably influence the effect of a miRNA.
The approach of identifying miRNA-target pairs by simultaneous analysis of miRNA and proteomic profiles under physiological conditions was developed by our laboratory recently (20). The approach may be less specific compared with studies using experimental overexpression of an individual miRNA (32–36). The advantage of our approach is that we are detecting physiological or pathophysiological up- and downregulation of miRNAs and their target proteins. High-throughput analysis of miRNA targets has been performed mostly at the mRNA level (9,29–31). Current technologies for quantitative proteomics provide only a partial coverage of a cellular proteome, but have the significant advantage of examining the final effect of miRNA action since some miRNAs could inhibit translation without altering mRNA abundance. The DIGE/mass spectrometry approach used in the present study is limited in its ability to detect membrane proteins, low-abundance proteins and proteins with extreme molecular weights or isoelectric points. However, the approach has exceptional power for comparative and quantitative analysis compared with other proteomic approaches.
The present study was the first to characterize miRNA and proteomic responses to TGFβ1 in renal epithelial cells. Responses of miRNAs or proteomes to TGFβ1 have been examined by several recent studies of metastatic epithelial–mesenchymal transition (38–44). Effect of TGFβ1 on miRNAs in glomerular mesangial cells has also been reported (45). There are some overlaps between the result of the present study and studies of metastatic models, including downregulation of miR-192 and mitochondrial 60 kD heat-shock protein (HSPD1) and upregulation of TPM1, meosin (MSN) and tubulin α1 chain (TUBA1B) (37,40,41,44). Most of the miRNAs and proteins identified in the present study, however, have not been reported as responsive to TGFβ1 in metastatic models. It suggests that much of the effect of TGFβ1 observed in the present study might be specific to renal tubulointerstitial fibrosis.
The role of miR-382 and SOD2 in TGFβ1-induced loss of epithelial characteristics was a novel finding. Mitochondrial oxidative stress has been reported to be involved in epithelial–mesenchymal transition in renal epithelial cells (46). Renal oxidative stress is a prominent feature of models of chronic renal injury including the Dahl salt-sensitive rat, which exhibits significant signs of epithelial–mesenchymal transition (23). We have now shown that TGFβ1 downregulates SOD2, an enzyme important for the protection against mitochondrial oxidative stress, while overexpression of SOD2 is protective against TGFβ1-induced loss of epithelial characteristics. Moreover, the downregulation of SOD2 induced by TGFβ1 is at least in part mediated by upregulation of miR-382. These novel findings reveal a possible, new mechanism of epithelial–mesenchymal transition that involves upregulation of miR-382, leading to SOD2 downregulation and oxidative stress. The interplay between this new mechanism and other molecular pathways known to be involved in epithelial mesenchymal transition (21,22), as well as the in vivo significance of this new pathway, warrants further investigation in future studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (HL077263, HL085267, and DK084405 to M.L., N01-HV-28182 to A.S.G., HL082798, HL029587, and HL081091). Funding for open access charge: National Institutes of Health grants.
Conflict of interest statement. None declared.
Supplementary Material
Supplementary Data
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 5.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 6.Meister G. miRNAs get an early start on translational silencing. Cell. 2007;131:25–28. doi: 10.1016/j.cell.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 7.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 8.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 9.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 10.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Taylor NE, Lu L, Usa K, Cowley AW, Jr, Ferreri NR, Yeo NC, Liang M. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension. 2010;55:974–982. doi: 10.1161/HYPERTENSIONAHA.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couzin J. MicroRNAs make big impression in disease after disease. Science. 2008;319:1782–1784. doi: 10.1126/science.319.5871.1782. [DOI] [PubMed] [Google Scholar]
- 13.Makeyev EV, Maniatis T. Multilevel regulation of gene expression by microRNAs. Science. 2008;319:1789–1790. doi: 10.1126/science.1152326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang M, Liu Y, Mladinov D, Cowley AW, Jr, Trivedi H, Fang Y, Xu X, Ding X, Tian Z. MicroRNA: a new frontier in kidney and blood pressure research. Am. J. Physiol. Renal Physiol. 2009;297:F553–F558. doi: 10.1152/ajprenal.00045.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang M. MicroRNA: A new entrance to the broad paradigm of systems molecular medicine. Physiol. Genomics. 2009;38:113–115. doi: 10.1152/physiolgenomics.00080.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krutzfeldt J, Poy MN, Stoffel M. Strategies to determine the biological function of microRNAs. Nat. Genet. 2006;38(Suppl.):S14–S19. doi: 10.1038/ng1799. [DOI] [PubMed] [Google Scholar]
- 17.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2008;38(Suppl.):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 20.Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M. microRNA-target pairs in rat kidneys identified through microRNA microarray, proteomic, and bioinformatic analysis. Genome Res. 2008;18:404–411. doi: 10.1101/gr.6587008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J. Clin. Invest. 2003;112:1776–1784. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y. Epithelial to mesenchymal transition in renal fibrogenesis: pathologic significance, molecular mechanism, and therapeutic intervention. J. Am. Soc. Nephrol. 2004;15:1–12. doi: 10.1097/01.asn.0000106015.29070.e7. [DOI] [PubMed] [Google Scholar]
- 23.Mori T, Polichnowski A, Glocka P, Kaldunski M, Ohsaki Y, Liang M, Cowley AW., Jr High perfusion pressure accelerates renal injury in salt-sensitive hypertension. J. Am. Soc. Nephrol. 2008;19:1472–1482. doi: 10.1681/ASN.2007121271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian Z, Greene AS, Usa K, Matus IR, Bauwens J, Pietrusz JL, Cowley AW, Jr, Liang M. Renal regional proteomes in young Dahl salt-sensitive rats. Hypertension. 2008;51:899–904. doi: 10.1161/HYPERTENSIONAHA.107.109173. [DOI] [PubMed] [Google Scholar]
- 25.Tian Z, Liu Y, Usa K, Mladinov D, Fang Y, Ding X, Greene AS, Cowley AW, Jr, Liang M. A novel role of fumarate metabolism in Dahl salt-sensitive hypertension. Hypertension. 2009;54:255–260. doi: 10.1161/HYPERTENSIONAHA.109.129528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang M, Pietrusz JL. Thiol-related genes in diabetic complications: a novel protective role for endogenous thioredoxin 2. Arterioscler. Thromb. Vasc. Biol. 2007;27:77–83. doi: 10.1161/01.ATV.0000251006.54632.bb. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Mladinov D, Pietrusz JL, Usa K, Liang M. Glucocorticoid response elements and 11β-hydroxysteroid dehydrogenases in the regulation of endothelial nitric oxide synthase expression. Cardiovasc. Res. 2009;81:140–147. doi: 10.1093/cvr/cvn231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hills CE, Willars GB, Brunskill NJ. Proinsulin C-peptide antagonizes the profibrotic effects of TGF-beta1 via up-regulation of retinoic acid and HGF-related signaling pathways. Mol. Endocrinol. 2010;24:822–831. doi: 10.1210/me.2009-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 31.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 32.Vinther J, Hedegaard MM, Gardner PP, Andersen JS, Arctander P. Identification of miRNA targets with stable isotope labeling by amino acids in cell culture. Nucleic Acids Res. 2006;34:e107. doi: 10.1093/nar/gkl590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc. Natl. Acad. Sci. USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 36.Grosshans H, Filipowicz W. Proteomics joins the search for microRNA targets. Cell. 2008;134:560–562. doi: 10.1016/j.cell.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Zheng Q, Safina A, Bakin AV. Role of high-molecular weight tropomyosins in TGF-beta-mediated control of cell motility. Int. J. Cancer. 2008;122:78–90. doi: 10.1002/ijc.23025. [DOI] [PubMed] [Google Scholar]
- 38.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 40.Keshamouni VG, Michailidis G, Grasso CS, Anthwal S, Strahler JR, Walker A, Arenberg DA, Reddy RC, Akulapalli S, Thannickal VJ, et al. Differential protein expression profiling by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype. J. Proteome Res. 2006;5:1143–1154. doi: 10.1021/pr050455t. [DOI] [PubMed] [Google Scholar]
- 41.Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zavadil J, Narasimhan M, Blumenberg M, Schneider RJ. Transforming growth factor-beta and microRNA:mRNA regulatory networks in epithelial plasticity. Cells Tissues Organs. 2007;185:157–161. doi: 10.1159/000101316. [DOI] [PubMed] [Google Scholar]
- 45.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc. Natl. Acad. Sci. USA. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang A, Jia Z, Guo X, Yang T. Aldosterone induces epithelial-mesenchymal transition via ROS of mitochondrial origin. Am. J. Physiol Renal Physiol. 2007;293:F723–F731. doi: 10.1152/ajprenal.00480.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data