A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila (original) (raw)
. Author manuscript; available in PMC: 2013 Dec 6.
Summary
A large fraction of our genome consists of mobile genetic elements. Governing transposons in germ cells is critically important, and failure to do so compromises genome integrity, leading to sterility. In animals, the piRNA pathway is the key to transposon constraint, yet the precise molecular details of how piRNAs are formed and how the pathway represses mobile elements remain poorly understood. In an effort to identify general requirements for transposon control and novel components of the piRNA pathway, we carried out a genome-wide RNAi screen in Drosophila ovarian somatic sheet cells. We identified and validated 87 genes necessary for transposon silencing. Among these were several novel piRNA biogenesis factors. We also found CG3893 (asterix) to be essential for transposon silencing, most likely by contributing to the effector step of transcriptional repression. Asterix loss leads to decreases in H3K9me3 marks on certain transposons but has no effect on piRNA levels.
Introduction
Transposable elements populate virtually every eukaryotic genome. Although their presence can impart some benefits, when not properly controlled, transposons can compromise the genomic integrity of their host and its offspring. Hence, in all higher animals there are control mechanisms in place, which prevent wholesale transposon mobilization (Malone and Hannon, 2009).
In Drosophila, the principal pathway that protects the inheritable genome is comprised of a catalogue of small RNAs that interact with an animal-specific clade of Argonaute family proteins, the Piwi proteins (Ishizu et al., 2012). This pool of Piwi-interacting RNAs (piRNAs), which contains millions of distinct sequences bearing homology to transposable elements, functions as a molecular memory of transposon identity (Brennecke et al., 2007). Using their bound piRNAs as a guide, Piwi proteins (Piwi, Aubergine and Argonaute3) recognize and silence their targets. Failure of this pathway leads to defects in germline development and to sterility (Khurana and Theurkauf, 2010).
Genetic studies have uncovered a number of loci that are essential for the proper function of the piRNA pathway. Besides the core proteins of the Piwi clade, flamenco has long been implicated in transposon control. This discrete locus on the X-chromosome of Drosophila was found to be a major determinant for silencing of the retroelement gypsy almost two decades ago, though its underlying molecular nature remained mysterious (Bucheton, 1995). Sequencing small RNAs bound to Piwi proteins and mapping these sequenced reads back to the genome revealed the true nature of the flamenco locus. Rather than being a protein coding gene, the flamenco transcript is the precursor to the majority of piRNAs expressed in the follicle cells of the ovary (Brennecke et al., 2007). This and other sites of abundant piRNA generation were termed piRNA clusters. What mechanisms mark flamenco and other cluster transcripts for processing into piRNAs remains largely unclear. Several studies have shed some light on this topic by identifying some of the protein factors that play a role in piRNA cluster transcription and transport. Rhino and Cutoff, as well as histone methylation marks deposited by Eggless (EGG), are necessary for cluster transcription (Klattenhoff et al., 2009; Pane et al., 2011; Rangan et al., 2011). In addition, UAP56, a helicase implicated in splicing and RNA export, was found to bind a germline piRNA cluster RNA and may escort the transcript from the nucleus to the nuage for processing (Zhang et al., 2012). Intriguingly, Rhino, Cutoff and UAP56 all were reported to be specific to germline piRNA clusters, leaving factors involved in somatic cluster determination a mystery.
Mutagenesis screens for sterility phenotypes in Drosophila also highlighted factors that later were found to be elements of the piRNA pathway (Schüpbach and Wieschaus, 1989, 1991). Molecular analyses have begun to place these factors at specific steps of the pathway, such as being required for biogenesis or downstream silencing of transposons. However, very little is known about specific biochemical functions or enzymatic activities of the proteins that act at each step. One major insight came from the discovery of a trimming activity in insect cell lysates, which shortens the 3′ end of putative piRNA precursors to their mature length (Kawaoka et al., 2011). However, the protein responsible for this activity remains unknown. Biochemical and structural studies of Zucchini (ZUC), which was previously implicated in the piRNA pathway, suggests it as a promising candidate for the nuclease that creates the 5′ ends of primary piRNAs (Ipsaro et al., 2012; Nishimasu et al., 2012). Whether Zucchini and a trimming enzyme together comprise the complete primary biogenesis machinery or whether other endo- or exonucleolytic cleavage events create intermediates that are further matured remains unknown.
Another enigmatic aspect of the pathway is precisely how Piwi-piRNA complexes silence their targets. In the case of somatic cells of the ovary it has become clear that control of transposons occurs at the transcriptional level through Piwi-directed deposition of epigenetic marks. Recently, three conclusive studies showed that upon Piwi depletion transposons engage in active transcription and show a depletion of H3K9 trimethylation (H3K9me3) (Le Thomas et al., 2013; Rozhkov et al., 2013; Sienski et al., 2012). In one of these studies, the authors place Maelstrom (MAEL) at the effector step of transcriptional repression (Sienski et al., 2012). Interestingly, mael derepresses transposons without preventing H3K9me3 deposition, indicating that this modification may not be the definitive silencing mark. What the final silencing mark may be, and what proteins are responsible for establishing these repressed states has yet to be determined.
Much of what has been learned of the piRNA pathway that operates in follicle cells relied on the use of a cultured ovarian somatic sheet (OSS) cell line (Niki et al., 2006). This cell line only expresses Piwi but not AUB or AGO3 (Lau et al., 2009; Saito et al., 2009). With an active primary piRNA pathway in place, genetic requirements for primary piRNA biogenesis and transposon silencing can be investigated.
Here, we describe a genome-wide screen that builds a foundation for addressing many open questions relevant to piRNA biogenesis and effector functions. By individually assaying more than 41,000 long double-stranded RNAs (dsRNAs) targeting every annotated gene in the Drosophila genome and examining their effect on transposons levels we describe a comprehensive genetic framework for transposon control. We reveal novel piRNA biogenesis factors and place previously uncharacterized proteins at the effector step of the pathway.
Results
An RNAi Screen for elements of the somatic piRNA pathway
In order to assay transposon derepression upon knockdown of any given target gene, we established a sensitive assay for gypsy mRNA levels. Based on quantitative PCR (qPCR) with hydrolysis probes (Taqman), this assay specifically detects the spliced subgenomic transcript of the retrotransposon (Figure 1A). The expression of this transcript is known to be highly sensitive to disruption of the piRNA pathway, even more so than its unspliced counterpart (Pelisson et al., 1994).
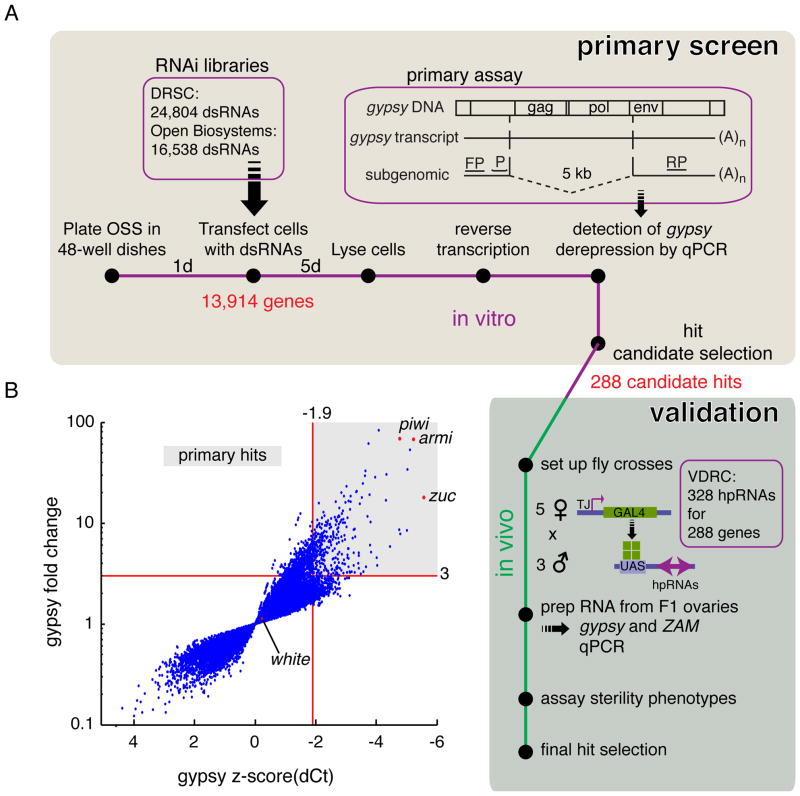
Figure 1. A genome-wide RNAi screen for piRNA pathway components acting in the somatic compartment of Drosophila ovaries.
A) A workflow of the primary RNAi screen in ovarian somatic sheet cells (OSS) and validation of primary hit candidates in vivo is shown. Each gene in the Drosophila genome was knocked down with one or more dsRNAs. 5 days after transfection, cells were tested for increased levels of the gypsy retrotransposon. The primers and the hydrolysis probe used for the qPCR are shown (FP: forward primer, P: hydrolysis probe, RP: reverse primer). The dashed line indicates the ~5kb segment not present in the subgenomic transcript. 288 genes were further tested in vivo using the Gal4/UAS system to drive hairpin RNAs (hpRNAs) within the traffic jam (TJ) expression domain.
B) All transfected wells were assayed for levels of gypsy and one reference gene for normalization. Levels of gypsy expression are displayed as z-scores and fold change. The cutoffs for both z-score (<−1.9) and fold change (>3) are indicated as red lines. The shaded area shows the selection of primary hit candidates. Three positive controls (piwi, armi, zuc) and one negative control (white) are marked as red dots. Only wells that passed the filter for primary datapoint selection are shown. For all primary datapoints see Table S1. See also Figure S1.
Knockdown of target genes was accomplished by transfecting dsRNAs from two independent genome-wide libraries with a total of 41,342 dsRNAs. The average count of dsRNAs per gene was 2.28, targeting 13,914 genes with valid IDs in Flybase (McQuilton et al., 2012). Additionally, the two libraries contained 1,045 negative controls, 2,097 dsRNAs without an annotated target and 2,301 dsRNAs targeting the Heidelberg collection of predicted genes.
We transfected OSS cells with individual dsRNAs in 48-well plates, lysed the cells five days later and used the lysate for reverse transcription (Figure 1A). The qPCR results were normalized to their respective plate using z-scores (Ramadan et al., 2007). As a secondary metric we calculated the fold change in relation to the median. The knockdown of piwi in this setting led to a gypsy mRNA signal that was detectable much earlier by qPCR than the average of the plate. In four biological replicates of piwi knockdown, the average normalized signal for gypsy was almost five standard deviations away from the median of its plate (Figure S1A). Hence, our assay for transposon derepression is both sensitive and robust, at least given that the RNAi trigger is of good quality. When comparing several independent dsRNAs, we saw that there was considerable variance in this respect. dsRNAs against known components of the pathway, such as armitage (armi), led to consistent derepression of gypsy, however, levels varied from 25 to 70 fold (Figure S1B). Since we assayed several dsRNAs against each annotated gene, we felt confident that the majority of pathway components could be identified.
Out of 41,342 tested dsRNAs, 33,780 met our criteria for inclusion in our analysis, of which 320 dsRNAs met the criteria for primary hit selection (Figure 1B, Table S1). Included in this list were 18 dsRNAs without annotated targets, which were disregarded. All genes that were previously implicated in gypsy control were outliers in the primary screen (Figure S1B). Knockdown of pathway components such as capsuleen, hen1, egg, or squash was not expected to cause strong gypsy derepression based on existing literature and indeed did not cause derepression of gypsy in our assay (Olivieri et al., 2010; Rangan et al., 2011). After choosing z-score and fold change cutoffs for hit selection based on 217 GFP negative controls, only 3 out of 645 (0.5%) additional negative controls scored as weak hits (Figure S1C).
To ask whether genes that affect transposon control show preference towards specific annotation groups, we performed functional enrichment analysis on our primary dataset. After multiple test correction, 215 functions were associated with transposon silencing (corrected p<0.05), many with strong potential relevance and very significant enrichment (the top 20 functions have a corrected p<1E-6, Table S2). Among the most significant, we find expected cellular components like the ‘Yb body’, but also more surprising functions like ‘regulation of growth of symbiont in host’. While genes implicated in the piRNA pathway drive several of these enrichments, all scoring highly in the primary screen, novel pathway components intersect with these in some of the enriched functions. For example under ‘dorsal appendage formation’ smt3 (SUMO) joins armi and zuc (Nie et al., 2009). Further work will be necessary to determine to what extent these unexpected intersections relate to biologically relevant connections.
Next, we compared protein interaction data to the full ranked fold change list. For every gene in the genome, the degree to which that gene’s interaction partners scored highly in the fold change list was measured (as ROC). Of the top 20 genes with interaction partners significantly elevated in the fold change list, 18 are annotated as belonging to the proteasomal complex (which had 65 genes in total). This observation was remarkably significant (p<1E-40 after multiple test correction), which may be partially due to the correlated interaction profiles of the proteasome complex. The two remaining genes not belonging to the proteasomal complex were bx42, a homolog of mammalian Skip (SKI-interacting protein), a protein implicated in splicing, and calypso, a Histone 2A deubiquitinase (Makarov et al., 2002; Scheuermann et al., 2010). Both genes are highly expressed in ovaries according to the modENCODE tissue expression data. Whether their interaction with particularly high scoring genes is indicative of any regulatory function remains to be tested.
In vivo validation of primary hits
We obtained 328 fly lines from the Vienna Drosophila Resource Center (VDRC) containing inducible hairpin RNAs (hpRNAs) against 288 of our primary hit candidates (Dietzl et al., 2007). When crossed to females expressing a follicle cell specific Gal4 driver (traffic jam), these hpRNAs can effectively knockdown any given target gene within the same expression domain (Tanentzapf et al., 2007). Using hpRNAs against aub as a negative control, we observed highly significant changes in gypsy expression by qPCR when knocking down armi (Figure S2A). We confirmed the effect on the piRNA pathway by measuring two additional transposons, ZAM and gypsy3. All known pathway components that were identified as primary hits were validated using this approach except for Piwi, which showed developmental defects upon knockdown. (Figure S2B). Harnessing this in vivo system, we validated 87 out of the 288 primary hit candidates (Figure 2A, Table S3). In order to be considered as validated, knockdown of the target gene had to result in upregulation of gypsy or ZAM by at least two fold. For crosses with male flies originating from the GD library from VDRC we used ZAM as a metric, for the KK library we measured gypsy levels. This decision was based on the finding that negative controls from the GD library already showed higher basal levels of gypsy subgenomic transcript when compared to the KK library negative controls (Figure S3A–C).
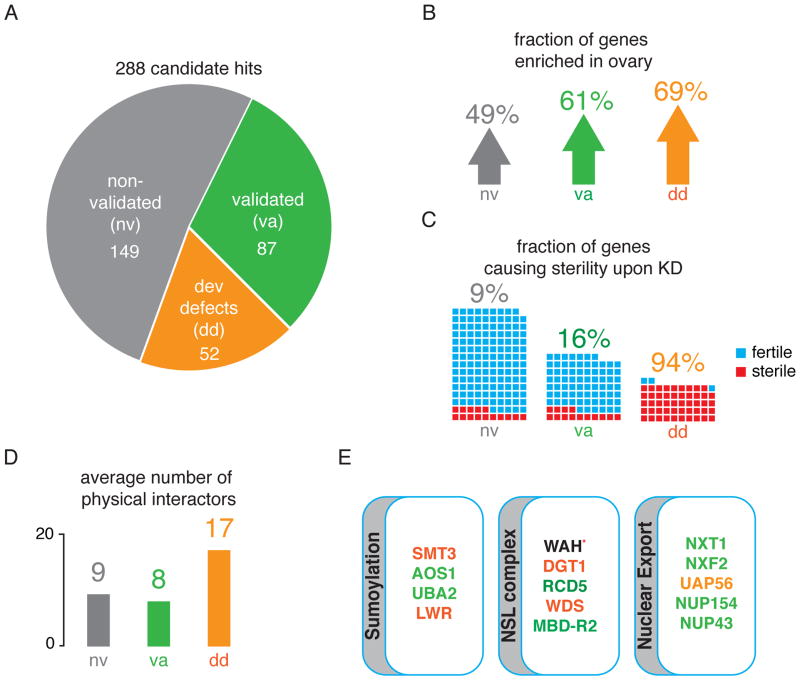
Figure 2. Primary candidates were validated in vivo.
A) The number of hit candidates that validated (va) or did not validate (nv) in vivo is shown. Genes that upon knockdown caused severe developmental defects and therefore could not be assayed are also indicated (dd). A full list of validated fly lines and corresponding transposon derepression information is available in Table S3.
B) Validated hits are preferentially expressed in ovaries. The percentage of genes that are enriched in ovaries compared to whole fly is shown for the three categories. This data is based on mRNA signals on Affymetrix expression arrays available from FlyAtlas (Chintapalli et al., 2007).
C) The fraction of genes causing sterility upon knockdown is shown. Each small box represents one gene, with blue and red indicating if flies were fertile or sterile upon knockdown.
D) The degree to which a gene may represent a node in a network in each of the classifications was measured by number of physical interactors. Interaction data from BIOGRID was used for this analysis.
E) Components of the Drosophila sumoylation pathway, the Nonspecific Lethal Complex, and proteins involved in nuclear export are primary hits that validate in vivo. WAH could not be validated in vivo because no RNAi fly was available at the time of submission (red asterisk). The text coloring of each gene indicates the result of the validation screen and is consistent with the categories in panel A.
See also Figure S2.
Out of the 288 candidate hits, knockdown of 52 genes, including Piwi, led to such severe developmental defects, that dissection of ovaries and confirmation of the initial screen result was not possible. However, several arguments can be made for this category to harbor a substantial number of ‘true’ hits. First, the genes in this category had an average gypsy fold change of 5.8 in the primary dataset, as compared to 3.3 for the primary hits that failed secondary validation. This average fold change was even higher than the validated subset (5.1 fold). Since primary fold changes as well as z-scores are a function of precision (the likelihood of a primary hit to be validated), this is indicative of the biological relevance of these hits (Figure S2C). Second, while the non-validated genes had a representation count in the dsRNA libraries of 2.84, the developmental defect set had a count of 2.42, which is significantly different for the two sets (p~0.0035, Wilcoxon test). Thus, the genes of the developmental defect category were disadvantaged according to their annotation class with respect to their possibility to be a primary hit by chance in comparison to the candidates that failed validation, yet they had a much higher average fold change.
Both the validated and developmental defect sets were significantly enriched for genes with higher expression levels in ovaries as compared to whole fly (Figure 2B). While knockdown of genes within the non-validated category only led to sterility of 9% of the crosses, the fraction was 16% for the validated and 94% for the developmental defect set (Figure 2C). The extreme phenotypes we observe in the developmental set could imply more generic functions for these genes than simply action in the piRNA pathway. Indeed, around 90% have an average of 17 physical interactors, which is significantly higher than the other categories (Figure 2D).
When ranked by their fold changes in the validation screen, the top 20 genes reveal expected and novel associations. The majority of the somatic piRNA pathway components were among the strongest hits (Table 1). nxt1, a nuclear export factor, ranks first with gypsy levels almost 2,500 fold higher than the negative control (Figure 2E) (Herold et al., 2001). In addition, depletion of NXT1 also led to sterility. Knockdown of the RNA helicase uap56, which acts in the same export pathway (Herold et al., 2003), showed derepression of gypsy of up to 8-fold in the primary screen. First implicated in splicing, uap56 was recently shown to be involved in transport of the primary piRNA transcript of dual-stranded clusters to the nuclear pore (Zhang et al., 2012). The knockdown of uap56 in follicle cells affected germline development to such an extent that in vivo verification of the primary screen results was not possible. We were able to validate another mRNA export factor, nxf2, which interacts with nxt1 (Table S3) (Herold et al., 2001).
Table 1.
Top 20 validated hits
| FlyBaseID | Symbol | Primary screen average fold change | Validation screen fold change | Fertility | Comments | ||
|---|---|---|---|---|---|---|---|
| Gypsy | Zam | Gypsy3 | |||||
| FBgn0028411 | nxt1 | 2 | 2452 | 3566 | 41 | − | Involved in mRNA export from nucleus |
| FBgn0000928 | fs(1)Yb | 11 | 96 | 700 | 335 | + | piRNA pathway component |
| FBgn0041164 | armi | 48 | 197 | 846 | 112 | + | piRNA pathway component |
| FBgn0261266 | zuc | 19 | 809 | 549 | 9 | + | piRNA pathway component |
| FBgn0263143 | vret | 4 | 74 | 315 | 22 | + | piRNA pathway component |
| FBgn0036826 | CG3893 (asterix) | 42 | 80 | 207 | 10 | + | Contains two CHHC zinc fingers |
| FBgn0016034 | mael | 3 | 159 | 452 | 16 | + | piRNA pathway component |
| FBgn0033273 | CG2183 | 4 | 173 | 158 | 11 | + | Fly homolog of GASZ |
| FBgn0029800 | lin-52 | 5 | 153 | 153 | 8 | + | dREAM complex subunit |
| FBgn0038016 | MBD-R2 | 16 | 85 | 48 | 4 | + | NSL complex subunit |
| FBgn0029113 | uba2 | 3 | 84 | 12 | 8 | + | Sumoylation E1 ligase |
| FBgn0034617 | CG9754 | 2 | 26 | 61 | 14 | + | No conserved domains |
| FBgn0027499 | wde | 3 | 40 | 120 | 12 | + | Co-factor of Eggless |
| FBgn0021761 | nup154 | 6 | 30 | 186 | 3 | + | Structural constituent of nuclear pore |
| FBgn0003612 | Su(var)2-10 | 2 | 9 | 20 | 4 | − | dPIAS, putative SUMO E3 ligase |
| FBgn0001624 | dlg1 | 2 | 16 | 2 | 1 | + | Guanylate kinase |
| FBgn0003401 | shu | 7 | 14 | 416 | 1 | + | piRNA pathway component |
| FBgn0038739 | CG4686 | 4 | 13 | 1 | 1 | + | Part of ribokinase/pfk B and DUF423 superfamilies |
| FBgn0038609 | nup43 | 3 | 12 | 3 | 1 | + | WD40-repeat-containing domain |
| FBgn0038925 | cchl | 0 | 12 | 1 | 2 | + | Cytochrome C heme lyase |
NXT1 was previously reported to affect interactions with the nuclear pore complex as well (Lévesque et al., 2001). Hence, the presence of several nuclear pore components within the top 20 hits is not surprising: Both nup154 and nup43 knockdowns caused similar derepression of gypsy (Table 1). Additionally, NUP154 deficient flies were sterile in our assay.
Another two genes ranking among the top 10 are as of yet uncharacterized: CG3893 and CG2183. CG3893 shows homology to mammalian GTSF1. Even though no direct link to the piRNA pathway has been shown, this germline specific factor also seems to be indispensible for transposon control in mice (Yoshimura et al., 2009). CG2183 is predicted to be a homolog of GASZ. This protein was previously implicated in the piRNA pathway in mice (Ma et al., 2009) and has now been validated as a piRNA pathway component in flies (Czech et al., submitted).
smt3 (SUMO), which was one of the highest scoring genes in the primary screen, could not be validated in vivo because of developmental defects that occur upon knockdown (Talamillo et al., 2008). However, the depletion of its E1 activating enzymes AOS1 and UBA2 caused consistent transposon derepression in the validation screen (Table 1, Figure 2E). Knockdown of the E2 conjugating enzyme lesswright also caused developmental defects and could not be validated.
Another notable validated hit is windei (wde), which was previously reported as a cofactor of EGG in histone 3 lysine 9 trimethylation (H3K9me3) (Koch et al., 2009). While EGG depletion had no effect on gypsy expression in our assay, knockdown of wde caused derepression of gypsy, although to a lower extend than ZAM (Table S3).
Additionally, two genes involved in transcriptional regulation were identified. MBD-R2 is part of the Nonspecific Lethal Complex (Figure 2E) (Raja et al., 2010). All members of this complex except for rcd1 scored in the primary screen. lin-52, which is part of the dREAM transcriptional regulator complex, also scored highly with gypsy and ZAM (Lewis et al., 2004).
Characterization of newly described piRNA pathway components
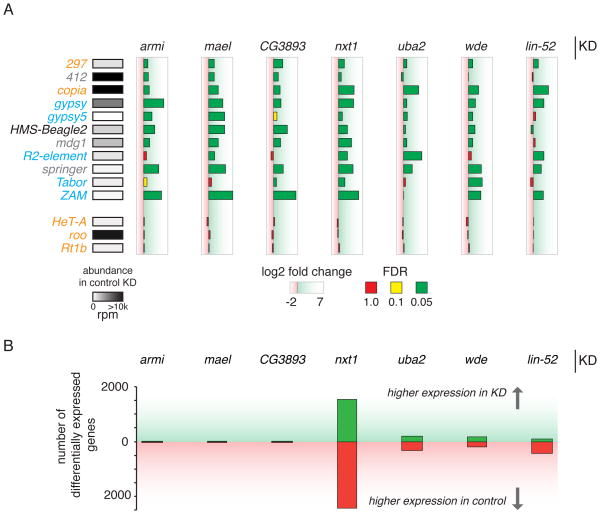
In order to place some of the validated hits at particular steps within the piRNA pathway, we constructed RNA-seq libraries from ovaries of biological replicates of _tj_-Gal4 driven hpRNA crosses. Mapping RNA-seq reads to transposon consensus sequences revealed the same levels of gypsy and ZAM derepression observed by qPCR (Figure 3A). When analyzed on a global scale, mainly transposons dominant within the somatic compartment of the ovary reacted significantly to the respective knockdown of the target gene in a _tj_-Gal4-dependent manner (FDR<0.05) (Malone et al., 2009). Transposons like HeT-A, roo, or Rt1b, which were previously shown to be germline dominant, did not change expression levels (Figure 3A) (Malone et al., 2009). The patterns of derepression that we observed in knockdowns of known components of the pathway (armi and mael) remarkably resembled those observed in CG3893 and wde knockdown.
Figure 3. RNA-seq shows changes in gene and transposon expression upon knockdown of top candidates in vivo.
A) A subset of somatically expressed transposons is derepressed in the indicated KD. The classification of transposons according to Malone et al. (2009), is indicated in orange (germline dominant), grey (intermediate), blue (soma dominant) and black (unclassified). The absolute abundance of reads in control knockdown mapping to each transposon is shown in shades of grey. The log2 fold change of each target gene versus a negative control (aub) is shown. Color of the bars represent the significance of these fold changes and are indicated as an adjusted p-value (FDR). Green indicates highly significant differences (p<=0.05), yellow moderately significant changes (0.05<p<=0.1) and red non-significant changes (0.1<p<=1), based on two biological replicates. Each knockdown is normalized to aub knockdown controls from their corresponding library (GD or KK). For differences in transposon abundance levels between both aub controls, see Figure S3.
B) The number of genes differentially expressed (p-adj<0.05) in each knockdown with respect to the control is shown. Green bars indicate the number of genes that have higher expression levels in the knockdown fly line, while red bars designate the number of genes with higher levels in the aub negative control.
We observed a similar behavior of CG3893 and known piRNA pathway components in their impacts on the expression of protein coding genes. While proteins like NXT1 or UBA2, with potentially more general functions, impacted the expression of hundreds to thousands of genes, this was not the case for ARMI, MAEL, or CG3893, which show effects that are much more restricted to transposon transcripts (Figure 3B). We interpreted these results as an indication that CG3893 might act specifically within the piRNA pathway.
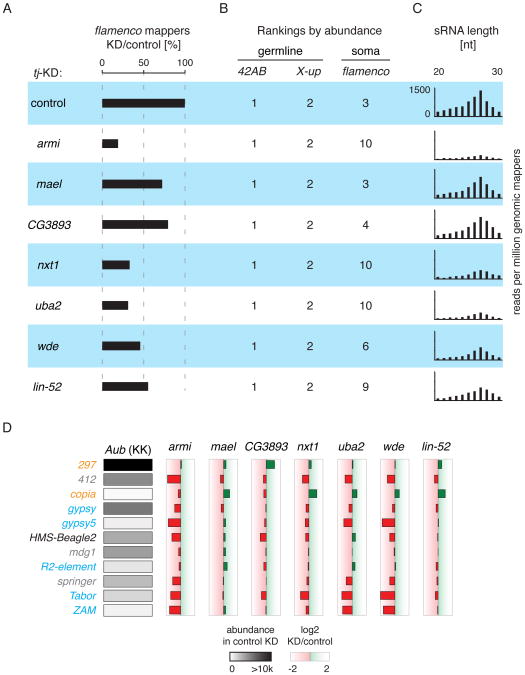
We further annotated validated hits based upon their impacts on piRNA levels. Interestingly, we saw a substantive drop in the abundance of piRNAs uniquely mapping to the soma-dominant flamenco piRNA cluster in the nxt1, uba2 and wde knockdowns (Figure 4A). To avoid skewing this result based on normalization to a piRNA producing locus, which theoretically should not change in soma specific knockdowns (i.e. cluster 1/42AB), we examined the rankings within each library of piRNA clusters based on the overall abundance of corresponding piRNAs. Given that we only knock down each gene in the somatic compartment, only clusters within that expression domain (i.e. flamenco) should change their ranking. Validating this approach, we observed that in the armi knockdown, flamenco was the 10th most abundant cluster while it was the third most abundant in total RNA libraries from negative-control ovaries. Conversely, 42AB and X-upstream remained at the top of the list in all tested knockdowns (Figure 4B). The two genes that mirrored armi are nxt1 and uba2. wde and lin-52 showed changes in flamenco piRNAs that altered its ranking; however, not to the same extent. In none of the knockdowns did the length profiles of the remaining piRNAs from flamenco change (Figure 4C). When compared to their negative controls, piRNA levels did not change in the mael and CG3893 knockdown. The same conclusions could be drawn when mapping to transposons consensus sequences: Antisense populations of piRNAs with homology to soma dominant transposons showed severe reductions in the nxt1, uba2, and wde knockdowns, which resembled patterns seen for the biogenesis factor armi (Figure 4D). Depletion of mael, lin-52, or CG3893 did not show the same effect. Intriguingly, in the case of lin-52 this did not coincide with the effects seen for flamenco mappers. None of the assayed knockdowns showed any changes in mature miRNA levels, indicating that the observed effects were specific to the piRNA pathway and not a general trend of all small RNA populations (Figure S4).
Figure 4. Biogenesis of small RNAs from somatic clusters and transposons is affected in knockdowns of a subset of top candidate genes.
A) Percentages of total unique mappers (sense species, >23nt) to flamenco in each knockdown (as indicated) in relation to the control knockdown are shown.
B) The internal rankings for three representative piRNA clusters based on their representation in piRNA populations are displayed. Expression bias towards either domain (soma or germline) is indicated. Cluster definitions are in concordance with Brennecke et al. (2007).
C) The size profiles of piRNAs mapping in sense orientation to flamenco in each knockdown (as in A) are plotted as total read count per million genomic mappers. As a control, we show that levels of microRNAs do not change in knockdowns versus negative control (Figure S4).
D) piRNAs mapping to a subset of somatically expressed transposons are reduced when gene expression of a subset of top hits is disrupted. The classification of transposons according to Malone et al. (2009), is indicated in orange (germline dominant), grey (intermediate), blue (soma dominant) and black (unclassified). The absolute abundance of antisense piRNAs in an aub control mapping to each transposon is shown in shades of grey. The log2 fold change of each target gene versus a negative control is shown.
See also Figure S4.
CG3893 is indispensible for transposon silencing in the germline
piwi and mael have recently been shown to silence transposons in the somatic compartment of the ovary through effects on transposon transcription (transcriptional gene silencing or TGS) (Le Thomas et al., 2013; Rozhkov et al., 2013; Sienski et al., 2012). CG3893, displaying patterns in global transposon derepression similar to mael and unaffected mature piRNA populations, appeared to be a promising candidate for a novel pathway component acting at the TGS effector step.
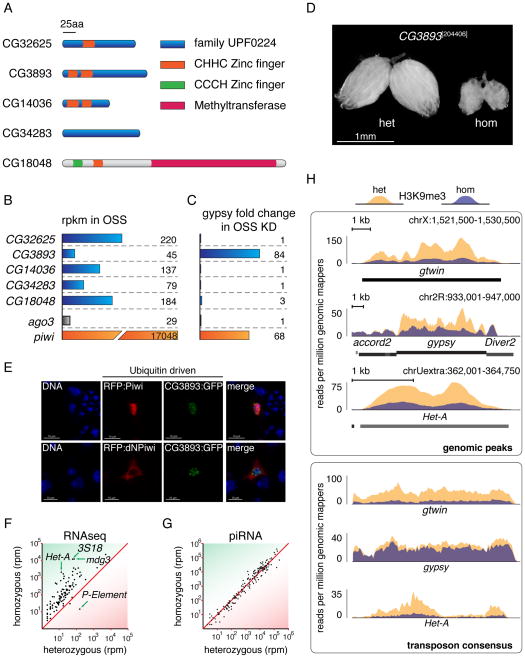
The ~20kDa protein, CG3893, is a member of a family of proteins with unknown function (UPF0224), characterized by the presence of highly conserved Zinc-finger domains (Figure 5A). All five proteins of this family are weakly expressed in OSS cells, and show germline-specific moderate to high expression in the ovary (Figure 5B, modENCODE Tissue Expression Data (Graveley et al., 2011)). Out of all five members of the family, only CG3893 showed strong effects on transposon mRNA abundance when knocked down in OSS cells (Figure 5C).
Figure 5. Disruption of CG3893 function has a severe impact on transposon silencing.
A) The five members of the Drosophila uncharacterized protein family UPF0224 and their domain structures are diagrammed. The conserved domains are highlighted as colored boxes.
B) All five family members are weakly expressed in OSS cells. piwi and ago3 expression levels are shown for comparison. Expression levels are based on the modENCODE cell line expression data and are displayed as reads per kilobase per million mapped reads (rpkm).
C) CG3893, but no other members of its protein family, has a strong impact on transposon silencing upon knockdown in OSS cells. Effects of knockdown of ago3 and piwi are shown for comparison. Numbers represent fold changes of gypsy levels in respect to the median fold change of the corresponding plate in the primary screen.
D) The ovarian morphology of flies heterozygous or homozygous for a P-element insertion in CG3893 is shown (204406, Kyoto Drosophila Genetic Resource Center). For a more detailed view of the insertion and expression levels see Figure S5.
E) Tagged CG3893 co-localizes with Piwi in the nucleus of OSS cells when overexpressed in transient transfections. Nuclear Hoechst staining is blue, GFP tagged CG3893 is green and RFP tagged Piwi or ΔNT-Piwi is shown in red (Saito et al., 2009).
F) Transposons are highly upregulated upon disruption of CG3893 in the P-element insertion line. A scatter plot of reads per million (rpm) is shown for RNA-seq of heterozygous versus homozygous flies. Each dot represents one transposon consensus sequence. Only sequences mapping in the sense orientation are taken into account.
G) piRNA levels are not affected by CG3893 disruption. The number of piRNA reads mapped to the same transposon consensus sequences as in F) is expressed in reads per million.
H) Levels of H3K9me3 decrease dramatically on a subset of transposons upon depletion of CG3893. Density plots for normalized H3K9me3 ChIP-seq reads over three transposons, gtwin, gypsy and Het-A are shown. Yellow distributions correspond to levels in heterozygous flies and blue distributions to the homozygous state. The upper box shows three distinct genomic peaks over transposon insertions, the lower box shows the corresponding consensus sequences. For all identified H3K9me3 peaks and their read densities see Figure S5C.
See also Figure S5.
In order to obtain an additional model for a loss of function of CG3893, we searched for available transposon insertion lines. We investigated two available lines (204406, DGRC Kyoto; 22464, Bloomington Drosophila Stock Center Indiana University). Line 204406 has a P-element insertion into the first exon of CG3893, disrupting its N-terminal CHHC Zinc-finger domain (Figure S5A). Consistent with the insertion site, we identified truncated CG3893 mRNAs by RNA-seq in libraries from homozygous animals. The levels of CG3893 mRNA expression were also reduced in animals homozygous for this mutation, when compared to heterozygous siblings. The results obtained by RNA-seq were confirmed by qPCR (Figure S5B). The second line (22462) has a P-element insertion upstream of the first exon in either the promoter or the 5′ UTR of CG3893. Our RNA-seq data in control flies indicated a slightly extended CG3893 transcript when compared to the gene model presented in Flybase. Even though this insertion is not in any coding sequence, the observed phenotypes were severe: homozygous females were completely sterile and were characterized by a complete absence of ovarian structures. This correlated with undetectable levels of CG3893 transcript when assayed by qPCR indicating a complete loss-of-function (Figure S5B). The phenotypes observed in females homozygous for the first insertion (204406) were milder with ovaries developing to a rudimentary stage (Figure 5D). Nevertheless, this potentially hypomorphic mutation caused females to be sterile, demonstrating the negative impact of the insertion on CG3893 function.
According to our current model of transposon silencing as a nuclear phenomenon, effectors at this step are expected to be nuclear as well. The mouse homolog of CG3893, Gtsf1, is reported to be mainly cytoplasmic in adult testes (Yoshimura et al., 2007). However, when overexpressed in OSS cells, GFP fusion proteins of CG3893 co-localized with Piwi in the nucleus (Figure 5E).
Our results to this point demonstrated the involvement of CG3893 in the somatic compartment of the ovary. In order to investigate its role in all tissues of the female germline, we generated RNA-seq and small RNA libraries from females heterozygous and homozygous for the exonic P-element insertion. RNA-seq revealed a remarkable change in global transposon expression. Almost all classes of annotated transposons populating the Drosophila genome except for the P-element itself showed upregulation in homozygous females when compared to their heterozygous sisters (Figure 5F). This derepression effect was equally strong for germline- and soma-dominant transposon classes. Yet, when mapping antisense piRNA reads to transposon consensus sequences, we see no change in the homozygous animals (Figure 5G).
Three recent publications not only demonstrate that piRNA mediated transposon silencing is a nuclear phenomenon occurring through TGS, but also that it acts through deposition of silencing H3K9 trimethyl marks on active copies of transposons (Le Thomas et al., 2013; Rozhkov et al., 2013; Sienski et al., 2012). Given its potential involvement in this step, we investigated the effects of CG3893 disruption on this histone mark by performing ChIP-seq analysis for H3K9me3 in ovaries from heterozygous and homozygous females. Strikingly, homozygous females showed a marked reduction of H3K9me3 levels over a subset of peaks identified in heterozygous animals. These differential peaks overlapped with full-length insertions of both germline and soma dominant transposons while neighboring peaks over transposon fragments did not change (Figure 5H; fragments correspond to accord2 and diver, center panel). We also observed the same changes when we mapped to consensus sequences of the corresponding transposon families, thereby aggregating signal from all genomic insertions (Figure 5H). 75% of all peaks showed lower levels of H3K9me3 deposition in homozygous flies, with 28 peaks exhibiting a reduction to less than 50% of the read density compared to heterozygous siblings (Figure S5C). These peaks corresponded mostly to retrotransposons (LTR, LINE), while none of the peaks over DNA elements were affected. However, the loss of H3K9me3 in homozygous mutant animals was not exclusive to transposable elements, as a limited number of protein coding genes, such as CG8964, showed similar patterns (Figure S5D). Our RNA-seq data was in accordance with this finding: the CG8964 transcript was increased ~26-fold in homozygote versus heterozygote flies. Whether this effect is due to effects on a transposon insertion within its genomic locus, or within another gene that controls CG8964 expression, or whether this represents a piRNA-independent function of CG3893 remains to be seen. Because of its small size yet powerful role in transposon silencing we name CG3893 asterix (arx).
Discussion
We are just beginning to understand the precise molecular steps necessary for piRNA biogenesis and successful silencing of transposons. In an effort to shed light on all aspects of the pathway from piRNA biogenesis to the effector mechanisms of transposon control, we performed an unbiased, genome-wide RNAi screen in cultured ovarian somatic cells. The primary in vitro screen proved to be a robust and specific assay for transposon derepression, with all expected piRNA components scoring strongly. To assess the validity of the primary dataset, we tested our top candidate hits for effects in vivo. In order to make this resource more accessible to the scientific community, we created a web-resource with all primary and validation datapoints (http://somatic-pirnascreen.cancan.cshl.edu/).
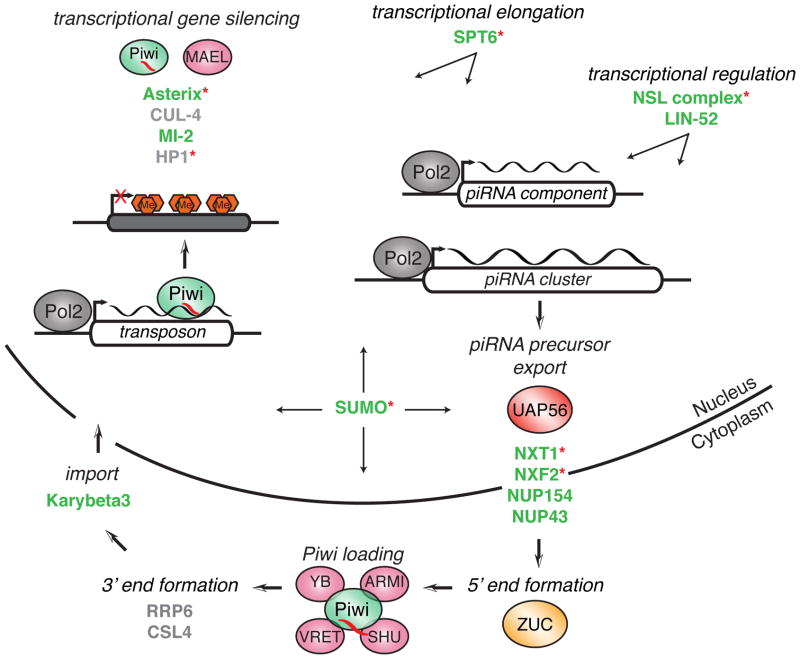
Within our list of 87 validated genes, we have promising candidates for filling almost every gap in our current understanding of the piRNA pathway (Figure 6). For example, the identified RNA export factors and nucleoporins could act in the export of primary cluster transcripts to the cytoplasm. We also identify genes that are likely to affect transcription of these piRNA precursors. WDE was shown to be a co-factor of eggless, a gene required for transcription of clusters (Koch et al., 2009; Rangan et al., 2011). While depletion of EGG did not result in mobilization of gypsy (as previously shown), wde knockdown led to high levels of gypsy expression, hinting towards a role for WDE independent of EGG. lin-52 was previously described as a transcriptional activator of Piwi (Georlette et al., 2007). However, Zamore and colleagues just recently showed that A-MYB, which provides activity orthologous to the dREAM complex in mouse, controls not only the expression of key pathway components but also of piRNA precursor transcripts (Li et al., 2013).
Figure 6. Potential roles for newly identified piRNA pathway components.
Known piRNA components are shown as bubbles. The newly identified genes are shown in bold text colored according to their validation status (color code as in figure 2; Pol2, RNA polymerase 2; red hexagons represent H3K9me3). Red asterisks denote genes that validated in the germline screen done in parallel (Czech et al., submitted).
Following nuclear export, piRNA precursors have to be further processed by an endonuclease to create the 5′ end of the mature piRNA. ZUC, which was recently shown to be a cytoplasmic, single-stranded RNA specific endonuclease, is the most likely candidate for this function (Ipsaro et al., 2012; Nishimasu et al., 2012). The fact that we do not identify any other annotated endonuclease with comparable derepression phenotypes in our screen supports the role for ZUC in this step. RNase P and RNase Z, both endonucleases implicated in tRNA processing, did score in the primary screen, but could not be validated in vivo because of their severe developmental defects (Dubrovsky et al., 2004; Frank and Pace, 1998). After 5′ end formation and loading into Piwi, each piRNA is proposed to be further trimmed to its mature length. The only genes exhibiting exonuclease activity and scoring highly in our screen were csl4 and rrp6, both components of the exosome (Andrulis et al., 2002). However, neither of the two genes could be validated in vivo due to arrested gonadal development in knockdowns.
Transcriptional silencing of transposons through Piwi is a nuclear process and previous data have demonstrated that unloaded Piwi remains in the cytoplasm (Saito et al., 2010). One protein possibly involved in reimportation of loaded Piwi is Karybeta3, a homolog of Importin 5 (Mosammaparast and Pemberton, 2004), which emerged as a hit from our screen.
Upon reentry into the nucleus Piwi is able to recognize transcription of active transposons through its bound piRNA and consequently silence them (Le Thomas et al., 2013; Rozhkov et al., 2013; Sienski et al., 2012). So far, only Piwi itself and MAEL have been implicated in this step. With Asterix, we present a new component of the nuclear piRNA silencing machinery that is indispensible for transcriptional repression. However, even though we see lower levels of H3K9me3 in mutant animals compared to heterozygous siblings, Asterix most likely is not directly responsible for depositing these marks. The only conserved domains within the protein are predicted to be RNA binding. This still leaves a need for identifying chromatin remodelers and histone methyltransferases (HMTs) that act as piRNA effectors in TGS. No annotated HMTs were hit in the screen; specifically, disruption of Su(var)3-9, which was recently implicated in epigenetic programming directed by piRNAs, did not lead to any significant transposon derepression, suggesting a possible redundancy in proteins that act in histone methylation (Huang et al., 2013).
Elongation factors have been shown to also play a role in chromatin modification. The elongation factor SPT6, which interacts with the nuclear exosome, emerged as another validated hit of the screen (Andrulis et al., 2002). Indeed, data from fission yeast implicated Spt6 in silencing of heterochromatic repeats (Kiely et al., 2011). In the spt6 mutants, decreased recruitment of the CLRC complex led to loss of H3K9 trimethylation and as a consequence lower occupancy of the HP1 homolog Swi6. Cul-4, a homolog of a CLRC member in yeast that is involved in histone methylation in Drosophila, was a primary hit in the screen, but led to developmental defects during validation (Higa et al., 2006; Hong et al., 2005).
Another validated hit involved in chromatin remodeling was mi-2, yet its knockdown only led to modest effects on transposon derepression (Brehm et al., 2000). Heterochromatin protein 1 (HP1), which interacts with Piwi, could not be validated due to developmental defects (Brower-Toland et al., 2007). Intriguingly, MI-2 and HP1 are both sumoylation substrates, implying a function for SUMO beyond the one in piRNA biogenesis demonstrated herein (Nie et al., 2009).
In summary, our unbiased, genome-wide approach was successful in identifying likely candidates to fill in many of the gaps in our understanding of the molecular mechanisms of transposon control in Drosophila. Together with the findings of a transcriptome-wide screen for germline piRNA pathway components done in parallel (Czech et al., submitted), which showed substantial overlap with the top hits of our screen, we are confident to have identified a comprehensive set of novel pathway components. Our data support the current view that the piRNA pathway is the major pathway exerting transposon control, given that both the primary and validation screen were dominated by known components of the piRNA machinery. Our meta-analysis on the primary dataset as a whole, as well as the list of validated genes, will provide a resource for the field in efforts toward a greater depth of understanding of piRNA production and of the mechanisms by which piRNAs silence transposons.
Experimental Procedures
Cell culture
OSS cell were cultured as previously described and transfected using Xfect transfection reagent according to manufacturer’s guidelines (Niki et al., 2006) (Clontech 631317).
DNA plasmids
Expression vectors of CG3893:GFP, RFP:Piwi and RFP:ΔNTPiwi were made using the Drosophila Gateway Collection.
Imaging of fluorescent fusion proteins in OSS
OSS cells were co-transfected with plasmids expressing the indicated fusion proteins using Cell Line Nucleofector kit V (Amaxa Biosystems; program T-029). Fixed cells were stained with Hoechst 33342 (Invitrogen, R37601).
RNAi libraries
Two Drosophila dsRNA libraries were used in this study, the Open Biosystems Drosophila RNAi Collection and the Drosophila RNAi Screening Center Genome-wide RNAi library (DRSC 2.0).
RNAi screening
A detailed description of the primary screen can be found in extended experimental procedures. A basic workflow is shown in Figure 1A. All primers used are listed in Table S4.
Drosophila stocks and husbandry
Fly stocks are listed in Extended Experimental Procedures. A description of husbandry and validation screen procedures can be found in extended experimental procedures.
RNA isolation and qPCR assays
Total RNA from 10 ovaries was extracted with Trizol and purified by organic extraction followed by isopropanol precipitation. After DNase treatment, cDNA was synthesized from 800ng RNA using oligo dT primers and Superscript III Reverse Transcriptase (Life Technologies). qPCR was performed to assay levels of gypsy, ZAM, gypsy3 and rp49. Fold changes for transposons were calculated using the delta Ct method (Livak and Schmittgen, 2001). All primers used are listed in Table S4.
RNA-seq and analysis
For RNA-seq libraries, 2.5–5μg of total RNA was depleted of ribosomal RNA using the Epicenter Ribo-Zero rRNA Removal Kits (Human/Mouse/Rat), following the manufacturer’s directions. Libraries were prepared using the Illumina Script Seq v2 RNA-Seq library preparation kit and were sequenced on an Illumina HiSeq platform. Details on Analysis can be found in extended experimental procedures.
Small RNA cloning and analysis
For small RNA libraries, total RNA was depleted of 2S rRNA and libraries were constructed using the Illumina Tru Seq small RNA sample Prep kit following the manufacturer’s protocol. Details on analysis can be found in extended experimental procedures.
ChIP-seq
ChIP from 50 ovaries was done as described in Ram et al and Garber et al, with some modifications (Garber et al., 2012; Ram et al., 2011). Details on the methodology and analysis can be found in extended experimental procedures.
Statistical procedures
Details on enrichment analysis and statistical procedures can be found in extended experimental procedures.
Accession Numbers
RNA-seq, ChIP-seq and small RNA data were deposited in the Gene Expression Omnibus Database under accession number GSExxxxx (pending).
Supplementary Material
01
02
03
Highlights.
- Nearly 100 genes contribute to transposon control in the Drosophila ovary
- RNA export factors and the sumoylation pathway may be involved in piRNA biogenesis
- Asterix is indispensible for transposon silencing but does not affect piRNA levels
Acknowledgments
We are greatly indebted to Norbert Perrimon (DGRC) for giving us reagents. We are grateful to Sabrina Boettcher who managed all logistics and supplies. We would like to thank Alon Goren for invaluable help with ChIP-seq cloning. Molly Hammell aided in analyzing RNA-seq and ChIP-seq data. Leah Sabin provided helpful insight and advice. We wish to thank Stephanie Muller, Assaf Gordon and Astrid Haase for help with robotics and library normalization. We would like to thank Ben Czech, Jon Preall, Sho Goh, and Julius Brennecke for sharing data prior to publication. Assistance with sequencing was provided by Emily Lee. We also would like to thank Jo Leonardo and all members of the Hannon lab for vital support. P.M.G. is a NIH trainee on CSHL WSBS NIH Kirschstein-NRSA pre-doctoral T32 GM065094, and is a William Randolph Hearst Scholar and Leslie Quick Junior Fellow. A grant from T. and V. Stanley supported J.G.’s work. Work in the Hannon laboratory is supported by grants from the National Institutes of Health (5R01GM062534) and by a kind gift from Kathryn W. Davis. G.J.H. is an investigator of the HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- Brehm A, Längst G, Kehle J, Clapier CR, Imhof A, Eberharter A, Müller J, Becker PB. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. The EMBO Journal. 2000;19:4332–4341. doi: 10.1093/emboj/19.16.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton A. The relationship between the flamenco gene and gypsy in Drosophila: how to tame a retrovirus. Trends in genetics: TIG. 1995;11:349–353. doi: 10.1016/s0168-9525(00)89105-2. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nature genetics. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Czech B, Preall JB, McGinn JT, Hannon GJ. A transcriptome-wide RNAi screen in the Drosophila ovary reveals novel factors of the germline piRNA pathway. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dubrovsky EB, Dubrovskaya VA, Levinger L, Schiffer S, Marchfelder A. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo. Nucleic acids research. 2004;32:255–262. doi: 10.1093/nar/gkh182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, Pace NR. Ribonuclease P: unity and diversity in a tRNA processing ribozyme. Annual review of biochemistry. 1998;67:153–180. doi: 10.1146/annurev.biochem.67.1.153. [DOI] [PubMed] [Google Scholar]
- Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, Robinson J, Minie B, Chevrier N, Itzhaki Z, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Molecular cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georlette D, Ahn S, Macalpine DM, Cheung E, Lewis PW, Beall EL, Bell SP, Speed T, Manak JR, Botchan MR. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes & Development. 2007;21:2880–2896. doi: 10.1101/gad.1600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold A, Klymenko T, Izaurralde E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA (New York, NY) 2001;7:1768–1780. [PMC free article] [PubMed] [Google Scholar]
- Herold A, Teixeira L, Izaurralde E. Genome-wide analysis of nuclear mRNA export pathways in Drosophila. The EMBO Journal. 2003;22:2472–2483. doi: 10.1093/emboj/cdg233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- Hong EJ, Villen J, Gerace EL, Gygi SP, Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA biology. 2005;2:106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H. A Major Epigenetic Programming Mechanism Guided by piRNAs. Developmental cell. 2013;24:502–516. doi: 10.1016/j.devcel.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491:279–283. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes & Development. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y. 3′ end formation of PIWI-interacting RNAs in vitro. Molecular cell. 2011;43:1015–1022. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- Khurana JS, Theurkauf W. piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol. 2010;191:905–913. doi: 10.1083/jcb.201006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiely CM, Marguerat S, Garcia JF, Madhani HD, Bahler J, Winston F. Spt6 is required for heterochromatic silencing in the fission yeast Schizosaccharomyces pombe. Molecular and cellular biology. 2011;31:4193–4204. doi: 10.1128/MCB.05568-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. The Drosophila HP1 Homolog Rhino Is Required for Transposon Silencing and piRNA Production by Dual-Strand Clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CM, Honemann-Capito M, Egger-Adam D, Wodarz A. Windei, the Drosophila Homolog of mAM/MCAF1, Is an Essential Cofactor of the H3K9 Methyl Transferase dSETDB1/Eggless in Germ Line Development. PLoS genetics. 2009;5:e1000644. doi: 10.1371/journal.pgen.1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome research. 2009;19:1776–1785. doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque L, Guzik B, Guan T, Coyle J, Black BE, Rekosh D, Hammarskjöld ML, Paschal BM. RNA export mediated by tap involves NXT1-dependent interactions with the nuclear pore complex. The Journal of biological chemistry. 2001;276:44953–44962. doi: 10.1074/jbc.M106558200. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes & Development. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, et al. An Ancient Transcription Factor Initiates the Burst of piRNA Production during Early Meiosis in Mouse Testes. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, Han DY, Harris RA, Coarfa C, Gunaratne PH, et al. GASZ Is Essential for Male Meiosis and Suppression of Retrotransposon Expression in the Male Germline. PLoS genetics. 2009;5:e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Lührmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science (New York, NY) 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, Mccombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA Pathways Act in Germline and Somatic Tissues of the Drosophila Ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuilton P, St Pierre SE, Thurmond J. FlyBase 101--the basics of navigating FlyBase. Nucleic acids research. 2012;40:D706–714. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N, Pemberton LF. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends in cell biology. 2004;14:547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Nie M, Xie Y, Loo JA, Courey AJ. Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS ONE. 2009;4:e5905. doi: 10.1371/journal.pone.0005905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki Y, Yamaguchi T, Mahowald AP. Establishment of stable cell lines of Drosophila germ-line stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16325–16330. doi: 10.1073/pnas.0607435103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010;29:3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Jiang P, Zhao DY, Singh M, Schüpbach T. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. The EMBO Journal. 2011;30:4601–4615. doi: 10.1038/emboj.2011.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Song SU, Prud’homme N, Smith PA, Bucheton A, Corces VG. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. The EMBO Journal. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja SJ, Charapitsa I, Conrad T, Vaquerizas JM, Gebhardt P, Holz H, Kadlec J, Fraterman S, Luscombe NM, Akhtar A. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Molecular cell. 2010;38:827–841. doi: 10.1016/j.molcel.2010.05.021. [DOI] [PubMed] [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan N, Flockhart I, Booker M, Perrimon N, Mathey-Prevot B. Design and implementation of high-throughput RNAi screens in cultured Drosophila cells. Nature Protocols. 2007;2:2245–2264. doi: 10.1038/nprot.2007.250. [DOI] [PubMed] [Google Scholar]
- Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R. piRNA production requires heterochromatin formation in Drosophila. Current biology: CB. 2011;21:1373–1379. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, Hannon GJ. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013;27:400–412. doi: 10.1101/gad.209767.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes & Development. 2010;24:2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Müller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics. 1989;121:101–117. doi: 10.1093/genetics/121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamillo A, Sánchez J, Barrio R. Functional analysis of the SUMOylation pathway in Drosophila. Biochemical Society transactions. 2008;36:868–873. doi: 10.1042/BST0360868. [DOI] [PubMed] [Google Scholar]
- Tanentzapf G, Devenport D, Godt D, Brown NH. Integrin-dependent anchoring of a stem-cell niche. Nature cell biology. 2007;9:1413–1418. doi: 10.1038/ncb1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Miyazaki T, Toyoda S, Miyazaki S, Tashiro F, Yamato E, Miyazaki J-i. Gene expression pattern of Cue110: A member of the uncharacterized UPF0224 gene family preferentially expressed in germ cells. Gene Expression Patterns. 2007;8:27–35. doi: 10.1016/j.modgep.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Yoshimura T, Toyoda S, Kuramochi-Miyagawa S, Miyazaki T, Miyazaki S, Tashiro F, Yamato E, Nakano T, Miyazaki J-i. Gtsf1/Cue110, a gene encoding a protein with two copies of a CHHC Zn-finger motif, is involved in spermatogenesis and retrotransposon suppression in murine testes. Developmental biology. 2009;335:216–227. doi: 10.1016/j.ydbio.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, et al. UAP56 Couples piRNA Clusters to the Perinuclear Transposon Silencing Machinery. Cell. 2012;151:871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03