Nuclear microenvironments modulate transcription from low-affinity enhancers (original) (raw)
Abstract
Transcription factors bind low-affinity DNA sequences for only short durations. It is not clear how brief, low-affinity interactions can drive efficient transcription. Here, we report that the transcription factor Ultrabithorax (Ubx) utilizes low-affinity binding sites in the Drosophila melanogaster shavenbaby (svb) locus and related enhancers in nuclear microenvironments of high Ubx concentrations. Related enhancers colocalize to the same microenvironments independently of their chromosomal location, suggesting that microenvironments are highly differentiated transcription domains. Manipulating the affinity of svb enhancers revealed an inverse relationship between enhancer affinity and Ubx concentration required for transcriptional activation. The Ubx cofactor, Homothorax (Hth), was co-enriched with Ubx near enhancers that require Hth, even though Ubx and Hth did not co-localize throughout the nucleus. Thus, microenvironments of high local transcription factor and cofactor concentrations could help low-affinity sites overcome their kinetic inefficiency. Mechanisms that generate these microenvironments could be a general feature of eukaryotic transcriptional regulation.
Research organism: D. melanogaster
Introduction
Genomic regions near coding genes, called enhancers, direct specific patterns of gene expression (Spitz and Furlong, 2012; Reiter et al., 2017; Long et al., 2016). Enhancers contain short DNA sequences that bind sequence-specific activating and repressive transcription factor proteins, and the integration of these positive and negative signals directs gene expression (Crocker et al., 2016a). Protein-DNA binding is often an ephemeral event; studies in mammalian cells demonstrate that transcription factors disassociate within seconds of binding to DNA (Liu et al., 2014; Chen et al., 2014; Izeddin et al., 2014; Voss et al., 2011; Normanno et al., 2015; Morisaki et al., 2014). Furthermore, recent studies in animals ranging from fruit flies to mammals have revealed that low-affinity DNA-binding sites are critical to confer specificity between related transcription factors having binding sites with similar DNA sequences (Crocker et al., 2015; Farley et al., 2015; Farley et al., 2016; Lorberbaum et al., 2016; Antosova et al., 2016; Rister et al., 2015; Crocker et al., 2010; Crocker et al., 2016b; Tanay, 2006; Lebrecht et al., 2005; Rowan et al., 2010; Gaudet and Mango, 2002; Jiang and Levine, 1993). Increasing the affinity of binding sites to more stably recruit transcription factors activates promiscuous gene expression (Farley et al., 2015; Ramos and Barolo, 2013), which leads to developmental defects. It is unclear how brief protein-DNA contacts can mediate efficient transcription from enhancers containing low-affinity binding sites.
One possible mechanism that could mitigate their kinetic inefficiency is to increase the local concentrations of transcription factors. At the scale of a single enhancer over a few hundred base pairs long, multiple low-affinity binding sites for the same transcription factor in close proximity could increase the frequency of binding events by trapping the protein. Furthermore, interactions between transcription factors and cofactors with multiple binding sites within an enhancer could generate ‘microenvironments’ (Reiter et al., 2017) of high factor concentrations.
We have explored this problem using the shavenbaby (svb) locus, which contains multiple enhancers that drive specific patterns of svb gene expression in developing Drosophila embryos. Each of three characterized svb enhancers contains clusters of low-affinity binding sites for the Hox gene Ultrabithorax (Ubx). These enhancers also require a Ubx cofactor Homothorax (Hth) to function (Crocker et al., 2015). We have exploited robust transgenic tools in Drosophila, new fluorescent dyes, and new approaches to prepare embryos for microscopy to systematically perturb these svb enhancers and directly image the results at a sub-nuclear level. We find that microenvironments of high Ubx and Hth concentrations mediate transcription from low-affinity enhancers.
Results
Ubx is present in microenvironments of varying local concentrations
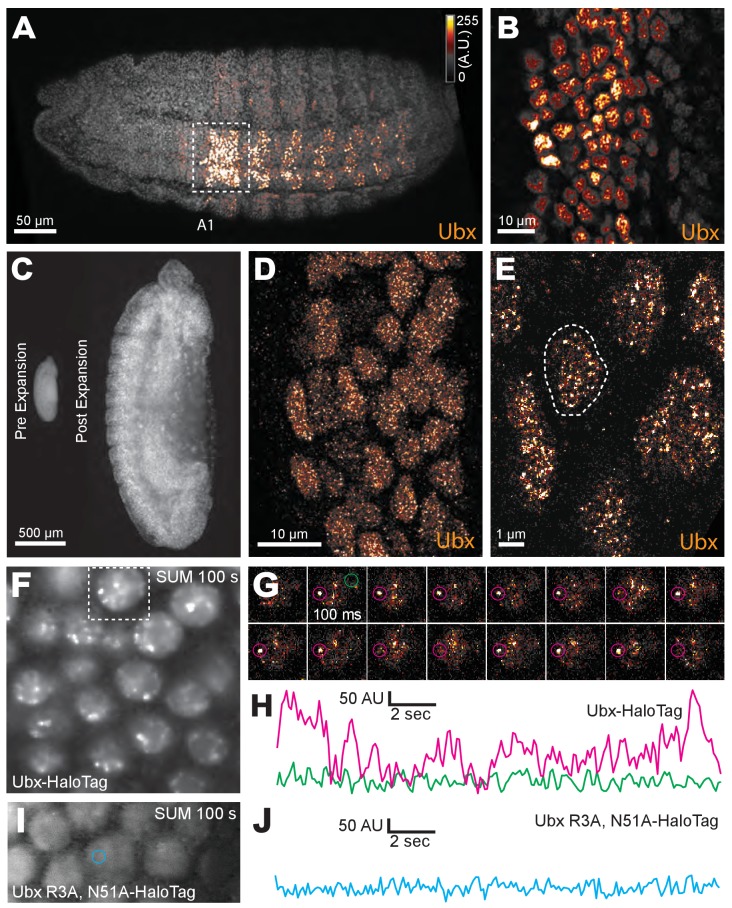
We first examined whether nuclei in Drosophila melanogaster embryos possess Ubx microenvironments by performing immunofluorescence (IF) staining in fixed embryos and high-resolution confocal imaging using Airyscan (Carl Zeiss Microscopy, Jena, Germany). We found that Ubx protein was not distributed uniformly, but rather exhibited regions of high and low Ubx intensities (Figure 1A,B). To observe Ubx distribution at higher resolution, we expanded the size of the embryos (Tillberg et al., 2016) by approximately four-fold in each dimension (Figure 1C). Nuclei of expanded embryos revealed distinct regions of high Ubx intensity separated by regions of low Ubx intensity. We observed, on average, 185 ± 25 (n = 12, three embryos) clusters per nucleus that were stronger than one-quarter of the maximum Ubx intensity within that nucleus (Figure 1D,E, and Figure 1—figure supplement 1).
Figure 1. Ubx is present in microenvironments with varying local concentrations.
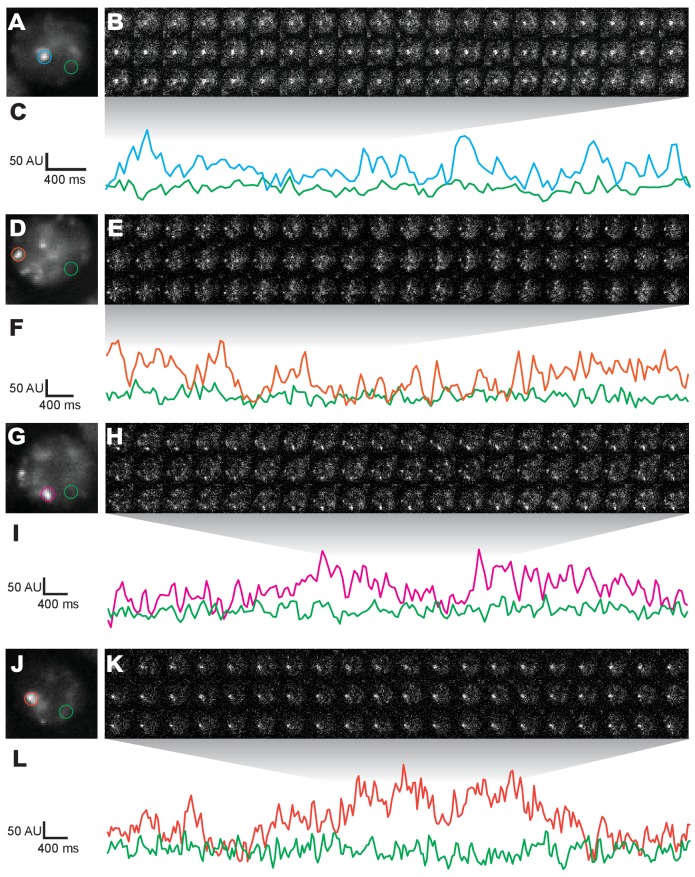
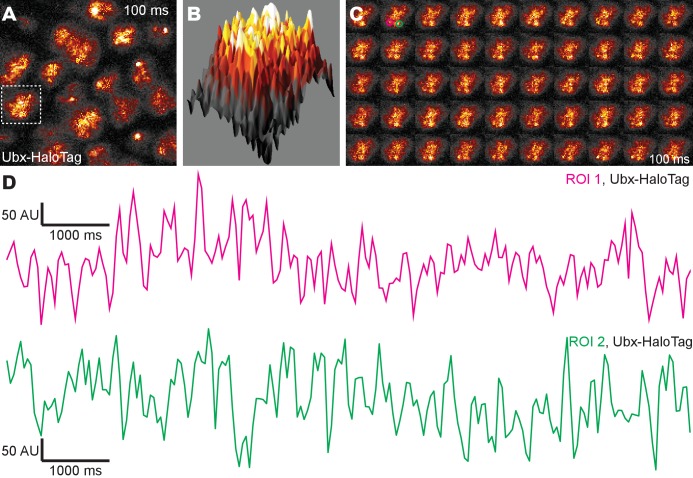
(A) Stage 15 embryos stained for Ubx protein with a bounding box indicating a ventral region of abdominal segment one (A1). (B) Higher magnification, Airyscan image of the region indicated in panel (A). (C) Stage 15 embryo pre- and post-expansion. (D, E) Expanded stage 15 embryos stained for Ubx protein. The dashed line encircles a single nucleus in (E). (F, I) Projections of summed pixel intensity over 100 s from videos of nos::GAL4, UAS::HaloTag-Ubx for either a wild-type Ubx (F) or a binding deficient Ubx (I), imaged with JF635 dye. (G) Sixteen individual, 100 millisecond video frames of the nucleus surrounded by a dashed box in panel (F). (H, J) Temporal traces of the signal intensity of the regions noted in panel (G) or (I). The color of each trace in (H) and (J) corresponds to the colors of the circles in panels (G) and (I), respectively. AU indicates Arbitrary Units of fluorescence intensity.
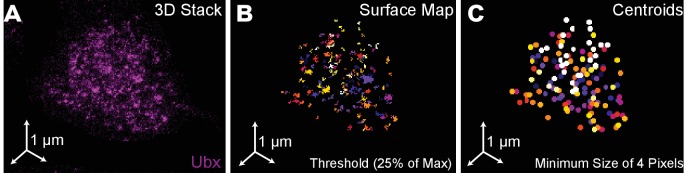
Figure 1—figure supplement 1. Quantification of Ubx microenvironments in single nuclei.
(A) 3D projection of a single nucleus stained for Ubx protein. (B) Surface plot of contiguous Ubx regions containing a minimum of four pixels with signal intensity greater than 25% of the maximum intensity. (C) Centroids of the Ubx regions found in panel (B). Colors denote different segmented regions.
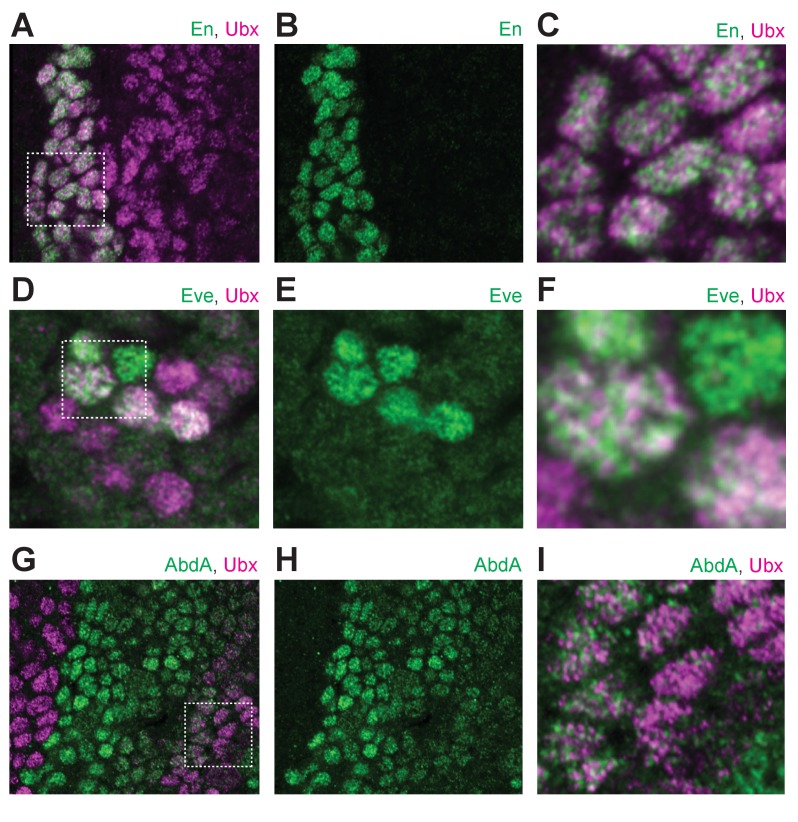
Figure 1—figure supplement 2. Ubx distribution compared to other transcription factors.
(A) Stage 15 embryo stained with an antibody against Engrailed (En) and Ubx protein. (B) Same image as (A) with only En shown. (C) A higher magnification of the area in the bounding box of panel (A). (D) Stage 15 embryo stained with an antibody against Eve and Ubx protein. (E) Same image as (D) with only Eve shown. (F) A higher magnification of the area in the bounding box of panel (D). (G) Stage 15 embryo stained with an antibody against AbdA and Ubx protein. (H) Same image as (G) with only AbdA shown. (I) A higher magnification of the area in the bounding box of panel (G).
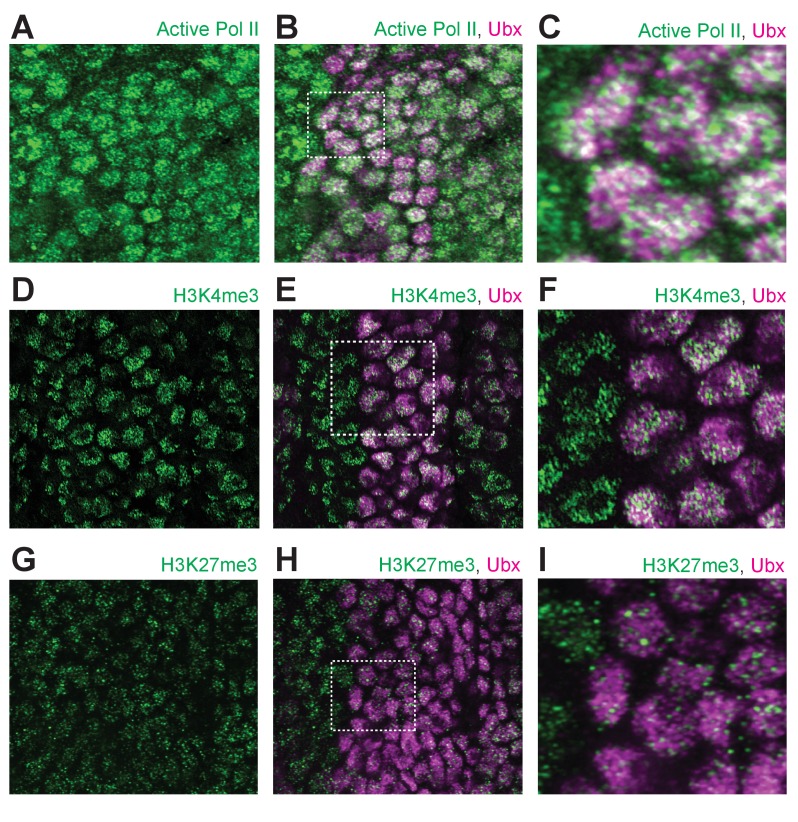
Figure 1—figure supplement 3. Ubx distribution compared to general markers for transcriptional activity.
(A) Stage 15 embryo stained with an antibody against transcriptionally active RNA Polymerase II RPB1 (Ser5 phosphorylated) and Ubx protein, with only active Pol II shown. (B) Same image as (A) with both active Pol II and Ubx shown. (C) A higher magnification of the area in the bounding box of panel (B). (D) Stage 15 embryo stained with an antibody against histone H3K4me3 and Ubx protein, with only H3K4me3 shown. (E) Same image as (D) with both H3K4me3 and UBx shown. (F) A higher magnification of the area in the bounding box of panel (E). (G) Stage 15 embryo stained with an antibody against histone H3K27me3 and Ubx protein, with only H3K27me3 shown. (H) Same image as (G) with both H3K27me3 and Ubx shown. (I) A higher magnification of the area in the bounding box of panel (H).
Figure 1—figure supplement 4. Control experiments for HaloTag-Ubx.
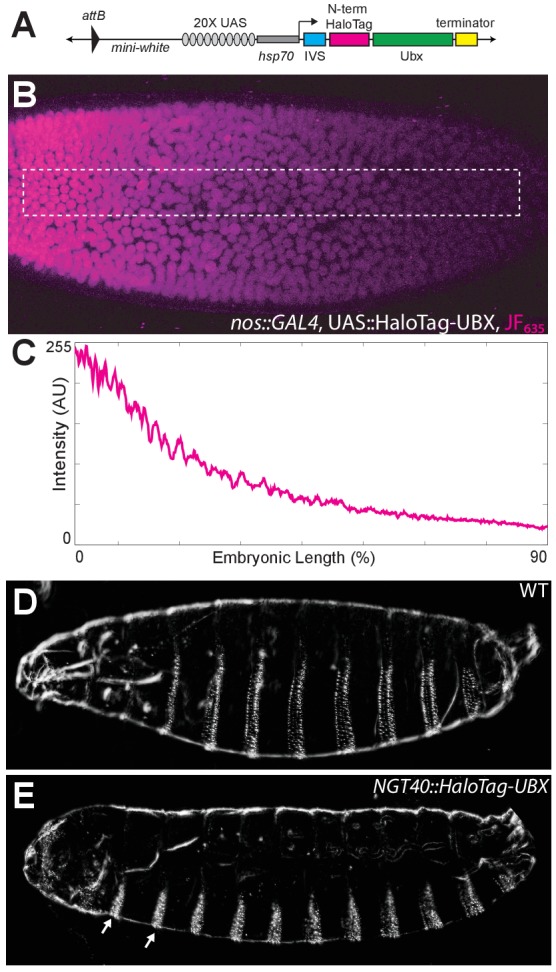
(A) Schematic of the N-terminal HaloTag construct with HaloTag-Ubx under the control of 20x UAS (responsive to GAL4) and the hsp70 prometer. Ubx is inserted at the MCS. (B) Stage 5 embryo resulting from a cross of a homozygous nos::GAL4 line with a homozygous 20x UAS::HaloTag-Ubx line. The embryo was injected with JF635 dye (magenta) at the anterior end. (C) Quantification of the signal intensity of the bounded region in panel (B) along the embryonic axis. A.U. indicates Arbitrary Units of fluorescence intensity. (D, E) Cuticle preps of first instar larva from either a wild-type (WT) (D) or nos::GAL4, UAS::HaloTag-Ubx embryo. (E) Arrows point to the nos::GAL4, UAS::HaloTag-Ubx induced transformation of anterior segments.
Figure 1—figure supplement 5. Additional nuclei from live imaging of Halo-Ubx.
(A, D, G, J) Projections of summed pixel intensities over 100 s of nos::GAL4, UAS::HaloTag-Ubx nuclei imaged with JF635 dye. (B, E, H, K) Individual 100 millisecond video frames of the nuclei in panels (A, D, G, J). (C, F, I, L) Signal traces of the signal intensity of the regions noted in panels (A, D, G, J), where color of trace corresponds to color of circle. AU indicates Arbitrary Units of fluorescence intensity.
Figure 1—figure supplement 6. Live imaging at higher concentrations of HaloTag-Ubx in late stage 6 embryos.
(A) One hundred millisecond video frame from video of nos::GAL4, UAS::HaloTag-Ubx in late stage 6 embryos with higher overall nuclear concentrations of HaloTag-Ubx molecules compared to stage 5 embryos Figure 1, imaged with JF635 dye. (B) 3D surface plot of the nucleus indicated in panel (A). (C) Individual, 100 millisecond video frames of the nucleus from panel (A). (D) Signal traces of the signal intensity of the regions noted with red and green circles in panel (C). AU indicates Arbitrary Units of fluorescence intensity.
Figure 1—figure supplement 7. DNA-binding deficient Ubx is stable and localized in the nucleus.
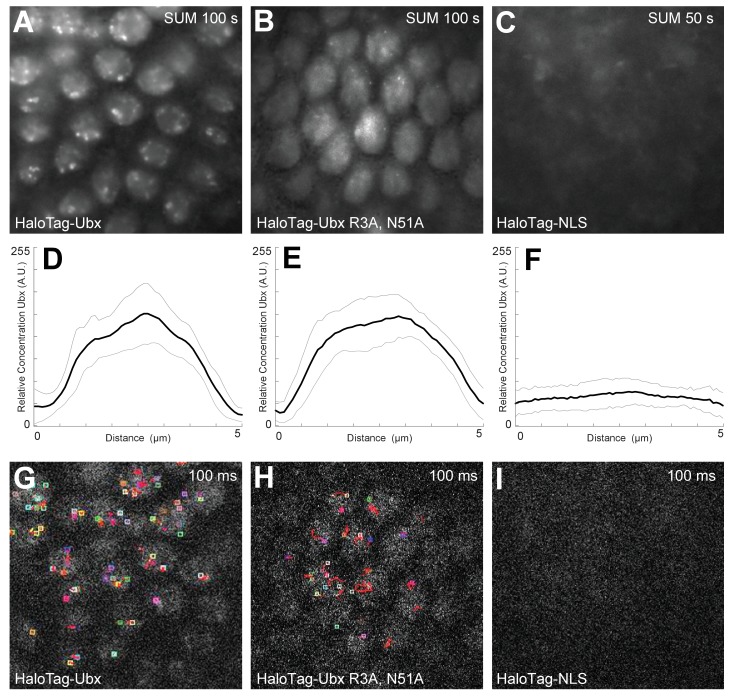
(A, B, C) Projections of summed pixel intensity over 100 or 50 seconds (as indicated in the top right corner of the panels) from videos of (A) HaloTag-Ubx, (B) HaloTag-Ubx(R3A, N51A), and (C) HaloTag-NLS (from H2B). (D, E, F) Average intensity profiles across the nuclei in the field of view of the previous panels, showing nuclear enrichment for both variants of HaloTag-Ubx fusion proteins but no enrichment for the NLS-HaloTag construct, which is unstable. The light grey lines show the variance. (G, H, I) Sample frame from single-molecule localization using Fiji, showing (G) short but localized binding of HaloTag-Ubx, (H) very short and fast moving single-molecule traces from DNA-binding deficient HaloTag-Ubx, and (I) no localization at all for the unstable HaloTag-NLS.
One explanation for the observed distribution of Ubx is that transcription factors localize generally to accessible regions of the nucleus that have high levels of transcriptional activity. This mechanism, if shared by transcription factors in general, should yield Ubx distributions that mostly overlap with that of other transcription factors. Engrailed (En), a transcription factor unrelated to Ubx, displayed non-uniform sub-nuclear concentrations, but its distribution only partially overlapped with that of Ubx (Figure 1—figure supplement 2A–C, white regions indicate overlap). We similarly observed only partial overlap between Ubx and Even-skipped (Eve) (Figure 1—figure supplement 2D–F). Abdominal-A (AdbA), a paralog of Ubx that is expressed mainly in separate cells from Ubx and that has similar DNA-binding specificity as Ubx, was excluded from Ubx regions in the few nuclei where both were expressed (Figure 1—figure supplement 2G–I). These results indicate that the distributions of these transcription factors do not result from a shared mechanism that limits the distribution of all transcription factors to the same sub-nuclear regions.
We also examined whether Ubx simply occupies regions containing actively transcribed DNA. Both active RNA Polymerase II (Pol II, Ser5 phosphorylated CTD) and the methylated histone H3K4me3, which marks actively transcribed DNA, only partially overlapped with Ubx (Figure 1—figure supplement 3A–F). In contrast, the histone mark H3K27me3, which marks regions of repressed chromatin, displayed almost no overlap with the distribution of Ubx (Figure 1—figure supplement 3G–I). Thus, Ubx is not merely restricted to regions inside the nucleus that are available to transcription factors or to regions of high transcriptional activity.
Ubx repeatedly binds to specific regions in nuclei of live embryos
To understand if the heterogeneous distribution of Ubx is dynamic or stable over the timescale of seconds to minutes, as well as to rule out the possibility that our observations of Ubx microenvironments are an artifact of the fixation protocol (Teves et al., 2016), we examined the spatiotemporal dynamics of single Ubx molecules in live Drosophila embryos. Single-molecule imaging has been mostly performed in cell lines previously because live-imaging studies of transcription factor dynamics in embryos requires overcoming several new challenges, including imaging at lower signal-to-noise ratios, compensating for rapid morphological changes during embryonic development, and determining how to deliver fluorescent dyes. We overcame these challenges by generating a HaloTag-Ubx transgene that allowed precise control of fusion protein levels (Figure 1—figure supplement 4A) and coupling HaloTag-Ubx in vivo to new, strongly fluorescent dyes (Grimm et al., 2017). The transgene we built can be expressed either from a heat-shock promoter (hsp70) or from a 20x UAS promoter by crossing with a GAL4 driver line.
Over-expression of the HaloTag-Ubx transgene by incubating the embryos at 30°C transformed anterior segments to the fate of more posterior segments, indicated by the presence of additional trichomes. This result indicates that the HaloTag-Ubx protein retains the expected Ubx behavior (Figure 1—figure supplement 4D and E). We then expressed HaloTag-Ubx from the 20x UAS promoter with the nos::GAL4 (nanos promoter driving GAL4) driver line, which drives HaloTag-Ubx expression in all cells at early developmental stages. We injected the HaloTag ligand of Janelia Fluor 635 (JF635) (Grimm et al., 2017) into these live embryos. JF635 is minimally fluorescent in solution but its fluorescence increases by over 100-fold when bound to a HaloTag protein, allowing the detection of labeled Ubx molecules against a background of dim freely diffusing dyes. The fluorescence intensity of labeled Ubx scaled with distance from the site of dye injection (Figure 1—figure supplement 4B and C), consistent with dye diffusion from the site of injection. To measure the time-averaged density of HaloTag-Ubx in specific locations of nuclei in live embryos in early stage 5, we calculated the summed intensity over 100 s (1000 frames at 100 ms per frame). We observed regions of Ubx signal (3-10x background) similar to the high-intensity clusters observed in fixed embryos (Figure 1F). We examined the dynamics of HaloTag-Ubx in nuclei by plotting fluorescence intensity over time (Figure 1G and H and Figure 1—figure supplement 5). We found that fluorescence signals over time changed in discrete up or down steps, indicating that individual HaloTag-Ubx molecules bind to specific nuclear domains with residence times on the order of a second before dissociation. Most unbound Ubx molecules move too quickly to be captured with the 100 ms exposure time; they move in and out of a diffraction-limited region in significantly less than 100 ms on average. These timescales are consistent with transcription factor-DNA binding dynamics measured in live-cell imaging experiments using mammalian cell lines (Liu et al., 2014; Izeddin et al., 2014; Voss et al., 2011; Normanno et al., 2015; Morisaki et al., 2014; Gebhardt et al., 2013). These repeated binding events produced the high intensities observed in the time-averaged projections and indicate that Ubx concentrates and remains within specific nuclear regions.
Observation of embryos at late stage 6 showed that total HaloTag-Ubx concentration continued to increase as the embryo ages (Figure 1—figure supplement 6A and B). Embryos at late stage 6 had nuclei containing high background concentrations of Ubx that masked single-molecule events, as well as displaying larger sites (>4 × 4 pixels) that constantly remained bright, possibly indicating the presence of multiple molecules or protein aggregation (Figure 1—figure supplement 6C and D). In contrast, the embryos observed during stage 5 did not contain areas that remained constantly bright, suggesting that we observed single molecule dynamics in stage 5 embryos.
To determine whether regions of high Ubx concentration depended on DNA binding, we performed the same experiments with a version of the HaloTag-Ubx transgene where Arg3 and Asn51 of the homeodomain were mutated to Ala (R3A and N51A), abrogating DNA binding (Slattery et al., 2011b). Both the wild-type and DNA-binding deficient Ubx were expressed and imported into the nucleus (Figure 1—figure supplement 7A,B,D,E,G, and H), suggesting that the protein is stable. In contrast, an unstable HaloTag-NLS construct (NLS from H2B) serving as a negative control, neither increased JF635 fluorescence post injection nor became enriched into the nucleus (Figure 1—figure supplement 7C,F, and I). The mutant HaloTag-Ubx (R3A N51A) did not display spatial heterogeneity and exhibited only extremely brief fluctuations in intensity inconsistent with transcription-factor DNA-binding events (Figure 1I and J). These results suggest that binding of Ubx to DNA is required to generate restricted nuclear distributions of Ubx.
Transcriptionally active svb loci and enhancers correlate with regions of high Ubx concentration
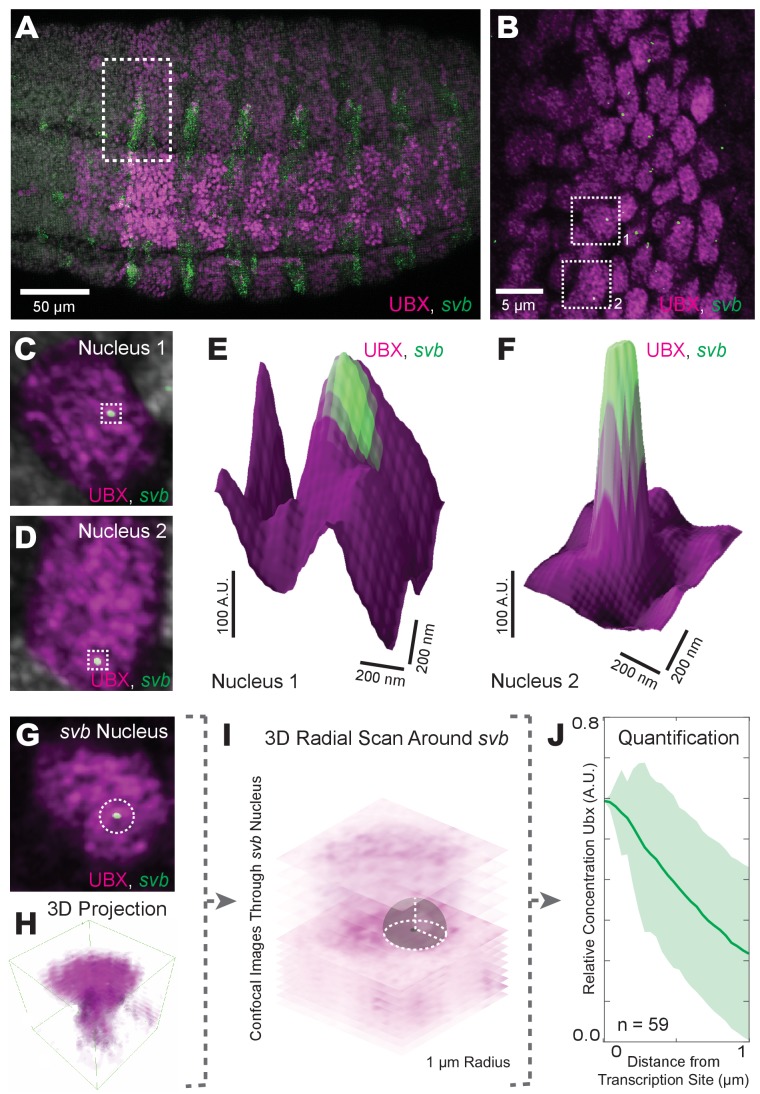
The heterogeneous distributions of Ubx we observed are consistent with the hypothesis of nuclear ‘microenvironments’ (Reiter et al., 2017), whereby high local concentrations of transcription factors may drive transcription. Therefore, we examined whether these regions of high Ubx concentration co-localized with sites of active transcription. The svb gene is directly regulated by Ubx protein through binding of Ubx to low-affinity sites in multiple svb enhancers (Crocker et al., 2015). We marked sites of active svb transcription by fluorescence in situ hybridization (FISH) and compared the localization of actively transcribed svb loci to Ubx protein concentration (Figure 2A and B). We observed high local Ubx concentrations surrounding active svb transcription sites (Figure 2C–F). To quantify Ubx distributions around these sites, we calculated the radially averaged Ubx intensity as a function of distance r from the point of maximum FISH intensity for each svb transcription site (Figure 2G–I). Ubx intensity was normalized to one at r = 0 (maximum FISH intensity) and averaged across all sites measured. To adjust for background fluorescence, we located the minimum intensity in the averaged Ubx distribution (r = 2–4 μm) and subtracted that value from the distribution. The first micrometer of the radially averaged 3D distribution is shown, with the shaded area representing the variance (Figure 2J). Within the first micrometer, svb transcription sites showed a relative enrichment of Ubx. Because these sites are on average within 200 nm of a local intensity maximum, Ubx intensity decreased monotonically away from the transcription sites, leading to a relatively constant variance after 200 nm. The normalized Ubx intensity after background subtraction at the site of svb transcription was 0.60 ± 0.17 (n = 59, four embryos, uncertainty is the variance of the background) and decreased approximately 250 nm away from the site. Thus, active svb transcription sites colocalized with areas of high Ubx concentration spanning approximately a few hundred nanometers.
Figure 2. Transcriptionally active svb loci reside in regions of high Ubx concentration.
(A) Embryos co-stained for both Ubx protein (magenta) and shavenbaby (svb) intronic mRNA (green). Bright spots of svb intronic nascent mRNA mark actively transcribed svb loci. Regions with high levels of both svb transcription and Ubx appear white (the sum of the two colors). (B) Higher magnification, Airyscan image of the region noted in panel (A), revealing sites of svb transcription (green). (C, D) Higher magnification, Airyscan images of the nuclei noted in panel (B). (E, F) 3D surface plots of the images in panels (C) and (D), centered on the sites of svb transcription (green), where height represents Ubx intensity. (G) A representative nucleus used for quantifying Ubx distribution around a svb transcription site. (H) 3D view of the confocal stack from the nuclei in panel (G). (I) Schematic outlining the method of Ubx quantification surrounding svb transcriptional sites. A 3D radial distribution of the average Ubx intensity on the surface of a sphere centered at the site of svb transcription was calculated. The gray sphere and white outlines is an example of the sphere with a radius r = 1 μm. (J) Quantification of the average relative concentration of Ubx and the distance from svb transcription sites (n = 59, see method supplements ‘settings for extracting radially averaged distributions’ for how relative concentration is computed). The shaded region indicates the variance. A.U. indicates Arbitrary Units of fluorescence intensity.
Figure 2—figure supplement 1. Transcription sites of minimal svb enhancers and the endogenous svb locus localize close to each other.
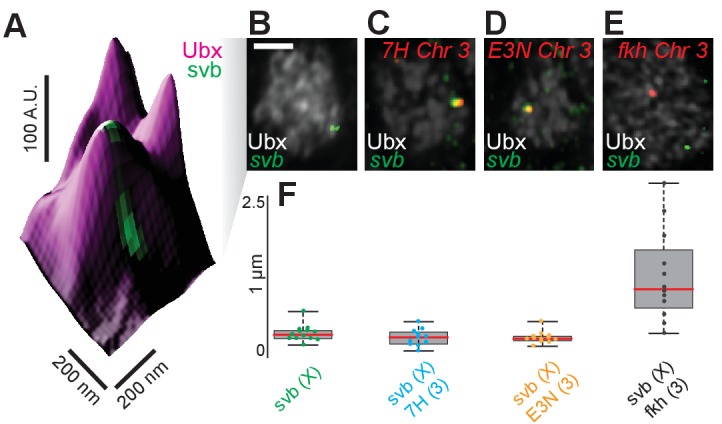
(A) 3D surface plot of a nuclear region showing two transcriptionally active svb loci close by and possibly within the same Ubx environment. The embryo is co-stained for Ubx (magenta) and the introns of nascent svb mRNA (green). The height of the plot indicates the Ubx intensity. A.U. indicates Arbitrary Units of fluorescence intensity. (B) The whole nucleus shown in (A), with Ubx in white and svb intronic mRNA in green. The svb locus is on the X chromosome. The scale bar is 2 µm. (C, D, E) The same plot as (B) stained additionally for either a reporter mRNA (lacZ) under the control of 7H (C) or E3N (D) minimal enhancers derived from svb or introns of the nascent mRNA of the endogenous forkhead locus (fkh, E). All are on chromosome 3. (F) Quantification of the distance between pairs of transcription sites. The red line is the mean, the grey box is the standard error, and the black bars are the maxima and minima of the distribution. The number of transcription site pairs are: n = 13 (svb only), n = 11 (svb & 7H), n = 12 (svb & E3N), and n = 12 (svb & fkh).
If Ubx protein co-localizes with actively transcribed svb loci because Ubx drives svb expression, then we would expect that transcription at a locus not regulated by Ubx should not co-localize with high Ubx concentrations. Indeed, we observed that active transcription sites driven by a synthetic enhancer containing binding sites for a TALEA transcription factor (Crocker et al., 2016a; Crocker and Stern, 2013; Crocker et al., 2017) did not show Ubx enrichment on average despite wide fluctuations in Ubx levels, with a relative enrichment of Ubx at TALEA-driven enhancers of 0.02 ± 0.63 (Figure 3A–C, n = 29, three embryos). As these transcription sites are not close to maxima of Ubx intensity, the variance in these distributions incresed with distance from the site of transcription.
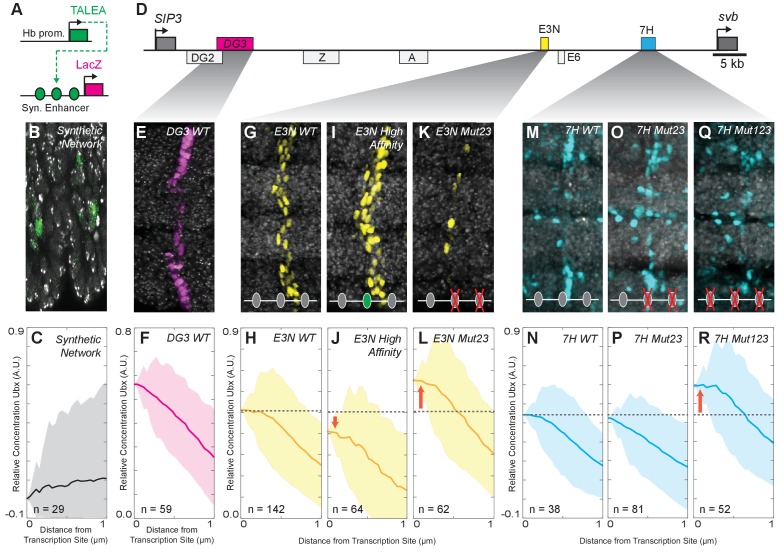
Figure 3. Manipulation of Ubx-binding site number and affinity alters the level of Ubx enrichment around svb enhancers.
(A) Schematic of the synthetic TALEA transcription network driven by the Hunchback (Hb) promoter, indicating TALEA-binding sites with green circles. (B) Early stage 15 embryos carrying the TALEA synthetic network stained with an antibody against ß-Galactosidase. (C) Quantification of the relative concentration of Ubx based on the distance from synthetic network transcription sites. (D) Schematic of the shavenbaby locus, indicating embryonic _cis_-regulatory enhancers in boxes. The ventral embryonic enhancers DG3, E3N and 7H are highlighted in magenta, yellow and blue boxes, respectively. (E, G, I, K, M, O, Q) Early stage 15 embryos carrying the reporter constructs DG3-lacZ (E), E3N-lacZ (G, I, K), or 7H-lacZ (M, O, Q) stained with an antibody against ß-Galactosidase, with Ubx-Exd sites altered as indicated. (F, H, J, L, N, P, R) Quantification of the relative concentration of Ubx versus the distance from svb transcription sites. The shaded regions in panels (C, F, H, J, L, N, P, R) indicate the variance. A.U. indicates Arbitrary Units of fluorescence intensity.
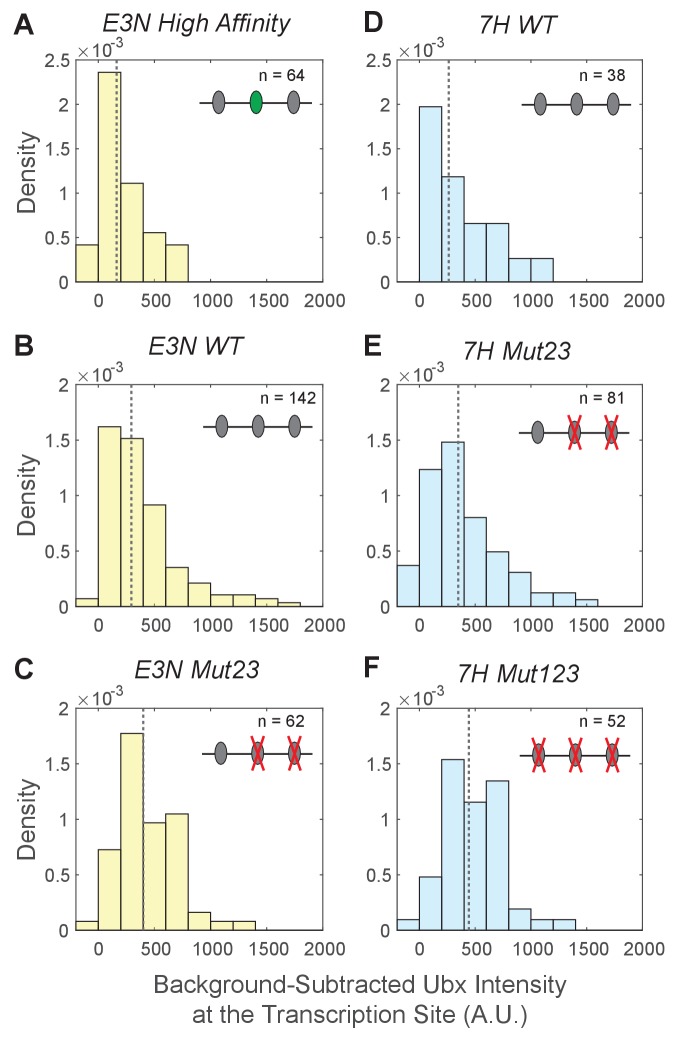
Figure 3—figure supplement 1. Background-subtracted Ubx intensity distributions at the transcription sites for E3N and 7H enhancers.
The Ubx intensity distributions at transcription sites were plotted for the E3N and 7 hr enhancers after subtraction of background fluorescence from raw Ubx intensities. The naming convention of the enhancers follows that of Figure 3. The density (_y_-axis) for each distribution is calculated by the count per bin divided by a normalization factor. The normalization factor is the bin size multiplied by the number of transcription sites (n) in the dataset. The dotted gray line is the median of the distributions, which are: (A) 160 for E3N High Affinity, (B) 290 for E3N WT, (C) 400 for E3N Mut23, (D) 260 for 7H WT, (E) 350 for 7H Mut23, and (F) 440 for 7H Mut123.
Transcription sites of svb and svb enhancers co-localize
In numerous nuclei actively transcribing svb on the X chromosome, we observed what appeared to be two transcription sites within 200 nm of each other (Figure 2—figure supplement 1A and B). This indicates that the svb locus on homologous X chromosomes often co-localizes to the same Ubx microenvironment. There are several possible mechanisms that could explain this observation. We consider two broad classes of mechanism. First, a unique chromosomal signature specific to the region containing the svb locus could facilitate localization of homologous alleles to the same transcriptional microenvironments. Second, microenvironments contain distinct combinations of transcription factors and enhancers localize to the relevant microenvironments to enable transcription. To distinguish between these alternative hypotheses, we examined the spatial distribution of the native svb locus, located on the X chromosome, and a single svb enhancer driving lacZ expression which we placed on chromosome 3.
Double-FISH experiments revealed that the native svb locus and the ectopic svb enhancer co-localized often in nuclei in which both were transcribed (Figure 2—figure supplement 1C and D). In contrast, the transcription sites of forkhead (fkh, also on chromosome 3) did not colocalize with the svb locus (Figure 2—figure supplement 1E). The average distance between pairs of related transcription sites (svb-svb, svb-7H, and svb-E3N) within single nuclei is approximately 250 nm, near the resolution limit of AiryScan images (Figure 2—figure supplement 1F). On the other hand, fkh and svb transcription sites are on average 1 µm apart. These results indicate that related enhancers co-localize in transcriptional microenvironments independently of their chromosomal location. This suggests that transcription factor microenvironments are highly differentiated and that related enhancers often exploit the same transcriptional microenvironments.
Manipulation of binding site number and affinity inversely changes the concentration of Ubx required to activate svb enhancers
The experiments described so far showed that the actively transcribed native svb locus co-localizes with local concentration maxima of Ubx in the nucleus. We wondered whether the position of actively transcribed enhancers within Ubx microenvironments depended on Ubx binding site affinity. To address this question, we examined transcription driven by the individual svb enhancers DG3, E3N, and 7 hr, each of which contains a cluster of low-affinity Ubx-binding sites and can independently drive transcription of a reporter gene when moved from their native location (Crocker et al., 2015). Transcription sites driven by these relocated enhancers also colocalized with regions of high Ubx concentration (Figure 3D). The relative Ubx enrichment for each of the three enhancers was 0.56 ± 0.16 for DG3 (n = 61, three embryos), 0.51 ± 0.19 for E3N (n = 142, 11 embryos), and 0.68 ± 0.10 for 7 hr (n = 38, three embryos) (Figure 3E–H,M,N). These results indicate that low-affinity enhancers actively transcribed far from the native svb locus also co-localize with microenvironments of high Ubx concentrations.
Increasing the binding affinity of a site should increase its sensitivity to Ubx and allow transcriptional activation at lower Ubx concentrations. We found previously that replacing a single low-affinity Ubx site with one of a higher affinity led to higher levels of expression and sometimes drove promiscuous transcription (Crocker et al., 2015), suggesting that more stable Ubx-DNA interactions allowed higher transcriptional activation. Consistent with these previous results, we observed that increasing the affinity of a single low-affinity binding site in the E3N enhancer decreased Ubx enrichment near transcription sites to 0.44 ± 0.27 (Figure 3I and J, E3N High Affinity, n = 36, three embryos).
In contrast, we reported previously that deletion of low-affinity binding sites reduced transcription (Crocker et al., 2015). Removing some Ubx-binding sites should lower the effective affinity of the enhancer, and we hypothesized that this might result in transcription only when genes are localized to areas of higher Ubx concentrations. Consistent with this model, when we deleted two low-affinity sites in E3N, active transcription was observed in regions of increased Ubx enrichment (0.65 ± 0.18, Figure 3K and L, E3N Mut23, n = 62, five embryos). Deletion of two low-affinity Ubx sites from the 7 hr enhancer did not alter Ubx enrichment around transcription sites (0.63 ± 0.37, Figure 3O and P, 7H Mut23 n = 81, six embryos). But, deletion of three Ubx-binding sites in the 7H enhancer increased relative Ubx enrichment, consistent with the pattern we observed for the E3N enhancer (0.91 ± 0.27, Figure 3Q and R, 7H Mut123, n = 52, eight embryos).
Across all manipulations, we observed an inverse correlation between binding site affinity and the distribution of Ubx intensities at transcription sites (Figure 3—figure supplement 1). Thus, the number of Ubx-binding sites and their affinities determine the response of svb enhancers to local Ubx concentration. Lower affinity enhancers require higher Ubx concentrations to drive transcription. Conversely, higher affinity enhancers can drive transcription at lower local Ubx concentrations.
Taken together, these data suggest that enhancers may be dynamically sampling local nuclear environments. A lower fraction of nuclei showing transcription from enhancers with binding site deletions (Figure 3K,O,Q) may occur because there are fewer areas of the nucleus in which peak Ubx levels are sufficient for weakened svb elements.
The Ubx cofactor Homothorax (Hth) is co-enriched around transcription sites with Ubx
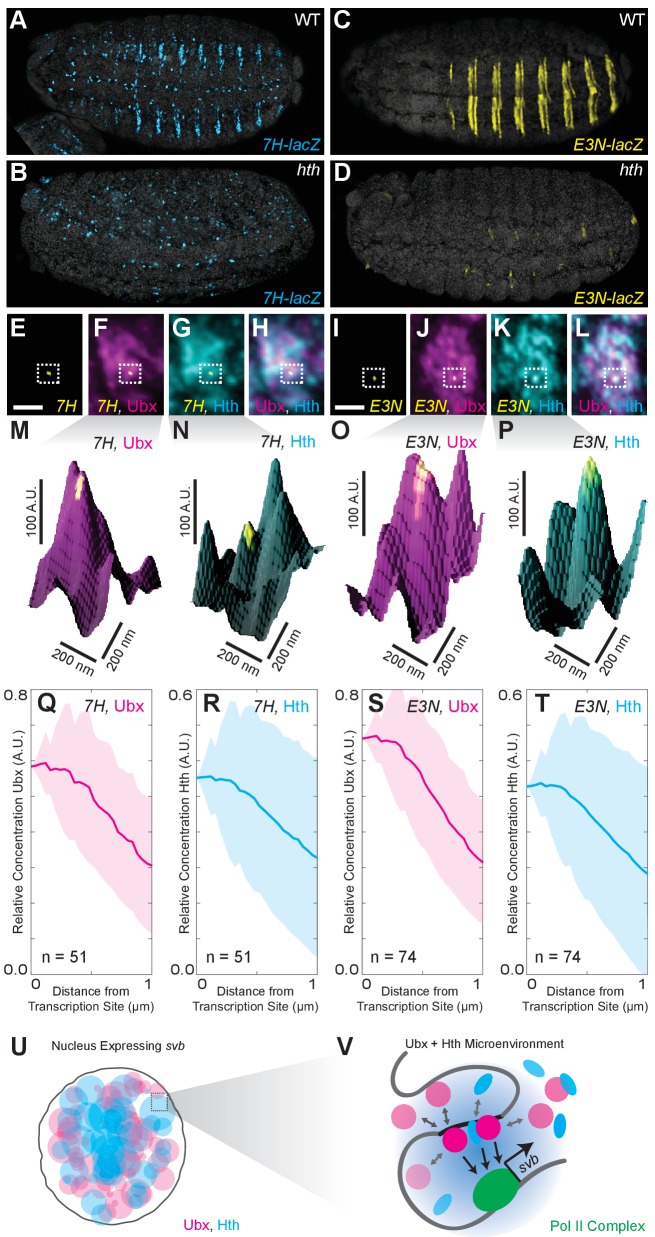
Co-factors can stabilize low-affinity binding interactions through cooperative and scaffolding interactions with transcription factors. A co-factor-dependent enhancer would require sufficient concentrations of both the factor and the co-factor to drive transcription. The homeodomain proteins Extradenticle (Exd)/Pbx and Homothorax (Hth)/MEIS (Slattery et al., 2011b; Rieckhof et al., 1997; Ryoo and Mann, 1999; Lelli et al., 2011) interact with Ubx during DNA binding, and Ubx and Hth regulate a partially overlapping set of genes (Choo et al., 2011; Slattery et al., 2011a). In vitro, Ubx requires Hth/Exd to bind to the low-affinity sites in 7H and E3N (Crocker et al., 2015). In vivo, Hth deficiency led to the loss of expression for both 7H and E3N (Figure 4A–D). Consistent with this requirement for both Ubx and Hth, Hth was co-enriched with Ubx around active transcription sites driven by 7H or E3N (Figure 4E–T). The relative enrichment for Ubx and Hth, respectively, was 0.58 ± 0.14 and 0.41 ± 0.16 for 7H (n = 51, seven embryos) and 0.66 ± 0.13 and 0.39 ± 0.24 for E3N (n = 74, five embryos). These results suggest that transcription from co-factor-dependent enhancers requires microenvironments that contain high concentrations of both transcription factors and their co-factors. This observation provides further support for the model that transcription factor microenvironments are present as multiple highly differentiated transcription domains containing unique combinations of transcription factors.
Figure 4. Ubx and its cofactor Hth are co-enriched around transcription sites.
(A–D) Early stage 15 embryos with 7H-lacz (A–B) or E3N-lacZ reporter constructs (C–D) stained with an antibody against ß-Galactosidase in either wild-type (WT) (A and C) or hthP2 mutant embryos (B and D). (E–H) A nucleus displaying active transcription of the 7H-lacZ reporter construct denoted by a bounding box (E–H) and co-stained for Ubx protein (F), Hth protein (G), or both Ubx and Hth proteins (H). (I–L) A nucleus displaying active transcription of the E3N-lacZ reporter construct denoted by a bounding box (I–L) and co-stained for Ubx protein (J), Hth protein (K), or both Ubx and Hth proteins (L). (M–P) 3D surface plots of the images in panels (F, G, J, K), centered on the sites of enhancer activity (yellow). The height of the plot is Ubx intensity in panels (M) and (O) and Hth intensity in panels (N) and (P). (Q–T) Quantification of the relative concentration of Ubx (Q, S) and Hth (R, T) versus distance from active enhancer sites. The shaded regions indicate the variance. A.U. indicates Arbitrary Units of fluorescence intensity. (U, V) A conceptual model showing nuclei with multiple regions of high local concentrations of Ubx or Hth (U) and high local concentrations of both Ubx and Hth that allow rapid ON rates (V, grey arrows) and collectively may recruit RNA pol II complexes.
Discussion
Biological systems often generate locally high concentrations of interacting molecules to increase the efficiency of biochemical reactions (Dueber et al., 2009; Oehler and Müller-Hill, 2010). This appears to be true also for transcription from low-affinity enhancers. Microenvironments (Reiter et al., 2017) of high local concentrations of transcription factors and their co-factors may circumvent the instability of low-affinity interactions by promoting more frequent DNA binding and cooperative interactions when enhancers are located within these domains (Farley et al., 2016) (Figure 4U and V). These microenvironments may be relatively stable domains generated by rapid dynamics of individual molecules. For example, we observed interactions between transcription factors and DNA on the timescale of seconds, with transcription factors continuously arriving to and departing from specific loci. From the perspective of gene expression, transcription likely occurs intermittently, switching on and off as the gene locus samples different nuclear regions. These rapid dynamics ensure that, once the gene locus moves outside of a microenvironment, or the conditions to form microenvironments are no longer satisfied, then the transcription factors needed to sustain expression quickly depart from low-affinity binding sites. In contrast, the fact that svb enhancers placed on the third chromosome often co-localized with the native svb locus on the X chromosome suggests that unique microenvironments may have relatively long half-lives. One challenge for the future is to determine how rapid dynamics of individual molecules generates apparently stable sub-nuclear domains.
Many mechanisms might work in concert to create these observed microenvironments. First, clustered binding sites for the same transcription factor (Crocker et al., 2016b) could lengthen the dwell time of proteins near enhancers and increase effective local protein concentrations (Yao et al., 2006; Zhang et al., 2006; Elf et al., 2007; Kabata et al., 1993; Leith et al., 2012; Ruusala and Crothers, 1992). Second, cooperative and scaffolding interactions between transcription factors and co-factors, each of which may bind independently to enhancers, can stabilize transcription factors at low-affinity sites (Farley et al., 2016; Junion et al., 2012). Finally, clustering of enhancers could trap transcription factors over longer length scales (Noordermeer et al., 2014; de Laat and Duboule, 2013; Symmons et al., 2016; Williamson et al., 2016; Giorgetti et al., 2016), perhaps generating the ~200 nm microenvironments that we observed. This last model is supported by recent findings that multiple promoters can share the same enhancer in a common local environment (Fukaya et al., 2016).
Transcription factor microenvironments may be a general feature of eukaryotic transcription, as supported by studies showing mouse and human cells exhibiting RNA polymerase II crowding (Cisse et al., 2013; Cho et al., 2016), transcription factors using local clustering to efficiently find their binding sites (Liu et al., 2014; Izeddin et al., 2014), and chromatin packaging in Drosophila cells generating distinct chromatin environments at the kilobase-to-megabase scale (Boettiger et al., 2016). Collectively, these findings are consistent with a phase-separated model of transcriptional regulation (Hnisz et al., 2017) whereby distinct microenvironments contain different combinations of proteins inside the nucleus. These localized regions impose a spatial constraint on the expression of genes, allowing transcriptional activation from enhancers only when they are physically in regions with the correct combinations of transcription factors and co-factors. Multiple enhancers acting as DNA scaffolds for protein binding could provide the anchoring interactions that form transcriptional microenvironments. These microenvironments would, in turn, provide a mechanism to allow both efficient and specific transcription from low-affinity enhancers.
Materials and methods
Preparing fixed Drosophila embryos
D. melanogaster strains were maintained under standard laboratory conditions. All enhancer constructs were cloned into the placZattB expression construct with a hsp70 promoter (Crocker et al., 2015). Transgenic fly lines were made by Rainbow Transgenic Flies Inc. E3 and 7H were integrated at the attP2 landing site. DG3 was integrated at ZH-86Fb.
Immuno-fluorescence staining of transcription factors and in situ hybridization to mRNA
Flies were reared at 25°C and embryos were fixed and stained according to standard protocols (Crocker et al., 2015). Primary antibodies were detected using secondary antibodies labeled with Alexa Fluor dyes (1:500, Invitrogen). In situ hybridizations were performed using DIG or biotin-labeled, antisense RNA-probes against a reporter construct RNA (lacZ) or the first intron of svb or fkh. DIG-labeled RNA products were detected with a DIG antibody: Invitrogen, 9H27L19 (1:200 dilution) and biotin-labeled RNA products are detected using a biotin antibody: Pierce, PA1-26792 (1:200).
The following primary antibodies for proteins were used at the indicated concentrations:
Ubx: Developmental Studies Hybridoma Bank, FP3.38-C (1:20)
Hth: Santa Cruz Biotechnology (dN-19), sc-26186 (1:50)
Eve: Developmental Studies Hybridoma Bank, 2B8-C (1:20)
AbdA: Santa Cruz Biotechnology (dN-17), sc-27063 (1:50)
En: Santa Cruz Biotechnology (d-300), sc-28640 (1:50)
RNA PolII RPB1 (Ser5 phosphorylated): BioLegend, (920304), (1:200)
Histone H3K27me3: Active Motif, 39157 (1:200)
Histone H3K4me3: Cell-signaling technology C42D8 (1:200)
LacZ: Promega anti-ß-Gal antibody (1:1000)
Imaging fixed embryos with Airyscan
Fixed Drosophila embryos mounted in ProLong Gold mounting media (Molecular Probes, Eugene, OR) were imaged on a Zeiss LSM 880 confocal microscope with Airyscan (Carl Zeiss Microscopy, Jena, Germany) using 3D Airyscan in SR mode to obtain images with 1.7-fold higher resolution compared to diffraction-limited confocal imaging (Sheppard et al., 2013) (method supplements: imaging setup for Airyscan). Images presented in the figures were processed with ImageJ (Schindelin et al., 2015).
Expanding fixed embryos
To expand embryos, after fixation and staining, embryos were embedded into poly-acrylate gels and expended according to a previously published protocol (Tillberg et al., 2016) (method supplements: handling expansion gels).
Imaging expanded embryos
Expanded gels containing embryos were imaged in 6-well glass bottom plates (Cellvis, Mountain View, CA) using a Zeiss LSM 800 confocal microscope (Carl Zeiss Microscopy, Jena, Germany) using standard settings (method supplements: imaging setup for expanded embryos).
HaloTag-Ubx transgene construct for live imaging and overexpression assay
Transgenic fly lines containing HaloTag-Ubx under the control of both a hsp70 and a 20x UAS promoter was made by Rainbow Transgenic Flies Inc. The lines were made homozygous for the transgene.
Preparing embryos for live imaging
Embryos resulting from crossing the homozygous line with the HaloTag-Ubx transgene with a nos::GAL4 driver line were injected following previously established protocols (Rubin and Spradling, 1982) with the HaloTag ligand of JF635. Briefly, embryos were collected for 30 min at 25°C and placed in oxygen permeable Halocarbon 27 oil. The stock dye solution of 1 mM JF635 with a HaloTag ligand in DMSO was diluted 1:100 into fly injection buffer and injected into the posterior end of the embryos. The embryos were then aged to stage 5 or late stage 6 and imaged in oxygen permeable Halocarbon 27 oil.
Live imaging of Drosophila embryos
Injected embryos were imaged on a customized inverted Nikon Ti-Eclipse (Nikon Instruments, Tokyo, Japan) with the appropriate settings (method supplements: imaging setup for live embryos).
Embryos for HaloTag-Ubx overexpression assay
Embryos from the homozygous HaloTag-Ubx transgene line were exposed to 30°C to induce the heat shock promoter and cuticle preps were prepared following previously established protocols (Crocker et al., 2015).
Radially averaged distributions centered around transcription sites
To obtain the distributions of Ubx and Hth around a transcription site, the processed Airyscan stacks obtained from the Zeiss LSM 880 confocal microscope were analyzed in Fiji (Schindelin et al., 2012) using native functions and the 3D ImageJ Suite plugin (Schmid et al., 2010). Radially averaged distributions for individual transcription sites were computed using the 3D ImageJ Suite Plugin. Distributions for all sites were averaged and background offset in Matlab (MathWorks, Natick, MA) using a custom script (method supplements: settings for extracting radially averaged distributions).
Method supplements
Imaging setup for Airyscan
All Airyscan images were acquired using a Zeiss Plan-Apochromat 63x/1.4 Oil DIC M27 objective due to its well-characterized point spread function. First an embryo at the appropriate developmental stage (stage 15 for most embryos) and proper orientation was located. The band of mRNA expression in high Ubx regions of the first abdominal (A1) segment was then found. Within that band, areas containing transcription sites in nuclei of high Ubx expression were imaged. Images with both Ubx and Hth were acquired in the same manner by locating the proper area using the mRNA and Ubx. When Ubx was imaged together with RNA polymerase II, a histone marker, or other transcription factors, Ubx expression levels were used to locate the region of interest.
The optimal setting suggested by Zeiss for the number of pixels in the x-y direction (40 nm per pixel) and displacement in the z-stack (190 nm) were used for all Airyscan images. The images from different fluorophores were acquired sequentially with the appropriate laser lines (405 nm, 488 nm, 561 nm, or 633 nm) and spectral filters. The laser power and gain were adjusted to maximize the signal to noise ratio within the dynamic range of the Airyscan detector. The acquired stacks were processed with Zen 2.3 SP1 (Carl Zeiss Microscopy GmbH, Jena, Germany) in 3D mode to obtain super-resolved images.
Handling expansion gels
To allow easier handling of expanded gels, the gels containing embryos were cast into eight-well silicone isolators without adhesives (eight round chambers with a diameter of 9 mm and a thickness of 0.5 mm, Grace Bio-Labs (Bend, OR)) and allowed to polymerize. The gels were transferred into a six-well glass-bottom cell culture plate (Cellvis, Mountain View, CA) and expanded using ultrapure water containing 500 nM DAPI. Before imaging, the water was removed and the gel encased in 3% low melting temperature agarose (NuSieve GTG Agarose, Lonza Group Ltd, Basel, Switzerland), taking care not to allow the agarose to flow under the gel and float the gel away from the cover glass bottom. Water was then added back into the wells to prevent drying.
Imaging setup for expanded embryos
A long working-distance water immersion objective, the Zeiss LCI Plan-Neofluar 25x/0.8 Imm Korr DIC M27, was selected for index-matching with the gel and its ability to image up to 400 µm above the surface of the coverslip. Stage 15 embryos in the correct orientation were located using the DAPI and Ubx staining. Regions of low to high Ubx expression were imaged sequentially using the appropriate laser lines (405 nm, 488 nm, or 561 nm) with the proper spectral filters. The laser power settings and the gain were selected to maximize signal to noise within the dynamic range of the detector. The full field of view of the microscope was imaged with 2048 × 2048 pixels and with a z-step of 1 μm. The final images presented were processed in ImageJ (Schindelin et al., 2015).
Imaging setup for live embryos
All videos were collected under a Nikon CFI Plan Apo NCG 100X Oil NA 1.41 objective with an Andor iXon 897 EMCCD camera (Andor Technology Ltd., Belfast, UK). Embryos at stage 5 and late stage 6 in the correct orientation were found and imaged. We selected an area in the middle of the embryo with enough dye-labeled Ubx molecules to observe single molecules and we avoided regions close to the injection site to avoid oversaturating the camera (compare with Figure S5A-D where there are too many labeled Ubx). The samples were illuminated with a 633 nm laser to image the JF635 tagged Halo-Ubx molecules with laser power and camera gain set to maximize signal from individual Ubx molecules without oversaturating the EMCCD detector. The 512 × 512 pixel videos were acquired at an exposure time of 100 ms per frame for up to 200 s. Images were processed using ImageJ to generate the time-averaged images and the intensity-over-time traces presented in the figures.
Settings for extracting radially averaged distributions
To extract radially averaged protein distributions, we used Fiji to identify transcription sites inside nuclei by thresholding at a level that is roughly 50-fold above the background intensity. The center of a transcription site was defined as the pixel of maximum intensity in 3D in the mRNA channel inside a nucleus with high levels of Ubx expression. The radially averaged distribution out to a radius of 4 μm from transcription site for the transcription factor in 3D was computed using the 3D ImageJ Suite. The suite generates the distribution by computing the average intensity on the surface of a sphere with a radius r from the center in three dimensions for all the values of r ranging from zero to a desired outer limit (4 μm in this case).
The individual distributions from each transcription site were normalized to have the intensity at the center (r = 0) equal to 1. The distributions were averaged and background offset in Matlab. To adjust for background Ubx intensity outside of the nucleus, the entire averaged distribution was offset by a constant value to bring the minimum intensity present in the distribution to zero to generate the distribution plots. The shaded area around the line represents the variance. The first μm of the distributions, where contributions from outside of the nucleus were minimal, are shown in the figures. The relative enrichment of Ubx or Hth for each enhancer variant is the intensity at r = 0 in the distribution and the cited uncertainty is the variance at the location of zero Ubx or Hth intensity (the site of minimum intensity before offsetting, between 2 and 4 μm from the transcription site).
The initial dataset for 7H enhancers contained only a part of the deletion series. A subsequent dataset contained all the 7H deletion mutants. The 7H mutants present in both sets were compared and the distributions of Ubx intensity between the sets were found to differ by a multiplicative factor. When such factor was computed for each overlapping 7H mutant present in both datasets, the results were similar, indicating that there was a systematic shift in background noise. This could have resulted from differences in embryo handling during fixation, antibody staining, and other steps in sample preparation. Other characteristics such as the functional form of the distributions between the two sets and the trends between 7H mutants within each set remained unchanged after correcting for the difference in intensity. The wild-type 7H data from the first set with a correction factor and the rest of the deletion series uncorrected from the second set were used to minimize the normalization employed.
Acknowledgements
We thank Richard Mann, Timothée Lionnet, Paul Tillburg, and Brian English for advice and assistance on experimental design. We thank François Payre for advice on data presentation. We thank all members of the Stern and Singer labs for discussion. Albert Tsai is a Damon Runyon Fellow of the Damon Runyon Cancer Research Foundation (DRG 2220–15). Robert H Singer is supported by the 4D Nucleome Award U01-EB21236. Howard Hughes Medical Institute supported Albert Tsai, Anand K Muthusamy, Luke D Lavis, Robert H Singer, David L Stern, and Justin Crocker Mariana R P Alves and Justin Crocker are supported by the European Molecular Biological Laboratory (EMBL).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Albert Tsai, Email: tsaia@janelia.hhmi.org.
Justin Crocker, Email: justin.crocker@embl.de.
David N Arnosti, Michigan State University, United States.
Funding Information
This paper was supported by the following grants:
- Damon Runyon Cancer Research Foundation DRG 2220-15 to Albert Tsai.
- National Institutes of Health U01-EB21236 to Robert H Singer.
- Howard Hughes Medical Institute to Albert Tsai, Anand K Muthusamy, Luke D Lavis, Robert H Singer, David L Stern, Justin Crocker.
- European Molecular Biology Laboratory to Mariana RP Alves, Justin Crocker.
Additional information
Competing interests
Reviewing editor, eLife.
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Validation, Investigation, Visualization, Methodology, Writing—original draft, Writing—review and editing, Conceived and designed the experiments, Executed the experiments, Analyzed the data.
Resources, Provided reagents and design for live imaging experiments, Writing—review and editing.
Investigation, Executed the experiments, Analyzed the data, Writing—review and editing.
Resources, Writing—review and editing, Provided reagents and design for live imaging experiments.
Resources, Supervision, Funding acquisition, Writing—review and editing, Conceived of and designed the experiments.
Conceptualization, Supervision, Funding acquisition, Investigation, Visualization, Methodology, Writing—original draft, Writing—review and editing, Conceived and Designed the experiments.
Conceptualization, Supervision, Funding acquisition, Investigation, Visualization, Methodology, Writing—original draft, Writing—review and editing, Conceived and designed the experiments, Executed the experiments, Analyzed the data.
Additional files
Source code 1. Matlab script to average radially averaged intensity distributions from individual transcription sites and offset background fluorescence.
Transparent reporting form
Major datasets
The following dataset was generated:
Tsai A, author; Muthusamy A, author; Alves M, author; Lavis L, author; Singer R, author; Stern D, author; Crocker J, author. Data from: Nuclear microenvironments modulate transcription from low-affinity enhancers. 2017 http://dx.doi.org/10.5061/dryad.q96g6 Available at Dryad Digital Repository under a CC0 Public Domain Dedication.
References
- Antosova B, Smolikova J, Klimova L, Lachova J, Bendova M, Kozmikova I, Machon O, Kozmik Z. The gene regulatory network of lens induction is wired through meis-dependent shadow enhancers of Pax6. PLOS Genetics. 2016;12:e1006441. doi: 10.1371/journal.pgen.1006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Bintu B, Moffitt JR, Wang S, Beliveau BJ, Fudenberg G, Imakaev M, Mirny LA, Wu CT, Zhuang X. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016;529:418–422. doi: 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, Tjian R, Liu Z. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WK, Jayanth N, English BP, Inoue T, Andrews JO, Conway W, Grimm JB, Spille JH, Lavis LD, Lionnet T, Cisse II. RNA Polymerase II cluster dynamics predict mRNA output in living cells. eLife. 2016;5:e13617. doi: 10.7554/eLife.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo SW, White R, Russell S, Mi H, Diemer K. Genome-wide analysis of the binding of the Hox protein Ultrabithorax and the Hox cofactor Homothorax in Drosophila. PLoS One. 2011;6:e14778. doi: 10.1371/journal.pone.0014778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, Darzacq X. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013;341:664–667. doi: 10.1126/science.1239053. [DOI] [PubMed] [Google Scholar]
- Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, Wang S, Alsawadi A, Valenti P, Plaza S, Payre F, Mann RS, Stern DL. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell. 2015;160:191–203. doi: 10.1016/j.cell.2014.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J, Ilsley GR, Stern DL. Quantitatively predictable control of Drosophila transcriptional enhancers in vivo with engineered transcription factors. Nature Genetics. 2016a;48:292–298. doi: 10.1038/ng.3509. [DOI] [PubMed] [Google Scholar]
- Crocker J, Noon EP, Stern DL. The soft touch: low-affinity transcription factor binding sites in development and evolution. Current Topics in Developmental Biology. 2016b;117:455–469. doi: 10.1016/bs.ctdb.2015.11.018. [DOI] [PubMed] [Google Scholar]
- Crocker J, Potter N, Erives A. Dynamic evolution of precise regulatory encodings creates the clustered site signature of enhancers. Nature Communications. 2010;1:99. doi: 10.1038/ncomms1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J, Stern DL. TALE-mediated modulation of transcriptional enhancers in vivo. Nature Methods. 2013;10:762–767. doi: 10.1038/nmeth.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J, Tsai A, Stern DL. A fully synthetic transcriptional platform for a multicellular eukaryote. Cell Reports. 2017;18:287–296. doi: 10.1016/j.celrep.2016.12.025. [DOI] [PubMed] [Google Scholar]
- de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nature Biotechnology. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, Levine MS. Suboptimization of developmental enhancers. Science. 2015;350:325–328. doi: 10.1126/science.aac6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Rokhsar DS, Levine MS. Syntax compensates for poor binding sites to encode tissue specificity of developmental enhancers. PNAS. 2016;113:6508–6513. doi: 10.1073/pnas.1605085113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T, Lim B, Levine M. Enhancer control of transcriptional bursting. Cell. 2016;166:358–368. doi: 10.1016/j.cell.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Gebhardt JC, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, Maniatis T, Xie XS. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nature Methods. 2013;10:421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, Dekker J. Structural organization of the inactive X chromosome in the mouse. Nature. 2016;535:575–579. doi: 10.1038/nature18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm JB, Muthusamy AK, Liang Y, Brown TA, Lemon WC, Patel R, Lu R, Macklin JJ, Keller PJ, Ji N, Lavis LD. A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nature Methods. 2017;14:987–994. doi: 10.1038/nmeth.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeddin I, Récamier V, Bosanac L, Cissé II, Boudarene L, Dugast-Darzacq C, Proux F, Bénichou O, Voituriez R, Bensaude O, Dahan M, Darzacq X. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. eLife. 2014;3:e02230. doi: 10.7554/eLife.02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72:741–752. doi: 10.1016/0092-8674(93)90402-C. [DOI] [PubMed] [Google Scholar]
- Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, Furlong EE. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell. 2012;148:473–486. doi: 10.1016/j.cell.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Kabata H, Kurosawa O, Arai I, Washizu M, Margarson SA, Glass RE, Shimamoto N. Visualization of single molecules of RNA polymerase sliding along DNA. Science. 1993;262:1561–1563. doi: 10.1126/science.8248804. [DOI] [PubMed] [Google Scholar]
- Lebrecht D, Foehr M, Smith E, Lopes FJ, Vanario-Alonso CE, Reinitz J, Burz DS, Hanes SD. Bicoid cooperative DNA binding is critical for embryonic patterning in Drosophila. PNAS. 2005;102:13176–13181. doi: 10.1073/pnas.0506462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leith JS, Tafvizi A, Huang F, Uspal WE, Doyle PS, Fersht AR, Mirny LA, van Oijen AM. Sequence-dependent sliding kinetics of p53. PNAS. 2012;109:16552–16557. doi: 10.1073/pnas.1120452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelli KM, Noro B, Mann RS. Variable motif utilization in homeotic selector (Hox)-cofactor complex formation controls specificity. PNAS. 2011;108:21122–21127. doi: 10.1073/pnas.1114118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Legant WR, Chen BC, Li L, Grimm JB, Lavis LD, Betzig E, Tjian R. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. eLife. 2014;3:e04236. doi: 10.7554/eLife.04236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J. Ever-changing landscapes: Transcriptional enhancers in development and evolution. Cell. 2016;167:1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorberbaum DS, Ramos AI, Peterson KA, Carpenter BS, Parker DS, De S, Hillers LE, Blake VM, Nishi Y, McFarlane MR, Chiang AC, Kassis JA, Allen BL, McMahon AP, Barolo S. An ancient yet flexible cis-regulatory architecture allows localized Hedgehog tuning by patched/Ptch1. eLife. 2016;5 doi: 10.7554/eLife.13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki T, Müller WG, Golob N, Mazza D, McNally JG. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nature Communications. 2014;5:4456. doi: 10.1038/ncomms5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Schorderet P, Joye E, Chabaud F, Duboule D. Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. eLife. 2014;3:e02557. doi: 10.7554/eLife.02557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanno D, Boudarène L, Dugast-Darzacq C, Chen J, Richter C, Proux F, Bénichou O, Voituriez R, Darzacq X, Dahan M. Probing the target search of DNA-binding proteins in mammalian cells using TetR as model searcher. Nature Communications. 2015;6:7357. doi: 10.1038/ncomms8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehler S, Müller-Hill B. High local concentration: a fundamental strategy of life. Journal of Molecular Biology. 2010;395:242–253. doi: 10.1016/j.jmb.2009.10.056. [DOI] [PubMed] [Google Scholar]
- Ramos AI, Barolo S. Low-affinity transcription factor binding sites shape morphogen responses and enhancer evolution. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368:20130018. doi: 10.1098/rstb.2013.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter F, Wienerroither S, Stark A. Combinatorial function of transcription factors and cofactors. Current Opinion in Genetics & Development. 2017;43:73–81. doi: 10.1016/j.gde.2016.12.007. [DOI] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/S0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Rister J, Razzaq A, Boodram P, Desai N, Tsanis C, Chen H, Jukam D, Desplan C. Single-base pair differences in a shared motif determine differential Rhodopsin expression. Science. 2015;350:1258–1261. doi: 10.1126/science.aab3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Siggers T, Lachke SA, Yue Y, Bulyk ML, Maas RL. Precise temporal control of the eye regulatory gene Pax6 via enhancer-binding site affinity. Genes & Development. 2010;24:980–985. doi: 10.1101/gad.1890410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Ruusala T, Crothers DM. Sliding and intermolecular transfer of the lac repressor: kinetic perturbation of a reaction intermediate by a distant DNA sequence. PNAS. 1992;89:4903–4907. doi: 10.1073/pnas.89.11.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Mann RS. The control of trunk Hox specificity and activity by Extradenticle. Genes & Development. 1999;13:1704–1716. doi: 10.1101/gad.13.13.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: An open platform for biomedical image analysis. Molecular Reproduction and Development. 2015;82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B, Schindelin J, Cardona A, Longair M, Heisenberg M. A high-level 3D visualization API for Java and ImageJ. BMC Bioinformatics. 2010;11:274. doi: 10.1186/1471-2105-11-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard CJ, Mehta SB, Heintzmann R. Superresolution by image scanning microscopy using pixel reassignment. Optics Letters. 2013;38:2889–2892. doi: 10.1364/OL.38.002889. [DOI] [PubMed] [Google Scholar]
- Slattery M, Ma L, Négre N, White KP, Mann RS. Genome-wide tissue-specific occupancy of the Hox protein Ultrabithorax and Hox cofactor Homothorax in Drosophila. PLoS One. 2011a;6:e14686. doi: 10.1371/journal.pone.0014686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, Mann RS. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell. 2011b;147:1270–1282. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nature Reviews Genetics. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Symmons O, Pan L, Remeseiro S, Aktas T, Klein F, Huber W, Spitz F. The Shh topological domain facilitates the action of remote enhancers by reducing the effects of genomic distances. Developmental Cell. 2016;39:529–543. doi: 10.1016/j.devcel.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Research. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, An L, Hansen AS, Xie L, Darzacq X, Tjian R. A dynamic mode of mitotic bookmarking by transcription factors. eLife. 2016;5:e22280. doi: 10.7554/eLife.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu CC, English BP, Gao L, Martorell A, Suk HJ, Yoshida F, DeGennaro EM, Roossien DH, Gong G, Seneviratne U, Tannenbaum SR, Desimone R, Cai D, Boyden ES. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nature Biotechnology. 2016;34:987–992. doi: 10.1038/nbt.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I, Lettice LA, Hill RE, Bickmore WA. Shh and ZRS enhancer colocalisation is specific to the zone of polarising activity. Development. 2016;143:2994–3001. doi: 10.1242/dev.139188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- Zhang C, Xuan Z, Otto S, Hover JR, McCorkle SR, Mandel G, Zhang MQ. A clustering property of highly-degenerate transcription factor binding sites in the mammalian genome. Nucleic Acids Research. 2006;34:2238–2246. doi: 10.1093/nar/gkl248. [DOI] [PMC free article] [PubMed] [Google Scholar]
In the interests of transparency, eLife includes the editorial decision letter and accompanying author responses. A lightly edited version of the letter sent to the authors after peer review is shown, indicating the most substantive concerns; minor comments are not usually included.
Thank you for submitting your article "Nuclear microenvironments modulate transcription from low-affinity enhancers" for consideration by eLife. Your article has been favorably evaluated by Jessica Tyler (Senior Editor) and four reviewers, one of whom is a member of our Board of Reviewing Editors. The following individuals involved in review of your submission have agreed to reveal their identity: Robert P Zinzen (Reviewer #2); Stephen J. Small (Reviewer #3).
The reviewers have discussed the reviews with one another and the Reviewing Editor has drafted this decision to help you prepare a revised submission.
The reviewers of your manuscript found that the observations on the inhomogeneity of intranuclear Ubx transcription factor concentration and the activation of homologous enhancers with different Ubx affinities were highly significant and convincingly demonstrated. A strength of this study is that the authors use complementary techniques on fixed and living cells to observe the distribution of Ubx and other TF, and see similar trends. The use of svb regulatory elements with carefully "tuned" Ubx affinities and output is another powerful tool that reveals the importance of the different Ubx domains. Overall, the reviewers found that this study uses a set of advanced imaging methods to fundamentally reshape our view of the nuclear environment in the context of developmental gene regulation.
Essential points to be addressed:
There was some uncertainty whether the authors are proposing that the change in affinity of the Ubx motifs reshapes the intranuclear concentration gradients, or whether they are just responding differently to the existing inhomogeneities in the nucleus. The statement "Manipulation of binding site number and affinity changes the level of Ubx enrichment around svb enhancers" might be interpreted to mean that the selected enhancers are shaping the nuclear gradients. A clearer understanding of the authors' interpretation is necessary, and better statement of what they propose is establishing the different concentration of Ubx. Another related point: are the authors proposing that the enhancers get "stuck" in areas of appropriate concentration, or do they randomly sample the different intranuclear environments, activating when they are experiencing sufficient Ubx levels?
Pertinent to the above point, the in vivo halo-tagged imaging was found to be a very helpful complement to the analysis of fixed material, but the paper should do a better job of explicitly tying the results from the in vivo imaged spots to later work with the fixed, expanded embryos. The halo-tagged construct was not adequately described in the paper (only a brief outline in Figure 1—figure supplement 4). Finally, an important control for the in vivo expression was testing of a DNA-binding deficient form of Ubx, which did not show the inhomogeneous distributions, suggesting that DNA binding to something is essential for formation of the gradients. It was not clear that this negative control protein was expressed, however; a Western blot would demonstrate that the lack of signal is not due to trivial lack of stability.
An intriguing point of this paper is that as Ubx motifs are degraded, the active spots become localized only to regions of higher Ubx activity. In addition to the shown mean values that are higher for the low affinity enhancers, it is also clear from the embryo images that the weaker site enhancers are found in fewer nuclei altogether. Is this because the low affinity enhancer is able to respond to Ubx concentrations that are only found in a small percentage of nuclei? The authors are asked to show how the distribution of active enhancers in all nuclei changes as a function of Ubx motif affinity, as well as the mean values in nuclear regions where it is active. In addition, the mean levels of Ubx found in proximity to the active transcription loci have a very small variation at close range, and increase with increasing distance for a small range. Then the variation in concentration appears to be constant (and not continue to increase, as might be expected for searches of larger and larger spaces). The authors are asked to explain the shape of the Ubx distributions noted for the reporter constructs.
The reporter constructs with different Ubx affinities were derived from svb enhancer sequences. Do the reporters activate and colocalize with the endogenous svb locus in the fixed specimens? Double FISH with intronic probes should be able to discern this point.
Essential points to be addressed:
- There was some uncertainty whether the authors are proposing that the change in affinity of the Ubx motifs reshapes the intranuclear concentration gradients, or whether they are just responding differently to the existing inhomogeneities in the nucleus. The statement "Manipulation of binding site number and affinity changes the level of Ubx enrichment around svb enhancers" might be interpreted to mean that the selected enhancers are shaping the nuclear gradients. A clearer understanding of the authors' interpretation is necessary, and better statement of what they propose is establishing the different concentration of Ubx.
We agree that the original statement could imply that changing the enhancer architecture changes the distribution of Ubx in the nucleus. We have modified this statement to clarify that we do not expect this to happen. Our interpretation is that the mutated enhancers respond to existing Ubx concentration gradients within the nucleus. Even if the aggregated effects of transcription factor interacting with binding sites could shape their overall distribution, changing only one to three low affinity binding sites out of the thousands of possible Ubx binding sites in the genome of D. melanogaster is unlikely to change Ubx distribution in general.
Another related point: are the authors proposing that the enhancers get "stuck" in areas of appropriate concentration, or do they randomly sample the different intranuclear environments, activating when they are experiencing sufficient Ubx levels?
Our interpretation is that enhancers sample various areas of the nuclear environment and initiate transcription only when Ubx and cofactor concentrations are sufficient, but this is only our working model. In the future, we plan to characterize the dynamics of these interactions using additional live imaging strategies, but this technology is not yet fully operational.
- Pertinent to the above point, the in vivo halo-tagged imaging was found to be a very helpful complement to the analysis of fixed material, but the paper should do a better job of explicitly tying the results from the in vivo imaged spots to later work with the fixed, expanded embryos.
It is not entirely clear what the reviewers are requesting here. We first observed microenvironments in fixed embryos and then expanded fixed embryos to examine the microenvironments at higher resolution. We then used live imaging to characterize the temporal dynamics of transcription factor binding and found that the dynamics were consistent with the observations of fixed specimens. We explicitly state that the live imaging was performed, in part, to test whether the observations of microenvironments in fixed specimens was an artifact of fixation. The consistency between the live-imaging and fixed specimens suggests that microenvironments are real. All following experiments were performed with fixed, unexpanded embryos.
The halo-tagged construct was not adequately described in the paper (only a brief outline in Figure 1—figure supplement 4).
We have added a description of the HaloTag-Ubx construct when we first introduce it in the Results, referencing the construct diagram in the figure. We also added a more complete description in the Materials and methods section. We also updated the diagram in Figure 1—figure supplement 4 to indicate the location of the actual HaloTag-Ubx sequence to further clarify its architecture. Additionally, we will deposit the construct to Addgene to enable use by the community.
Finally, an important control for the in vivo expression was testing of a DNA-binding deficient form of Ubx, which did not show the inhomogeneous distributions, suggesting that DNA binding to something is essential for formation of the gradients. It was not clear that this negative control protein was expressed, however; a Western blot would demonstrate that the lack of signal is not due to trivial lack of stability.
The DNA-binding deficient Ubx involves only two mutations to alanine in the DNA binding pocket and has previously been reported by Richard Mann and colleagues. With the binding deficient mutant, we still observed that the mutant Ubx is labeled using JF635 with a HaloTag ligand based on the bright fluorescent signals we observed post dye injection. Without a folded and functional HaloTag domain, the dye would have remained dark. The mutant Ubx is also selectively localized into the nucleus, indicating a functional NLS that has not been degraded. We still occasionally observed single-molecules of this Ubx inside the nucleus, but their apparent dwell-time is very short (less than 100 ms). These observations suggest that the mutant protein is expressed and stable so that it is not degraded immediately post translation and can be imported into the nucleus.
We also performed an experiment with just an H2B-derived NLS fused to the HaloTag, which is unstable, and we observed no fluorescence signal beyond background noise and auto-fluorescence. There was no enrichment in the nucleus. This control experiment demonstrates the effect of an unstable protein that is degraded immediately, in sharp contrast with the Ubx binding-deficient mutant. We have added Figure 1—figure supplement 7 and accompanying text in the Results section summarizing this point.
- An intriguing point of this paper is that as Ubx motifs are degraded, the active spots become localized only to regions of higher Ubx activity. In addition to the shown mean values that are higher for the low affinity enhancers, it is also clear from the embryo images that the weaker site enhancers are found in fewer nuclei altogether. Is this because the low affinity enhancer is able to respond to Ubx concentrations that are only found in a small percentage of nuclei? The authors are asked to show how the distribution of active enhancers in all nuclei changes as a function of Ubx motif affinity, as well as the mean values in nuclear regions where it is active.
We agree with the interpretation that weaker enhancers require higher Ubx concentration to activate transcription. As a result, fewer microenvironments within a nucleus would contain sufficient concentrations of Ubx to activate transcription, lowering the probability that an enhancer exploring different areas within a nucleus would be transcriptionally active at any given moment. However, due to the heterogeneous distributions of Ubx in the nuclei of embryos at stage 15, the average concentration of Ubx over an entire nucleus is a poor representation of the specific conditions within microenvironments. While we observed that these weakened enhancers are active in microenvironments with higher Ubx concentrations, transcription sites are not preferentially located in nuclei of high average Ubx concentration. In fact, we did not observe transcription sites in most nuclei with some of the highest average levels of Ubx. It is known that other transcriptional inputs, both positive and negative, regulate svb transcription (Stern and Orgogozo, 2009). In this case, transcriptional repression may prevent svb expression at peak Ubx concentrations. It would be of interest to explore in greater detail the mechanisms of this activity as a part of our future work.
In addition, the mean levels of Ubx found in proximity to the active transcription loci have a very small variation at close range, and increase with increasing distance for a small range. Then the variation in concentration appears to be constant (and not continue to increase, as might be expected for searches of larger and larger spaces). The authors are asked to explain the shape of the Ubx distributions noted for the reporter constructs.
The convergence of the variance in Ubx intensity to zero at the transcription site is an effect of intensity normalization. The intensity of Ubx directly over the transcription site is normalized to one for each of the transcription sites. We average the distributions for all observed nuclei and then subtract the residual Ubx intensity 4 μm from the transcription site as the average background to generate the plots in Figures 2–4. Because the Ubx intensity at the transcription site is the point of normalization, its variance is zero.
The reviewers made an important observation concerning the relatively constant variance in the Ubx distributions after an initial increase. Because a transcription site may sit slightly off from the local maximum of Ubx concentration, the Ubx intensity distribution close to each site fluctuates within the first 200 nm (which is near the optical resolution of our images taken using AiryScan). This leads to an initial increase in variance as the distributions move away from the transcription sites. However, most transcriptions sites are close to a maximum of Ubx concentration, so Ubx intensity decreases monotonically after initial fluctuations. Because of the relatively uniform shapes of the individual distributions, the variance stops increasing after a short distance from the transcription site. We believe that there is no a priori reason that this must be the case. For example, the individual Ubx distributions around the transcription sites of the synthetic enhancer, which is not under the control of Ubx, fluctuate at random out to the full distance plotted in Figure 3C (out to 1 μm), leading to a variance that increases throughout the plot. We have added this observation into the Results.
- The reporter constructs with different Ubx affinities were derived from svb enhancer sequences. Do the reporters activate and colocalize with the endogenous svb locus in the fixed specimens? Double FISH with intronic probes should be able to discern this point.
We conducted double FISH experiments and measured the distances of transcription sites between the endogenous svb locus (on the X chromosome) and other transcription sites (on chromosome 3). Enhancers related to svb, which are 7H and E3N, preferentially colocalized close to the svb site in nuclei expressing both, despite being on different chromosomes. The average distance between transcription sites is about 200 nm. The transcription sites for forkhead (fkh), also on chromosome 3, did not preferentially colocalize with svb, with an average distance of 1 μm between transcription sites. In fact, nuclei showing two transcription sites from the svb locus (two bright spots next to each other) also have an average of 200 nm between those pairs of sites. We have added a new section in the Results and a new figure (Figure 2—figure supplement 1) describing this new result. We have also made additions in the Discussion highlighting this result. We would like to thank the reviewers for suggesting this experiment to improve our work.
Supplementary Materials
Source code 1. Matlab script to average radially averaged intensity distributions from individual transcription sites and offset background fluorescence.
Transparent reporting form