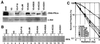Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining - PubMed (original) (raw)
Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining
A Kurimasa et al. Mol Cell Biol. 1999 May.
Abstract
The catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) is an enormous, 470-kDa protein serine/threonine kinase that has homology with members of the phosphatidylinositol (PI) 3-kinase superfamily. This protein contributes to the repair of DNA double-strand breaks (DSBs) by assembling broken ends of DNA molecules in combination with the DNA-binding factors Ku70 and Ku80. It may also serve as a molecular scaffold for recruiting DNA repair factors to DNA strand breaks. This study attempts to better define the role of protein kinase activity in the repair of DNA DSBs. We constructed a contiguous 14-kb human DNA-PKcs cDNA and demonstrated that it can complement the DNA DSB repair defects of two mutant cell lines known to be deficient in DNA-PKcs (M059J and V3). We then created deletion and site-directed mutations within the conserved PI 3-kinase domain of the DNA-PKcs gene to test the importance of protein kinase activity for DSB rejoining. These DNA-PKcs mutant constructs are able to express the protein but fail to complement the DNA DSB or V(D)J recombination defects of DNA-PKcs mutant cells. These results indicate that the protein kinase activity of DNA-PKcs is essential for the rejoining of DNA DSBs in mammalian cells. We have also determined a model structure for the DNA-PKcs kinase domain based on comparisons to the crystallographic structure of a cyclic AMP-dependent protein kinase. This structure gives some insight into which amino acid residues are crucial for the kinase activity in DNA-PKcs.
Figures
FIG. 1
Schematic presentation of the human DNA-PKcs cDNA and the process used in assembly of cDNA fragments. The full-length DNA-PKcs cDNA with the PI 3-kinase domain is shown just below the DNA size scale. The gray lines represent seven cDNAs identified from libraries and one PCR fragment, which were used for reconstruction of the full-length DNA-PKcs cDNA. The solid lines in the bottom groups represent assembled cDNAs that were cloned into plasmid vectors. The restriction enzyme sites (N, _Not_I; H, _Hin_dIII; Sa, _Sal_I; A, _Avr_II; St, _Stu_I; B, _Bgl_II; F, _Fse_I; X, _Xba_I; C, _Cla_I; N, _Nhe_I) shown at the bottom of figure were used for assembling the full-length cDNA.
FIG. 2
(A) Western blot analysis of recombinant DNA-PKcs protein expression. Eight micrograms each of protein extract from the human glioma cell line M059K (wild-type); M059J (DNA-PKcs mutant); MJ-L24, MJ-L35, and MJ-M6 (M059J cells transfected with the full-length DNA-PKcs cDNA expression vector); and MJ-NA (M059J cells transfected with the control plasmid pPur) were resolved by SDS-PAGE, and DNA-PKcs expression was detected by Western blot analysis. As a loading control, c-Abl and Ku80 expression was also examined. (B) For the CHO-derived cell line, AA8 (wild type); V3 (DNA-PKcs mutant); V3-F18, V3-H15, and V3-l1 (intact DNA-PKcs cDNA-transfected V3 clones); and V3-JM (control transfection V3 clone), 80 μg of each protein extract was used for Western blot analysis. The c-Abl loading control was also examined. (C) Complementation of radiation sensitivity by recombinant human DNA-PKcs. Human glioma cell lines (left) and CHO V3 cell lines (right) were assayed for radiation sensitivity.
FIG. 3
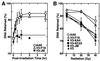
(A) Expression analysis of DNA-PKcs PI 3-kinase domain mutants in CHO V3 cells by Western blotting. AA8, wild type; V3, DNA-PKcs mutant; V3-F18, intact DNA-PKcs-transfected V3 cell line; V3-JM, transfection control V3 cell line; V3-KA4, kinase domain II mutant; V3-KB20, domain II mutant; V3-KC23, frameshift mutant which makes the truncated protein; V3-KD51, domain VIb mutant. As a loading control, c-Abl expression was also examined. (B) DNA-activated protein kinase activity of wild-type and mutant DNA-PKcs-expressing cells. DNA-PKcs was immunoprecipitated from whole-cell extracts. Protein kinase activity was analyzed in the absence or presence of added RPA, as indicated. The position of the 32-kDa subunit of recombinant human RPA is indicated. Phosphorylated RPA signals are observed in lane 1 (AA8) and lane 5 (V3-F18). (C) Radiation sensitivity of wild-type and mutant DNA-PKcs-expressing V3 cell lines. V3 cells expressing intact human DNA-PKcs (V3-F18) and each of the V3 cell lines expressing the four kinase domain mutant DNA-PKcs proteins were assayed for radiation sensitivity. Radiation sensitivity of the control pSV2neo transfectant V3-JM is also shown.
FIG. 4
DNA DSB repair capacities of wild-type and mutant DNA-PKcs-expressing cell lines. DNA DSB repair is expressed as the percentage of DNA retained in the agarose plug after pulsed-field gel electrophoresis (PFGE) analysis of irradiated cell cultures. (A) Time courses of DNA DSB repair activities for AA8 (wild type), V3-F18 (intact DNA-PKcs-transfected V3 cell line), and V3-JM (control pSV2neo-transfected V3 cell line). (B) Comparisons of DNA DSB repair capacities of wild-type and DNA-PKcs mutant cell lines at 4 h postirradiation with 20- and 40-Gy doses. V3-KA4, kinase domain II mutant; V3-KC23, frameshift mutant (truncated DNA-PKcs); V3, DNA-PKcs mutant; V3-JM, control pSV2neo transfectant.
FIG. 5
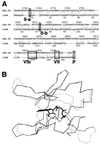
Three-dimensional modeling of the DNA-PKcs PI 3-kinase domain. (A) The sequence of the ATP binding pocket of the human DNA-PKcs was aligned with that of the catalytic subunit of cAPK (1APM). Each open box shows the serine/threonine protein kinase subdomain II homology domain within the PI 3-kinase superfamily (II-a), subdomain II from 1APM (II-b), and subdomain VIb and VII homology regions within DNA-PKcs and 1APM (VIb, VII). The phosphorylation site of threonine near the catalytic site is also shown as an open box (P). (B) Superimposed image of the ATP binding pocket of the modeled DNA-PKcs structure and the crystal structure of the 1APM active site. The thick black line in the center is ANP, the thin black line is the modeled structure of the DNA-PKcs, and the thin gray line is the crystal structure of 1APM. (C) Energy-minimized structure of the ATP binding pocket of the human DNA-PKcs. The ATP binding pocket is drawn in light gray, with ANP drawn in yellow and three conserved residues (K-3812, D-3921, and D-3940) drawn in magenta. (D) The backbone fold image of the binding pocket of the DNA-PKcs is shown together with ATP, the two conserved residues (K-3812 and D-3940) that are important for ATP binding, and another conserved residue, D-3921, that contributes to catalytic activity. The glycosylated T-3949, which may contribute to control kinase activity by phosphorylation, is also shown. (E) Structure model of the mutated residue D3921N of kinase subdomain VIb. The catalytic aspartate D-3921 is oriented with the carboxyl group facing the γ-phosphate of ATP (left). In the mutant molecule, N-3921 has an amino group pointing toward the γ-phosphate of ATP (right).
FIG. 5
Three-dimensional modeling of the DNA-PKcs PI 3-kinase domain. (A) The sequence of the ATP binding pocket of the human DNA-PKcs was aligned with that of the catalytic subunit of cAPK (1APM). Each open box shows the serine/threonine protein kinase subdomain II homology domain within the PI 3-kinase superfamily (II-a), subdomain II from 1APM (II-b), and subdomain VIb and VII homology regions within DNA-PKcs and 1APM (VIb, VII). The phosphorylation site of threonine near the catalytic site is also shown as an open box (P). (B) Superimposed image of the ATP binding pocket of the modeled DNA-PKcs structure and the crystal structure of the 1APM active site. The thick black line in the center is ANP, the thin black line is the modeled structure of the DNA-PKcs, and the thin gray line is the crystal structure of 1APM. (C) Energy-minimized structure of the ATP binding pocket of the human DNA-PKcs. The ATP binding pocket is drawn in light gray, with ANP drawn in yellow and three conserved residues (K-3812, D-3921, and D-3940) drawn in magenta. (D) The backbone fold image of the binding pocket of the DNA-PKcs is shown together with ATP, the two conserved residues (K-3812 and D-3940) that are important for ATP binding, and another conserved residue, D-3921, that contributes to catalytic activity. The glycosylated T-3949, which may contribute to control kinase activity by phosphorylation, is also shown. (E) Structure model of the mutated residue D3921N of kinase subdomain VIb. The catalytic aspartate D-3921 is oriented with the carboxyl group facing the γ-phosphate of ATP (left). In the mutant molecule, N-3921 has an amino group pointing toward the γ-phosphate of ATP (right).
Similar articles
- DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination.
Allen C, Kurimasa A, Brenneman MA, Chen DJ, Nickoloff JA. Allen C, et al. Proc Natl Acad Sci U S A. 2002 Mar 19;99(6):3758-63. doi: 10.1073/pnas.052545899. Proc Natl Acad Sci U S A. 2002. PMID: 11904432 Free PMC article. - Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair.
Fukushima T, Takata M, Morrison C, Araki R, Fujimori A, Abe M, Tatsumi K, Jasin M, Dhar PK, Sonoda E, Chiba T, Takeda S. Fukushima T, et al. J Biol Chem. 2001 Nov 30;276(48):44413-8. doi: 10.1074/jbc.M106295200. Epub 2001 Sep 27. J Biol Chem. 2001. PMID: 11577093 - DNA-dependent protein kinase stimulates an independently active, nonhomologous, end-joining apparatus.
DiBiase SJ, Zeng ZC, Chen R, Hyslop T, Curran WJ Jr, Iliakis G. DiBiase SJ, et al. Cancer Res. 2000 Mar 1;60(5):1245-53. Cancer Res. 2000. PMID: 10728683 - Mechanisms of DNA double strand break repair and chromosome aberration formation.
Iliakis G, Wang H, Perrault AR, Boecker W, Rosidi B, Windhofer F, Wu W, Guan J, Terzoudi G, Pantelias G. Iliakis G, et al. Cytogenet Genome Res. 2004;104(1-4):14-20. doi: 10.1159/000077461. Cytogenet Genome Res. 2004. PMID: 15162010 Review. - Ku, a DNA repair protein with multiple cellular functions?
Featherstone C, Jackson SP. Featherstone C, et al. Mutat Res. 1999 May 14;434(1):3-15. doi: 10.1016/s0921-8777(99)00006-3. Mutat Res. 1999. PMID: 10377944 Review.
Cited by
- DNA-PK: A synopsis beyond synapsis.
Goff NJ, Mikhova M, Schmidt JC, Meek K. Goff NJ, et al. DNA Repair (Amst). 2024 Sep;141:103716. doi: 10.1016/j.dnarep.2024.103716. Epub 2024 Jul 8. DNA Repair (Amst). 2024. PMID: 38996771 Review. - Non-homologous end joining shapes the genomic rearrangement landscape of chromothripsis from mitotic errors.
Hu Q, Espejo Valle-Inclán J, Dahiya R, Guyer A, Mazzagatti A, Maurais EG, Engel JL, Lu H, Davis AJ, Cortés-Ciriano I, Ly P. Hu Q, et al. Nat Commun. 2024 Jul 4;15(1):5611. doi: 10.1038/s41467-024-49985-5. Nat Commun. 2024. PMID: 38965240 Free PMC article. - BUB1 regulates non-homologous end joining pathway to mediate radioresistance in triple-negative breast cancer.
Sriramulu S, Thoidingjam S, Chen WM, Hassan O, Siddiqui F, Brown SL, Movsas B, Green MD, Davis AJ, Speers C, Walker E, Nyati S. Sriramulu S, et al. J Exp Clin Cancer Res. 2024 Jun 11;43(1):163. doi: 10.1186/s13046-024-03086-9. J Exp Clin Cancer Res. 2024. PMID: 38863037 Free PMC article. - DNA-PK controls Apollo's access to leading-end telomeres.
Sonmez C, Toia B, Eickhoff P, Matei AM, El Beyrouthy M, Wallner B, Douglas ME, de Lange T, Lottersberger F. Sonmez C, et al. Nucleic Acids Res. 2024 May 8;52(8):4313-4327. doi: 10.1093/nar/gkae105. Nucleic Acids Res. 2024. PMID: 38407308 Free PMC article. - Different Impacts of DNA-PK and mTOR Kinase Inhibitors in Combination with Ionizing Radiation on HNSCC and Normal Tissue Cells.
Klieber N, Hildebrand LS, Faulhaber E, Symank J, Häck N, Härtl A, Fietkau R, Distel LV. Klieber N, et al. Cells. 2024 Feb 6;13(4):304. doi: 10.3390/cells13040304. Cells. 2024. PMID: 38391917 Free PMC article.
References
- Anderson C W, Carter T H. The DNA-activated protein kinase—DNA-PK. Curr Top Microbiol Immunol. 1996;217:91–111. - PubMed
- Blier P R, Griffith A J, Craft J, Hardin J A. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem. 1993;268:7594–7601. - PubMed
- Blunt T, Finnie N J, Taccioli G E, Smith G C, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA006294/CA/NCI NIH HHS/United States
- CA06294/CA/NCI NIH HHS/United States
- R37 CA050519/CA/NCI NIH HHS/United States
- CA50519/CA/NCI NIH HHS/United States
- R01 CA050519/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous