A role for p53 in base excision repair - PubMed (original) (raw)
A role for p53 in base excision repair
J Zhou et al. EMBO J. 2001.
Abstract
Wild-type p53 protein can markedly stimulate base excision repair (BER) in vitro, either reconstituted with purified components or in extracts of cells. In contrast, p53 with missense mutations either at hot-spots in the core domain or within the N-terminal transactivation domain is defective in this function. Stimulation of BER by p53 is correlated with its ability to interact directly both with the AP endonuclease (APE) and with DNA polymerase beta (pol beta). Furthermore, p53 stabilizes the interaction between DNA pol beta and abasic DNA. Evidence that this function of p53 is physiologically relevant is supported by the facts that BER activity in human and murine cell extracts closely parallels their levels of endogenous p53, and that BER activity is much reduced in cell extracts immunodepleted of p53. These data suggest a novel role for p53 in DNA repair, which could contribute to its function as a key tumor suppressor.
Figures
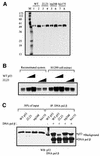
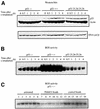
Fig. 1. p53 stimulates DNA BER in vitro. BER experiments with AP-DNA were performed as described in Materials and methods. Baculovirus-expressed purified p53 protein (200, 400, 800 ng) was added to BER reaction mixtures containing either purified APE and DNA pol β (A) or whole-cell extracts made from H1299 cells (B). As controls, the 21 bp U-DNA that was not treated with UDG [lane 1, (A)] and a 55 bp DNA that has a cytosine at the position of the uracil [lane 1, (B)] were used as templates. (C) Three hundred or 600 ng of bacterially expressed His-tagged p53 and p53 depletion mutants [p53 core domain (100–300), p53ΔC30 (1–363), p53ΔN96 (97–393) and the p53 C-terminus (300–393)] were added into reconstituted BER reaction mixtures. (D) Five hundred nanograms of each protein were run on an SDS gel, which was silver stained: full-length, lane 1; ΔN96, lane 2; ΔC30, lane 3; the core domain, lane 4; the C-terminus, lane 5. The numbers on the left show migration positions of the indicated molecular weight markers (in kDa) run in lane M.
Fig. 2. Tumor-derived p53 mutants have reduced stimulation of BER. p53 mutant proteins, R248W and R175H, and flu-tagged wild-type p53 protein were immunopurified as described in Materials and methods. (A) Silver-stained SDS gel with proteins (300 and 600 ng) as indicated. Lane M contains molecular weight marker proteins with sizes indicated on the left. (B) Wild-type, R248W or R175H p53 proteins (200, 400, 600 ng) were added to reconstituted BER reaction mixtures as in Figure 1A. (C) Three hundred or 600 ng of the indicated proteins were added to BER reaction mixtures containing H1299 whole-cell extracts as in Figure 1B.
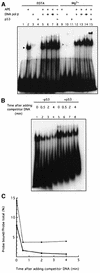
Fig. 3. p53 stabilizes the interaction between DNA pol β and AP-DNA. (A) p53 stimulates APE-facilitated loading of DNA pol β onto AP-DNA. EMSA with end-labeled 21 bp AP-DNA was performed as described. APE (20 ng), DNA pol β (20, 60 or 120 ng) and baculovirus-expressed p53 (80 ng) were added to EMSA reaction mixtures as indicated. The migrations of APE, DNA pol β and p53 are indicated by the black, gray and white arrowheads, respectively. (B) p53 slows the dissociation of DNA pol β from AP-DNA. EMSA reaction mixtures (in Mg2+ buffer), containing APE, DNA pol β and end-labeled AP-DNA, lacked (lanes 1–4) or contained (lanes 5–8) p53. Excess unlabeled competitor AP-DNA was added to each reaction mixture at time zero. After the indicated time, mixtures were loaded onto a running 8% polyacrylamide gel. (C) A graphic representation of the PhosphorImager-quantified result of the competition/dissociation assay.
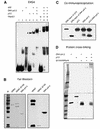
Fig. 4. p53 interacts with DNA pol β. (A) p53 and DNA pol β together supershift the 55 bp AP-DNA probe. EMSA with end-labeled 55 bp AP-DNA was performed as described. APE (20 ng), DNA pol β (20 ng), p53 (80 ng) and PAb421 (100 or 200 ng) were added to reaction mixtures (in Mg2+ buffer) as indicated. The migrations of p53 alone and the p53–DNA pol β complex are indicated by the black and white arrowheads, respectively. (B) Far-western blotting was performed as described. Full-length HDM2 and a truncated HDM2 (dl166), which does not bind to p53, were used as the positive and negative controls, respectively. Left panel shows Ponceau S-stained image of the nitrocellulose filter; right panel shows the result of incubation of filter with p53 followed by probing with PAb421. (C) Co-immunoprecipitation was performed as described in Materials and methods. Precipitations of PAb421-cross-linked beads were resolved by SDS–PAGE, followed by detection with PAb421 and antiserum against DNA pol β. (D) Protein cross-linking was performed as described in Materials and methods. Proteins were separated in a step-gradient SDS–PAGE gel. A silver-stained image of the gel is shown.
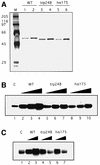
Fig. 5. BER activity correlates with p53 levels in cells. (A–C) Cell cultures at 70% confluence were exposed to 7 Gy γ-irradiation. At 0, 0.5, 1, 2, 3 and 4 h after irradiation, cells were collected and whole-cell extracts were prepared. Then BER was assayed using the 55 bp U-DNA. p53 protein in aliquots of extracts was detected by western blotting using PAb421. To monitor cell protein concentrations, actin was detected using an anti-actin antibody (Sigma). (D) Depletion of p53 reduces BER activity. Cell extracts (3 µg) made from LNCaP cells 2 h after γ-irradiation were pre-incubated with PAb421-coated magnetic Dynabeads (lanes 3–5) or beads alone (lane 2) and assayed for BER activity. Lane 1, untreated cell extract; lanes 4 and 5, 300 or 600 ng of baculovirus-expressed p53 were added into depleted extracts, respectively. (E) p53 protein levels in the cell extracts (15 µg) were detected by western blotting. Lane 1, untreated cell extract; lane 2, depleted with control beads; lane 3, extract depleted with PAb421-coated beads; lanes 4 and 5, 60 and 120 ng of p53 proteins were added to PAb421-depleted extracts, respectively.
Fig. 6. A mutant p53 (L22Q/W23S) does not interact with DNA pol β and loses the ability to stimulate BER. Baculovirus-expressed wild-type and mutant p53 proteins were immunopurified as described in Materials and methods. (A) Silver-stained SDS gel with proteins (300 and 600 ng) as indicated. Lane M contains molecular marker proteins with sizes indicated on the left. (B) Wild-type and L22Q/W23S p53 proteins (300 and 600 ng) were added to reconstituted BER reaction mixtures (lanes 2–5) and BER reaction mixtures containing H1299 whole-cell extracts (lanes 7–10). (C) The co-immunoprecipitation assay was performed as described in Materials and methods. Wild-type and mutant p53 proteins incubated with DNA pol β and Dynabeads coated with 18S monoclonal antibody against DNA pol β were resolved by SDS–PAGE, transferred to nitrocellulose and probed with PAb421 and 18S. Fifty percent of the inputs of p53 proteins are shown on the left.
Fig. 7. Extracts of cells with wild-type p53 have more BER activity than p53–/– or mutant p53 (L25Q/W26S) cell extracts. Cultures of MEF cells as indicated were γ-irradiated and extracts were prepared as described in Figure 5. (A) p53 protein in aliquots of extracts was detected by western blotting using a mixture of antibodies, including PAb421, PAb240 and PAb246. DNA pol β protein was detected using the 18S monoclonal antibody. In (B) and (C), BER activity was assayed using the 55 bp U-DNA fragment as in Figure 1B. Either aliquots of extracts prepared at the indicated times after γ-irradiation of MEFs were directly assayed for BER activity (B) or aliquots of the wild-type p53-containing samples above were either untreated (lanes 1–6) or incubated with PAb 421-coated Dynabeads (lanes 7–12) or Dynabeads alone (lanes 13–18). The BER activities in either the untreated samples or the different Dynabead supernatants were assayed (C).
Similar articles
- Structural and functional involvement of p53 in BER in vitro and in vivo.
Offer H, Milyavsky M, Erez N, Matas D, Zurer I, Harris CC, Rotter V. Offer H, et al. Oncogene. 2001 Feb 1;20(5):581-9. doi: 10.1038/sj.onc.1204120. Oncogene. 2001. PMID: 11313990 - Implication of p53 in base excision DNA repair: in vivo evidence.
Seo YR, Fishel ML, Amundson S, Kelley MR, Smith ML. Seo YR, et al. Oncogene. 2002 Jan 24;21(5):731-7. doi: 10.1038/sj.onc.1205129. Oncogene. 2002. PMID: 11850801 - The p53 pathway promotes efficient mitochondrial DNA base excision repair in colorectal cancer cells.
Chen D, Yu Z, Zhu Z, Lopez CD. Chen D, et al. Cancer Res. 2006 Apr 1;66(7):3485-94. doi: 10.1158/0008-5472.CAN-05-4103. Cancer Res. 2006. PMID: 16585172 - The potential roles of p53 tumor suppressor in nucleotide excision repair (NER) and base excision repair (BER).
Seo YR, Jung HJ. Seo YR, et al. Exp Mol Med. 2004 Dec 31;36(6):505-9. doi: 10.1038/emm.2004.64. Exp Mol Med. 2004. PMID: 15665582 Review. - Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA.
Sung JS, Demple B. Sung JS, et al. FEBS J. 2006 Apr;273(8):1620-9. doi: 10.1111/j.1742-4658.2006.05192.x. FEBS J. 2006. PMID: 16623699 Review.
Cited by
- The DNA repair function of BCL11A suppresses senescence and promotes continued proliferation of triple-negative breast cancer cells.
Vickridge E, Faraco CCF, Tehrani PS, Ramdzan ZM, Rahimian H, Leduy L, Gingras AC, Nepveu A. Vickridge E, et al. NAR Cancer. 2022 Sep 28;4(4):zcac028. doi: 10.1093/narcan/zcac028. eCollection 2022 Dec. NAR Cancer. 2022. PMID: 36186110 Free PMC article. - Age-Dependent Protective Effect of Selenium against UVA Irradiation in Primary Human Keratinocytes and the Associated DNA Repair Signature.
Favrot C, Beal D, Blouin E, Leccia MT, Roussel AM, Rachidi W. Favrot C, et al. Oxid Med Cell Longev. 2018 Feb 22;2018:5895439. doi: 10.1155/2018/5895439. eCollection 2018. Oxid Med Cell Longev. 2018. PMID: 29682159 Free PMC article. - Ubiquitination of mammalian AP endonuclease (APE1) regulated by the p53-MDM2 signaling pathway.
Busso CS, Iwakuma T, Izumi T. Busso CS, et al. Oncogene. 2009 Apr 2;28(13):1616-25. doi: 10.1038/onc.2009.5. Epub 2009 Feb 16. Oncogene. 2009. PMID: 19219073 Free PMC article. - p53 represses class switch recombination to IgG2a through its antioxidant function.
Guikema JE, Schrader CE, Brodsky MH, Linehan EK, Richards A, El Falaky N, Li DH, Sluss HK, Szomolanyi-Tsuda E, Stavnezer J. Guikema JE, et al. J Immunol. 2010 Jun 1;184(11):6177-87. doi: 10.4049/jimmunol.0904085. Epub 2010 May 5. J Immunol. 2010. PMID: 20483782 Free PMC article. - Oxidized base damage and single-strand break repair in mammalian genomes: role of disordered regions and posttranslational modifications in early enzymes.
Hegde ML, Izumi T, Mitra S. Hegde ML, et al. Prog Mol Biol Transl Sci. 2012;110:123-53. doi: 10.1016/B978-0-12-387665-2.00006-7. Prog Mol Biol Transl Sci. 2012. PMID: 22749145 Free PMC article. Review.
References
- Banin S. et al. (1998) Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science, 281, 1674–1677. - PubMed
- Barzilay G. and Hickson,I.D. (1995) Structure and function of apurinic/apyrimidinic endonucleases. BioEssays, 17, 713–719. - PubMed
- Bogenhagen D.F. and Pinz,K.G. (1998) The action of DNA ligase at abasic sites in DNA. J. Biol. Chem., 273, 7888–7893. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous