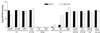Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes - PubMed (original) (raw)
Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes
E G Cormier et al. J Virol. 2001 Jun.
Abstract
CD4 and CCR5 mediate fusion and entry of R5 human immunodeficiency virus type 1 (HIV-1) strains. Sulfotyrosine and other negatively charged residues in the CCR5 amino-terminal domain (Nt) are crucial for gp120 binding and viral entry. We previously showed that a soluble gp120-CD4 complex specifically binds to a peptide corresponding to CCR5 Nt residues 2 to 18, with sulfotyrosines in positions 10 and 14. This sulfopeptide also inhibits soluble gp120-CD4 binding to cell surface CCR5 as well as infection by an R5 virus. Here we show that residues 10 to 18 constitute the minimal domain of the CCR5 Nt that is able to specifically interact with soluble gp120-CD4 complexes. In addition to sulfotyrosines in positions 10 and 14, negatively charged residues in positions 11 and 18 participate in this interaction. Furthermore, the CCR5 Nt binds to a CD4-induced surface on gp120 that is composed of conserved residues in the V3 loop stem and the C4 domain. Binding of gp120 to cell surface CCR5 is further influenced by residues in the crown of the V3 loop, C1, C2, and C3. Our data suggest that gp120 docking to CCR5 is a multistep process involving several independent regions of the envelope glycoprotein and the coreceptor.
Figures
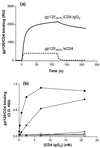
FIG. 1
gp120-CD4 complex binding to CCR5 Nt sulfopeptides. (a) Peptide 2-18 was bound to streptavidin-coated biosensor chips, and gp120JR-FL-sCD4 or gp120JR-FL-CD4–IgG2 was directed over the sensor chip surface. RU were measured as a function of time using a Biacore X and reflect complex-peptide binding. (b) Sulfopeptide 2-18 (solid symbols) or phosphopeptide 2-18(P) (open symbols) was immobilized on streptavidin-coated ELISA plates and incubated with gp120-CD4–IgG2 complexes. gp120 proteins were derived from the R5 isolate HIV-1JR-FL (squares), the R5X4 isolate HIV-1DH123 (circles), and the X4 isolate HIV-1LAI (diamonds). Complex-peptide binding was detected by an HRP-conjugated goat anti-human IgG antibody. OD450 was measured after the addition of HRP substrate and is expressed as a function of CD4-IgG2 concentration. Results shown are from representative experiments.
FIG. 2
Binding of anti-CCR5 MAbs to CCR5 Nt peptides. Sulfopeptides (a) or phosphopeptides (b) were immobilized on streptavidin-coated ELISA plates and incubated with anti-CCR5 MAb PA8 (solid squares), PA10 (open circles), PA11 (solid circles), PA12 (solid diamonds), or PA14 (solid triangles). Binding of the MAbs to the peptides was detected by an HRP-conjugated goat anti-mouse IgG antibody. OD450 was measured after addition of HRP substrate and expressed as a function of MAb concentration. (c) Biotinylated sulfopeptide 2-18 was immobilized on streptavidin-coated plates and incubated with gp120-CD4–IgG2 complex in the presence of increasing concentrations of PA8 (solid squares), TAK-779 (triangles), RANTES (inverted triangles), MIP-1α (diamonds), MIP-1β (circles), or SDF-1α (open squares). Binding of the complexes to the peptide was detected by incubation with HRP-conjugated goat anti-human IgG antibody. OD450 was measured after addition of HRP substrate, and the percentage of binding was expressed as a function of inhibitor concentration. Results shown are from representative experiments. Some symbols are not visible due to overlay.
FIG. 3
Binding of gp120JR-FL-CD4–IgG2 to different CCR5 Nt-based peptides. Streptavidin plates were coated with sulfopeptide 2-18 (solid squares), 10-18 (solid circles), 8-15 (solid diamonds), 6-16 (solid stars), 10-15 (open squares), or 10-18(11A/18A) (solid triangles). Plate contents were then incubated with gp120JR-FL-CD4–IgG2 complex. Binding of the complex to the peptide was detected by an HRP-conjugated goat anti-human IgG antibody. OD450 was measured after addition of HRP substrate and expressed as a function of CD4-IgG2 concentration. Results shown are from a representative experiment.
FIG. 4
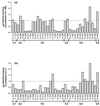
Inhibition of gp120-CD4–IgG2 complex binding to sulfopeptides by anti-gp120 MAbs. Biotinylated sulfopeptide 2-18 was bound to streptavidin-coated biosensor chips, and solutions of either gp120JR-FL-CD4–IgG2 complex (solid bars) or gp120DH123-CD4–IgG2 complex (open bars) were directed over the surface of the chip in the presence of different anti-gp120 MAbs. The names of the MAbs and the locations of their epitopes are indicated along the x axis. RU were measured as a function of time using a Biacore X and reflect complex-peptide binding in the presence of the MAbs. gp120-CD4–IgG2 binding was calculated using the following formula: (RU in the presence of MAbs)/(RU in the absence of MAbs) × 100%. The values shown are averages of three independent experiments. Standard deviation was no more than ±20% for each data set; standard deviations are not represented for the sake of clarity.
FIG. 5
Binding of gp120 mutants to sulfopeptide and wild-type CCR5. (a) Sulfopeptide 2-18 was immobilized on streptavidin-coated plates and incubated with a mixture of gp120-containing supernatants and CD4-IgG2. Peptide-complex binding was detected by an HRP-conjugated goat anti-human IgG antibody. OD450 was measured after addition of HRP substrate and normalized for binding of the gp120 mutants to CD4-IgG2. The dotted line represents the normalized value for the binding of wild-type gp120 to the peptide. The mutated amino acids and their locations in gp120 are indicated along the x axis. (b) L12-CCR5+ cells were incubated with a mixture of gp120-containing supernatants and CD4-IgG2. Binding of the complex was detected by fluorescence-activated cell sorter analysis after addition of streptavidin-phycoerythrin. The percentage of gp120-CD4–IgG2 binding to CCR5 was normalized for gp120 binding to CD4-IgG2. The dotted line represents the normalized value for the binding of wild-type gp120 to the L12-CCR5+ cells. The mutated amino acids and their locations in gp120 are indicated along the x axis. The values shown are averages of three independent experiments. Standard deviation was no more than ±20% for each data set; standard deviations are not represented for the sake of clarity.
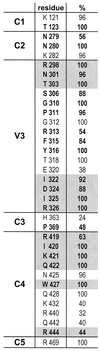
FIG. 6
Amino acid conservation among R5 isolates. The first column indicates the gp120 domains, the second column indicates the gp120 residues, and the third column indicates the percentage of conservation of the residues. Envelope sequences from 25 R5 strains described in the HIV Database and retrieved from the National Center for Biotechnology Information GenBank were aligned, and the percentage of conservation for the indicated residues was calculated and combined with results by Hwang et al. (19). Residues exhibiting a >50% decrease in binding to sulfopeptide 2-18 when changed to alanines are highlighted in gray. Residues exhibiting a >50% decrease in binding to cell surface CCR5 when changed to alanines are in boldface.
Similar articles
- Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120.
Cormier EG, Persuh M, Thompson DA, Lin SW, Sakmar TP, Olson WC, Dragic T. Cormier EG, et al. Proc Natl Acad Sci U S A. 2000 May 23;97(11):5762-7. doi: 10.1073/pnas.97.11.5762. Proc Natl Acad Sci U S A. 2000. PMID: 10823934 Free PMC article. - The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor.
Cormier EG, Dragic T. Cormier EG, et al. J Virol. 2002 Sep;76(17):8953-7. doi: 10.1128/jvi.76.17.8953-8957.2002. J Virol. 2002. PMID: 12163614 Free PMC article. - Interaction of human immunodeficiency virus type 1 envelope glycoprotein V3 loop with CCR5 and CD4 at the membrane of human primary macrophages.
Rabehi L, Seddiki N, Benjouad A, Gluckman JC, Gattegno L. Rabehi L, et al. AIDS Res Hum Retroviruses. 1998 Dec 20;14(18):1605-15. doi: 10.1089/aid.1998.14.1605. AIDS Res Hum Retroviruses. 1998. PMID: 9870313 - HIV gp120 interactions with coreceptors: insights from studies with CCR5-based peptides.
Berger EA, Alkhatib G. Berger EA, et al. Eur J Med Res. 2007 Oct 15;12(9):403-7. Eur J Med Res. 2007. PMID: 17933721 Review. - Human immunodeficiency virus and heparan sulfate: from attachment to entry inhibition.
Connell BJ, Lortat-Jacob H. Connell BJ, et al. Front Immunol. 2013 Nov 20;4:385. doi: 10.3389/fimmu.2013.00385. Front Immunol. 2013. PMID: 24312095 Free PMC article. Review.
Cited by
- Novel small synthetic HIV-1 V3 crown variants: CCR5 targeting ligands.
Anitha AK, Narayanan P, Ajayakumar N, Sivakumar KC, Kumar KS. Anitha AK, et al. J Biochem. 2022 Sep 5;172(3):149-164. doi: 10.1093/jb/mvac052. J Biochem. 2022. PMID: 35708645 Free PMC article. - Structure of CC Chemokine Receptor 5 with a Potent Chemokine Antagonist Reveals Mechanisms of Chemokine Recognition and Molecular Mimicry by HIV.
Zheng Y, Han GW, Abagyan R, Wu B, Stevens RC, Cherezov V, Kufareva I, Handel TM. Zheng Y, et al. Immunity. 2017 Jun 20;46(6):1005-1017.e5. doi: 10.1016/j.immuni.2017.05.002. Immunity. 2017. PMID: 28636951 Free PMC article. - A Prominent Site of Antibody Vulnerability on HIV Envelope Incorporates a Motif Associated with CCR5 Binding and Its Camouflaging Glycans.
Sok D, Pauthner M, Briney B, Lee JH, Saye-Francisco KL, Hsueh J, Ramos A, Le KM, Jones M, Jardine JG, Bastidas R, Sarkar A, Liang CH, Shivatare SS, Wu CY, Schief WR, Wong CH, Wilson IA, Ward AB, Zhu J, Poignard P, Burton DR. Sok D, et al. Immunity. 2016 Jul 19;45(1):31-45. doi: 10.1016/j.immuni.2016.06.026. Immunity. 2016. PMID: 27438765 Free PMC article. - Tyrosine-sulfated V2 peptides inhibit HIV-1 infection via coreceptor mimicry.
Cimbro R, Peterson FC, Liu Q, Guzzo C, Zhang P, Miao H, Van Ryk D, Ambroggio X, Hurt DE, De Gioia L, Volkman BF, Dolan MA, Lusso P. Cimbro R, et al. EBioMedicine. 2016 Aug;10:45-54. doi: 10.1016/j.ebiom.2016.06.037. Epub 2016 Jun 26. EBioMedicine. 2016. PMID: 27389109 Free PMC article. - Design and synthesis of small molecule-sulfotyrosine mimetics that inhibit HIV-1 entry.
Dogo-Isonagie C, Lee SL, Lohith K, Liu H, Mandadapu SR, Lusvarghi S, O'Connor RD, Bewley CA. Dogo-Isonagie C, et al. Bioorg Med Chem. 2016 Apr 15;24(8):1718-28. doi: 10.1016/j.bmc.2016.02.044. Epub 2016 Mar 3. Bioorg Med Chem. 2016. PMID: 26968647 Free PMC article.
References
- Allaway G P, Davis-Bruno K L, Beaudry G A, Garcia E B, Wong E L, Ryder A M, Hasel K W, Gauduin M C, Koup R A, McDougal J S, et al. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res Hum Retrovir. 1995;11:533–539. - PubMed
- Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–1926. - PubMed
- Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. - PubMed
- Binley J M, Sanders R W, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma D J, Maddon P J, Olson W C, Moore J P. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627–643. - PMC - PubMed
- Blanpain C, Doranz B J, Vakili J, Rucker J, Govaerts C, Baik S S W, Lorthioir O, Migeotte I, Libert F, Baleux F, Vassart G, Doms R W, Parmentier M. Multiple charged and aromatic residues in CCR5 amino-terminal domain are involved in high affinity binding of both chemokines and HIV-1 env protein. J Biol Chem. 1999;274:34719–34727. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous