Requirement for C3G-dependent Rap1 activation for cell adhesion and embryogenesis - PubMed (original) (raw)
Requirement for C3G-dependent Rap1 activation for cell adhesion and embryogenesis
Y Ohba et al. EMBO J. 2001.
Abstract
C3G is a guanine nucleotide exchange factor (GEF) for Rap1, and is activated via Crk adaptor protein. To understand the physiological role of C3G, we generated C3G knockout mice. C3G(-/-) homozygous mice died before embryonic day 7.5. The lethality was rescued by the expression of the human C3G transgene, which could be excised upon the expression of Cre recombinase. From the embryo of this mouse, we prepared fibroblast cell lines, MEF-hC3G. Expression of Cre abolished the expression of C3G in MEF-hC3G and inhibited cell adhesion-induced activation of Rap1. The Cre-expressing MEF-hC3G showed impaired cell adhesion, delayed cell spreading and accelerated cell migration. The accelerated cell migration was suppressed by the expression of active Rap1, Rap2 and R-Ras. Expression of Epac and CalDAG-GEFI, GEFs for Rap1, also suppressed the accelerated migration of the C3G-deficient cells. This observation indicated that Rap1 activation was sufficient to complement the C3G deficiency. In conclusion, C3G-dependent activation of Rap1 is required for adhesion and spreading of embryonic fibroblasts and for the early embryogenesis of the mouse.
Figures
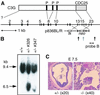
Fig. 1. Generation of C3G knockout mice. (A) Schematic structure of the mouse C3G gene and C3G protein. Coding exons 2–23 are shown at the top. A targeting vector, p836BL/R, consisted of 3.5 and 6.0 kb homologous DNA fragments, the PGK-neo cassette replacing exons 15 and 16, and the MC1-tk cassette at the 5′ end of the targeting vector. The diagnostic probe (probe B) and the _Eco_RV sites (vertical arrows) are indicated at the bottom. P, proline-rich Crk-binding regions; CDC25, CDC25 homology region. (B) Southern blot analysis of mouse genomic DNA. _Eco_RV-digested DNAs from ES cells were separated on a 0.5% agarose gel, transferred to a nylon membrane and hybridized with fluorescein isothiocyanate (FITC)-labeled probe B. The mouse genotype was identified by 9.4 and 6.5 kb fragments that were derived from the wild-type and mutant alleles, respectively. (C) Horizontal sections of uterus of a C3G+/– mouse crossed with a C3G+/– mouse and examined at E7.5. Embryos of _C3G_–/– mice did not conform to any histological structure, as shown on the right. A control section from a C3G+/– embryo is shown on the left.
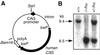
Fig. 2. Complementation of mouse C3G deficiency by the human C3G transgene. (A) Schematic illustration of an expression plasmid of the human C3G transgene. The human C3G gene, which is sandwiched with two loxP recombination sites, is placed downstream of the chicken β-actin promoter and the CMV immediate early enhancer. _Sal_I and _Bam_HI sites, which were used to isolate the transgene expression unit, are indicated by arrows. (B) Southern blot analysis of transgenic mice crossed with C3G knockout mice. The genotype of mouse C3G and the presence of the human C3G transgene are indicated at the top.
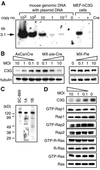
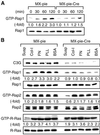
Fig. 3. Cre-dependent disruption of the C3G gene. (A) MEF-hC3G cells were infected with a recombinant retrovirus, MX-pie (indicated as Cre –) or MX-pie-Cre (Cre +), and selected in DMEM containing 2 µg/ml puromycin for 48 h. A 5 µg aliquot of DNA from the cells and from an irrelevant mouse tail biopsy sample containing the indicated copy numbers of the human C3G genes were digested with _Xho_I and analyzed by Southern blotting. (B) MEF-hC3G cells were infected with Cre-carrying adenovirus (AxCanCre), Cre-carrying retrovirus (MX-pie-Cre) and control virus (MX-pie) at the multiplicity of infection (MOI) indicated at the top of panels. Forty-eight hours after infection, cells were lysed in lysis buffer and separated by SDS–PAGE, followed by immunoblotting by use of anti-C3G antibody. The filter was reprobed with anti-tubulin monoclonal antibody to confirm that similar amounts of lysates were analyzed (lower panels). (C) Cell lysates of MEF-hC3G were separated by SDS–PAGE, followed by western blotting by use of anti-C3G polyclonal antibody, sc-869, anti-C3G serum 1A or 1B. (D) MEF-hC3G cells were infected with Cre-carrying retrovirus. After 48 h, cells were lysed in lysis buffer and GTP-bound G proteins were collected by use of either GST–Raf-RBD (for Ras and R-Ras) or GST–RalGDS-RBD (for Rap1 and Rap2). The resulting complexes were precipitated by glutathione–Sepharose beads and analyzed by SDS–PAGE and western blotting by use of antibodies. Small aliquots of lysates were analyzed by immunoblotting to confirm a similar level of expression of G proteins. For the detection of GTP-bound R-Ras, MEF-hC3G cells were infected with MSCV-R-Ras retrovirus and maintained in DMEM containing 2 µg/ml puromycin for 48 h. The cells were analyzed as described, except that anti-FLAG antibody was used to detect the expressed R-Ras.
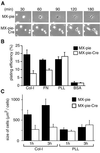
Fig. 4. Requirement for C3G in cell adhesion and cell spreading. (A) MEF-hC3G cells were infected with MX-pie-Cre or MX-pie. After 48 h, cells were trypsinized, kept in suspension for 1 h and plated on dishes coated with collagen type I. The cells were observed by time-lapse microscopy. We show representative photographs at the indicated time points. (B) MEF-hC3G cells were infected with MX-pie or MX-pie-Cre. After 48 h, cells were trypsinized and labeled with BCECF, AM. The cells were plated on 96-well black-colored plates coated with the reagents indicated at the bottom and incubated for 1 h at 37°C in a CO2 incubator. Cells were washed three times with HBSS, and the fluorescence intensity was measured at excitation and emission wavelengths of 488 and 530 nm, respectively. Plating efficiency is shown as the average for three wells, with the SE. (C) MEF-hC3G cells infected with MX-pie or MX-pie-Cre were plated to dishes coated with collagen (Col-I) or poly-
l
-lysine (PLL). Twenty EGFP-positive cells were photographed after 1 and 3 h and measured for size. Average and SE are shown.
Fig. 5. Rap1 activation induced by cell adhesion. (A) MEF-hC3G cells were infected with MX-pie-Cre or MX-pie. After 48 h, cells were trypsinized, kept in suspension for 1 h and plated on dishes coated with collagen type I. Cells were lysed at the indicated times. After normalizing on the protein quantity, the level of GTP-Rap1 was analyzed by Bos’s method. (B) MEF-hC3G cells were infected with MX-pie or MX-pie-Cre, maintained for 48 h, trypsinized, kept in suspension for 1 h and plated on dishes pre-coated as noted at the top. After 1 h, we harvested cells and examined the levels of GTP-bound Rap1, Rap2 and R-Ras. For the detection of GTP-R-Ras, we used MEF-hC3G cells infected with MSCV-R-Ras retrovirus as described in Figure 3C.
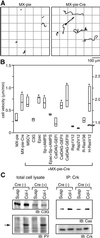
Fig. 6. Increased cell motility of C3G-deficient cells. (A) MEF-hC3G cells infected with MX-pie or MX-pie-Cre were trypsinized, kept in suspension for 1 h and plated on collagen-coated dishes. Starting after 1 h, cell images were collected every 3 min under time-lapse fluorescence microscopy equipped with a cooled CCD camera. Paths of the center of EGFP-positive cells during 8 h recording time were traced with MetaMorph2 software. (B) MEF-hC3G cells or MEF-hC3G cells expressing the proteins listed at the bottom were infected with MX-pie-Cre as indicated. We analyzed the cells as in (A) and obtained the mean velocities of 20 cells for each sample. Mid-line, top and bottom of each box indicate median, 75th quartile and 25th quartile, respectively. Cells that show a significant difference from the control, MX-pie infected cells, by _t_-test and Welch test (P <0.01) are marked with an asterisk at the top of the box. Note that scales shown on the right are for the cells expressing H-RasV12 used. (C) MEF-hC3G cells prepared as described in (A) were lysed and immunoprecipitated with anti-Crk monoclonal antibody and a mixture of protein G– and protein A–Sepharose. Total cell lysates and immunoprecipitated proteins were separated by SDS–PAGE, followed by immunoblotting with anti-C3G anbibody, anti-phosphotyrosine antibody (PY), anti-p130Cas antibody (Cas) or anti-Crk antibody.
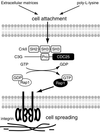
Fig. 7. A model for the role of C3G–Rap1 signaling pathway in cell attachment and cell spreading. In mouse embryonic fibroblasts, Rap1 is activated by C3G upon cell attachment, irrespective of the substratum. The activated Rap1 triggers an inside-out signal of integrin to induce cell adhesion, which requires specific interaction between integrin and extracellular matrices. SH2, SH3, Pro and CDC25 indicate Src homology 2, Src homology 3, proline-rich and yeast CDC25 homology domains, respectively.
Similar articles
- Activation of Rac by cadherin through the c-Src-Rap1-phosphatidylinositol 3-kinase-Vav2 pathway.
Fukuyama T, Ogita H, Kawakatsu T, Inagaki M, Takai Y. Fukuyama T, et al. Oncogene. 2006 Jan 5;25(1):8-19. doi: 10.1038/sj.onc.1209010. Oncogene. 2006. PMID: 16170364 - Interaction of Bcr/Abl with C3G, an exchange factor for the small GTPase Rap1, through the adapter protein Crkl.
Cho YJ, Hemmeryckx B, Groffen J, Heisterkamp N. Cho YJ, et al. Biochem Biophys Res Commun. 2005 Aug 12;333(4):1276-83. doi: 10.1016/j.bbrc.2005.06.030. Biochem Biophys Res Commun. 2005. PMID: 15982636 - Endothelin signaling via guanine exchange factor C3G in renal glomerular mesangial cells.
Rufanova VA, Alexanian A, Ostendorf T, Bokemeyer D, Prosser S, Miller B, Sorokin A. Rufanova VA, et al. Can J Physiol Pharmacol. 2010 Aug;88(8):808-16. doi: 10.1139/Y10-056. Can J Physiol Pharmacol. 2010. PMID: 20725139 - [Activation of Rap1, antagonist to ras, by Crk-C3G].
Hattori S, Matsuda M. Hattori S, et al. Gan To Kagaku Ryoho. 1997 Sep;24(11):1414-21. Gan To Kagaku Ryoho. 1997. PMID: 9309134 Review. Japanese. - Signalling to actin: role of C3G, a multitasking guanine-nucleotide-exchange factor.
Radha V, Mitra A, Dayma K, Sasikumar K. Radha V, et al. Biosci Rep. 2011 Aug;31(4):231-44. doi: 10.1042/BSR20100094. Biosci Rep. 2011. PMID: 21366540 Review.
Cited by
- C3G Is Upregulated in Hepatocarcinoma, Contributing to Tumor Growth and Progression and to HGF/MET Pathway Activation.
Sequera C, Bragado P, Manzano S, Arechederra M, Richelme S, Gutiérrez-Uzquiza A, Sánchez A, Maina F, Guerrero C, Porras A. Sequera C, et al. Cancers (Basel). 2020 Aug 14;12(8):2282. doi: 10.3390/cancers12082282. Cancers (Basel). 2020. PMID: 32823931 Free PMC article. - Defective angiogenesis, endothelial migration, proliferation, and MAPK signaling in Rap1b-deficient mice.
Chrzanowska-Wodnicka M, Kraus AE, Gale D, White GC 2nd, Vansluys J. Chrzanowska-Wodnicka M, et al. Blood. 2008 Mar 1;111(5):2647-56. doi: 10.1182/blood-2007-08-109710. Epub 2007 Nov 9. Blood. 2008. PMID: 17993608 Free PMC article. - C3G regulates the size of the cerebral cortex neural precursor population.
Voss AK, Krebs DL, Thomas T. Voss AK, et al. EMBO J. 2006 Aug 9;25(15):3652-63. doi: 10.1038/sj.emboj.7601234. Epub 2006 Jul 20. EMBO J. 2006. PMID: 16858399 Free PMC article. - Ras and calcium signaling pathways converge at Raf1 via the Shoc2 scaffold protein.
Yoshiki S, Matsunaga-Udagawa R, Aoki K, Kamioka Y, Kiyokawa E, Matsuda M. Yoshiki S, et al. Mol Biol Cell. 2010 Mar 15;21(6):1088-96. doi: 10.1091/mbc.e09-06-0455. Epub 2010 Jan 13. Mol Biol Cell. 2010. PMID: 20071468 Free PMC article. - Talin‑1 interaction network in cellular mechanotransduction (Review).
Zhao Y, Lykov N, Tzeng C. Zhao Y, et al. Int J Mol Med. 2022 May;49(5):60. doi: 10.3892/ijmm.2022.5116. Epub 2022 Mar 10. Int J Mol Med. 2022. PMID: 35266014 Free PMC article. Review.
References
- Birge R.B., Fajardo,J.E., Reichman,C., Shoelson,S.E., Songyang,Z., Cantley,L.C. and Hanafusa,H. (1993) Identification and characteriz ation of a high-affinity interaction between v-Crk and tyrosine-phosphorylated paxillin in CT10-transformed fibroblasts. Mol. Cell. Biol., 13, 4648–4656. - PMC - PubMed
- Bos J.L. (1997) Ras-like GTPases. Biochim. Biophys. Acta, 1333, M19–31. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous