The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila - PubMed (original) (raw)
The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila
Gábor Juhász et al. J Cell Biol. 2008.
Abstract
Degradation of cytoplasmic components by autophagy requires the class III phosphatidylinositol 3 (PI(3))-kinase Vps34, but the mechanisms by which this kinase and its lipid product PI(3) phosphate (PI(3)P) promote autophagy are unclear. In mammalian cells, Vps34, with the proautophagic tumor suppressors Beclin1/Atg6, Bif-1, and UVRAG, forms a multiprotein complex that initiates autophagosome formation. Distinct Vps34 complexes also regulate endocytic processes that are critical for late-stage autophagosome-lysosome fusion. In contrast, Vps34 may also transduce activating nutrient signals to mammalian target of rapamycin (TOR), a negative regulator of autophagy. To determine potential in vivo functions of Vps34, we generated mutations in the single Drosophila melanogaster Vps34 orthologue, causing cell-autonomous disruption of autophagosome/autolysosome formation in larval fat body cells. Endocytosis is also disrupted in Vps34(-/-) animals, but we demonstrate that this does not account for their autophagy defect. Unexpectedly, TOR signaling is unaffected in Vps34 mutants, indicating that Vps34 does not act upstream of TOR in this system. Instead, we show that TOR/Atg1 signaling regulates the starvation-induced recruitment of PI(3)P to nascent autophagosomes. Our results suggest that Vps34 is regulated by TOR-dependent nutrient signals directly at sites of autophagosome formation.
Figures
Figure 1.
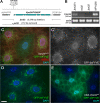
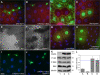
Reagents to disrupt and monitor Vps34 function in D. melanogaster. (A) Map of the Vps34 locus, showing the location and extent of the Δm22 deletion. (B) RT-PCR analysis of Vps34 mRNA expression in control and mutant third instar larvae. Vps34 transcript levels are strongly reduced in ex32 animals and undetectable in the Δm22 mutant. (C–E) Disruption of Vps34 function causes mislocalization of the PI(3)P reporter GFP-2xFYVE. In wild-type fat body cells (C and D), GFP-2xFYVE accumulates at the perinuclear endosomal compartment. This pattern of localization is lost in Vps34Δm22 clones (C; mutant cell marked by lack of dsRed) or in response to expression of Vps34KD (E). Bar, 10 μm. Genotypes: (C) hsflp; FRT42D UAS-GFP-2xFYVE Vps34Δm22/Cg-GAL4 FRT42D UAS-GFP-2xFYVE UAS-dsRed, (D) hsflp; UAS-GFP-2xFYVE/+; Act>CD2>GAL4/+, and (E) hsflp; UAS-GFP-2xFYVE/+ ; Act>CD2>GAL4 / UAS-Vps34KD.
Figure 2.
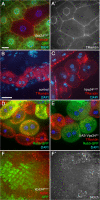
Starvation-induced autophagy in the larval fat body requires Vps34. All images depict live fat body tissue dissected from fed (C) or 4-h starved animals (all other panels). (A) Vps34Δm22 loss-of-function clones (marked by lack of GFP) fail to accumulate autolysosomes (labeled by punctate Lysotracker Red staining) after starvation. (B) Vps34KD-expressing cells (GFP positive) fail to accumulate punctate Lysotracker Red staining under starvation conditions. (C and D) Clonal expression of wild-type Vps34 (GFP-positive cells) does not induce autophagy under fed conditions (C) but increases the response to starvation (D). (E) Vps34Δm22 loss-of-function clones (marked by lack of myrRFP) fail to accumulate autophagosomes (marked by punctate GFP-Atg8) in response to starvation. (F and G) Starvation-induced accumulation of GFP-Atg8a punctae is observed in controls (F) but not in animals expressing Vps34KD (G). (H–K) TEM images reveal abundant autophagosomes (AP) and autolysosomes (AL) in control animals (H) but not Vps34Δm22 mutants (I) nor animals expressing Vps34KD (J). LD, lipid droplet. The graph in K shows autophagosome and autolysosome area ratios calculated from electron micrographs of five animals per genotype. Error bars show SD from the mean. P-values (Mann-Whitney U test): control versus Vps34Δm22, AP, 1.45e-11 and AL, 3.9e-9; and control versus Vps34KD, AP, 2.9e-11 and AL, 1.5 e-11. Bars: (A–G) 10 μm; (H–J) 1 μm. Genotypes: (A) hsflp; FRT42D Vps34Δm22/UAS-2xeGFP FRT42D fb-GAL4, (B) hsflp; Act>CD2>GAL4 UAS-GFP/UAS-Vps34KD, (C and D) hsflp; Act>CD2>GAL4 UAS-GFP/UAS-Vps34WT, (E) hsflp; FRT42D Vps34Δm22/Cg-GAL4 UAS-GFP-Atg8a FRT42D UAS-myrRFP, (F) hsflp; Act>CD2>GAL4 UAS-GFP-Atg8a/+, (G) hsflp; Act>CD2>GAL4 UAS-GFP-Atg8a/UAS-Vps34KD, (H) Vps34Δm22/CyO-GFP, (I) Vps34Δm22/Δm22, (J) Cg-GAL4 UAS-Vps34KD/Cg-GAL4 UAS-Vps34KD, and (K) as in H–J.
Figure 3.
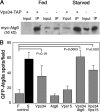
Physical and genetic interactions between D. melanogaster Vps34, Vps15, and Atg6. (A) Epitope-tagged Atg6 and Vps34 coimmunoprecipitate from larval extracts. myc-Atg6 and Vps15-FLAG were coexpressed with and without Vps34-TAP during larval development by heat shock–dependent GAL4 induction of UAS transgenes under either fed or starved conditions. Immunoprecipitation of larval extracts with hVps34 antibodies coprecipitates myc-Atg6. Levels of coprecipitated myc-Atg6 are higher in extracts from larvae overexpressing Vps34-TAP (lanes 2 and 6) than from larvae expressing only endogenous Vps34 (lanes 4 and 8). (B) Coexpression of Vps34-TAP with Atg6 or Vps15 results in cooperative induction of GFP-Atg8a–labeled autophagosomes under fed conditions. Number of GFP-Atg8a–positive spots per field in fat body of fed (F) and starved (S) control animals and in fed transgenic animals is indicated. Data represent mean ± SE of 10 animals per genotype. P-values were determined by two-tailed Student's t test.
Figure 4.
Vps34 and ESCRT components are required for distinct steps in autophagy. (A) Mutation of the ESCRT-II component Vps28 (mutant clone marked by lack of GFP) disrupts formation of Lysotracker Red punctae in response to 4-h starvation. Bar, 10 μm. (B–D) GFP-Atg8a–marked autophagosomes accumulate in clones of Vps28 mutant (B) and Vps34 Vps28 double mutant (D) cells but not in Vps34 mutant cells (C). Mutant clones are marked by lack of myrRFP. (E–G) Mosaic eye imaginal discs showing accumulation of ubiquitin-positive punctae in Vps25 (E) and Vps25 Vps34 (G) mutant clones but not in Vps34 mutant clones (F). Lack of GFP marks mutant clones. Bar, 10 μm (B–G). Genotypes: (A) hsflp; FRT42D Vps28k15603/UAS-2xeGFP FRT42D fb-GAL4, (B) hsflp; FRT42D Vps28k15603/Cg-GAL4 UAS-GFP-Atg8a FRT42D UAS-myrRFP, (C) hsflp; FRT42D Vps34Δm22/Cg-GAL4 UAS-GFP-Atg8a FRT42D UAS-myrRFP, (D) hsflp; FRT42D Vps28k15603 Vps34Δm22/Cg-GAL4 UAS-GFP-Atg8a FRT42D UAS-myrRFP, (E) hsflp; FRT42D Vps25A3/FRT42D Ubi-GFPnls, (F) hsflp; FRT42D Vps34Δm22/FRT42D Ubi-GFPnls, and (G) hsflp; FRT42D Vps25A3 Vps34Δm22/FRT42D Ubi-GFPnls.
Figure 5.
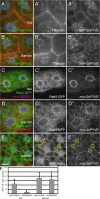
Vps34 is required for endocytosis. (A) Vps34Δm22 loss-of-function fat body clones (marked by lack of GFP) fail to incorporate the endocytic tracer TR avidin into the perinuclear early endosome (20-min pulse followed by 10-min chase). (B and C) TR avidin accumulates in Garland cells of heterozygous controls (B) but not Vps34Δm22 homozygous mutants (C; 2-min pulse followed by 20-min chase). (D and E) Mosaic larvae expressing Rab5-GFP (D) or Rab5-GFP and Vps34KD (E) in a subset of Garland cells. TR avidin uptake and cortical localization of Rab5-GFP are disrupted in Vps34KD-expressing cells (15-min pulse, no chase). Bar, 10 μm (B–E). (F) Clonal loss of Vps34 (marked by lack of GFP) in the eye imaginal disc results in accumulation of Notch-positive punctae. Bar, 10 μm (A and F). Genotypes: (A) hsflp; FRT42D Vps34Δm22/UAS-2xeGFP FRT42D fb-GAL4, (B) Vps34Δm22/CyO-GFP, (C) Vps34Δm22/Δm22, (D) hsflp; Act>CD2>GAL4 UAS-Rab5-GFP/+, (E) hsflp; Act>CD2>GAL4 UAS-Rab5-GFP/UAS-Vps34KD, and (F) hsflp; FRT42D Vps34Δm22/FRT42D Ubi-GFPnls.
Figure 6.
Vps34 does not influence TOR signaling or function. Transgene-expressing cells are marked by coexpression of GFP in A, B, D, and G–J. Loss-of-function mutant clones are marked by lack of GFP in C, E, and F. All images are from fed larvae except G and H, which are from 48-h starved animals. (A and B) Inhibition of TOR signaling by expression of Tor double-stranded RNA (dsRNA; A) or overexpression of Tsc1 and 2 (B) results in a reduction in fat body cell size. (C and D) Fat body cells lacking Vps34 (C) or expressing Vps34KD (D) are normal in size. (E and F) In the eye imaginal disc, cells mutant for Tor (E; GFP negative) proliferate more slowly than their wild-type “twin spot” control cells (two copies of GFP). Vps34Δm22 homozygous mutant cells (F) proliferate at a rate similar to that of twin spot controls. (G and H) Overexpression of Rheb activates TOR and increases cell growth both in the absence (G) or presence (H) of coexpressed kinase-defective Vps34. (I and J) Inhibition of TOR signaling by cooverexpression of Tsc1 and 2 reduces cell growth in control (I) and Vps34Δm22 (J) animals. (K) Ubiquitous hsGAL4-driven overexpression of wild-type Vps34 (lane 2) or kinase-defective Vps34 (lane 3) does not affect TOR-dependent phosphorylation of S6K-Thr398 or Akt-Ser505 relative to control extracts (lane 1). (L) Quantitation of the area of mutant or transgene-expressing cells in A–D relative to wild-type cells from the same tissue. Data represent mean ± SEM of 7–10 animals per genotype. Bar: 10 μm. Genotypes: (A) hsflp; Act>CD2>GAL4 UAS-GFP/UAS-TORdsRNA5092R-2, (B) hsflp; Act>CD2>GAL4 UAS-GFP/UAS-Tsc1 UAS-Tsc2, (C) hsflp; FRT42D Vps34Δm22/UAS-2xeGFP FRT42D fb-GAL4, (D) hsflp; Act>CD2>GAL4 UAS-GFP/UAS-Vps34KD, (E) hsflp; TorΔP FRT40A/Ubi-GFPnls FRT40A, (F) hsflp; FRT42D Vps34Δm22/FRT42D Ubi-GFPnls, (G) hsflp; Act>CD2>GAL4 UAS-GFP UAS-RhebEP50.084/+, (H) hsflp; Act>CD2>GAL4 UAS-GFP UAS-RhebEP50.084/UAS-Vps34KD, (I) hsflp; Act>CD2>GAL4 UAS-GFP/UAS-Tsc1 UAS-Tsc2, and (J) hsflp; Vps34Δm22/Vps34Δm22; Act>CD2>GAL4 UAS-GFP/UAS-Tsc1 UAS-Tsc2.
Figure 7.
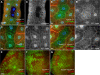
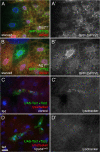
Recruitment of PI(3)P from early endosomes to autophagosomes in response to TOR/Atg1-dependent nutrient signaling. (A and B) Internalized TR avidin colocalizes with GFP-2xFYVE under fed conditions (A) but not after 4-h starvation (B). (C and D) Starvation causes dispersion of myc-2xFYVE but not Rab5-GFP. (E) Under starvation conditions, myc-2xFYVE displays extensive colocalization with GFP-Atg8a. Yellow circles in E′ and E′′ show overlap between GFP-Atg8a and myc-2xFYVE label. Bar, 10 μm. (F) Quantitation of the myc-2xFYVE–labeled compartment area in a 2-μm ring surrounding the nucleus (perinuclear) and the remaining peripheral cell area (cytoplasmic). Genotypes: (A and B) Cg-GAL4/UAS-GFP-2xFYVE, (C and D) hsflp; Act>CD2>GAL4 UAS-Rab5-GFP/UAS-myc-2xFYVE, and (E) Cg-GAL4 UAS-GFP-Atg8a/UAS-myc-2xFYVE.
Figure 8.
Vps34 functions downstream of TOR signaling. (A and B) Starvation-induced redistribution of Vps34 activity requires TOR-Atg1 signaling. 4-h starvation causes relocalization of GFP-2xFYVE in control cells but not in Tsc2 homozygous mutant cells (A) nor in cells mutant for Atg1 (B). Mutant clones are marked by lack of dsRed. Note also the changes in GFP-2xFYVE levels in the mutant cells. (C and D) Cooverexpression of Tsc1 and 2 in GFP-marked clones induces formation of autolysosomes under fed conditions in control (C) but not Vps34 mutant (D) animals. Bar: (A and B) 10 μm; (C and D) 25 μm. Genotypes: (A) hsflp; Cg-GAL4 UAS-GFP-2xFYVE/+; Tsc2192 FRT80B/UAS-dsRed FRT80B, (B) hsflp; Cg-GAL4 UAS-GFP-2xFYVE/+; Atg1Δ3D FRT80B/UAS-dsRed FRT80B, (C) hsflp; Vps34Δm22/+; Act>CD2>GAL4 UAS-GFP/UAS-Tsc1 UAS-Tsc2, and (D) hsflp; Vps34Δm22/Vps34Δm22 ; Act>CD2>GAL4 UAS-GFP/UAS-Tsc1 UAS-Tsc2.
Similar articles
- Atg6/UVRAG/Vps34-containing lipid kinase complex is required for receptor downregulation through endolysosomal degradation and epithelial polarity during Drosophila wing development.
Lőrincz P, Lakatos Z, Maruzs T, Szatmári Z, Kis V, Sass M. Lőrincz P, et al. Biomed Res Int. 2014;2014:851349. doi: 10.1155/2014/851349. Epub 2014 May 21. Biomed Res Int. 2014. PMID: 25006588 Free PMC article. - Nutrient-dependent regulation of autophagy through the target of rapamycin pathway.
Chang YY, Juhász G, Goraksha-Hicks P, Arsham AM, Mallin DR, Muller LK, Neufeld TP. Chang YY, et al. Biochem Soc Trans. 2009 Feb;37(Pt 1):232-6. doi: 10.1042/BST0370232. Biochem Soc Trans. 2009. PMID: 19143638 Review. - An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation.
Chang YY, Neufeld TP. Chang YY, et al. Mol Biol Cell. 2009 Apr;20(7):2004-14. doi: 10.1091/mbc.e08-12-1250. Epub 2009 Feb 18. Mol Biol Cell. 2009. PMID: 19225150 Free PMC article. - Vps34 and PLD1 take center stage in nutrient signaling: their dual roles in regulating autophagy.
Yoon MS. Yoon MS. Cell Commun Signal. 2015 Nov 21;13:44. doi: 10.1186/s12964-015-0122-x. Cell Commun Signal. 2015. PMID: 26589724 Free PMC article. Review.
Cited by
- Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila.
Shravage BV, Hill JH, Powers CM, Wu L, Baehrecke EH. Shravage BV, et al. Development. 2013 Mar;140(6):1321-9. doi: 10.1242/dev.089490. Epub 2013 Feb 13. Development. 2013. PMID: 23406899 Free PMC article. - Defects of Vps15 in skeletal muscles lead to autophagic vacuolar myopathy and lysosomal disease.
Nemazanyy I, Blaauw B, Paolini C, Caillaud C, Protasi F, Mueller A, Proikas-Cezanne T, Russell RC, Guan KL, Nishino I, Sandri M, Pende M, Panasyuk G. Nemazanyy I, et al. EMBO Mol Med. 2013 Jun;5(6):870-90. doi: 10.1002/emmm.201202057. Epub 2013 Apr 30. EMBO Mol Med. 2013. PMID: 23630012 Free PMC article. - Prognostic value and underlying mechanism of KIAA0101 in hepatocellular carcinoma: database mining and co-expression analysis.
Xu W, Wang X, Wu X, Yu S, Xiong J, Sang X, Zheng Y, Zhang Z. Xu W, et al. Aging (Albany NY). 2020 Aug 27;12(16):16420-16436. doi: 10.18632/aging.103704. Epub 2020 Aug 27. Aging (Albany NY). 2020. PMID: 32855364 Free PMC article. - Regulators of autophagosome formation in Drosophila muscles.
Zirin J, Nieuwenhuis J, Samsonova A, Tao R, Perrimon N. Zirin J, et al. PLoS Genet. 2015 Feb 18;11(2):e1005006. doi: 10.1371/journal.pgen.1005006. eCollection 2015 Feb. PLoS Genet. 2015. PMID: 25692684 Free PMC article. - Loss of the starvation-induced gene Rack1 leads to glycogen deficiency and impaired autophagic responses in Drosophila.
Erdi B, Nagy P, Zvara A, Varga A, Pircs K, Ménesi D, Puskás LG, Juhász G. Erdi B, et al. Autophagy. 2012 Jul 1;8(7):1124-35. doi: 10.4161/auto.20069. Epub 2012 May 7. Autophagy. 2012. PMID: 22562043 Free PMC article.
References
- Arsham, A.M., and T.P. Neufeld. 2006. Thinking globally and acting locally with TOR. Curr. Opin. Cell Biol. 18:589–597. - PubMed
- Britton, J.S., W.K. Lockwood, L. Li, S.M. Cohen, and B.A. Edgar. 2002. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell. 2:239–249. - PubMed
- Byfield, M.P., J.T. Murray, and J.M. Backer. 2005. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J. Biol. Chem. 280:33076–33082. - PubMed
- Chintapalli, V.R., J. Wang, and J.A. Dow. 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39:715–720. - PubMed
- Eskelinen, E.L. 2005. Maturation of autophagic vacuoles in Mammalian cells. Autophagy. 1:1–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM62509/GM/NIGMS NIH HHS/United States
- DK070679/DK/NIDDK NIH HHS/United States
- R01 DK070679/DK/NIDDK NIH HHS/United States
- R01 GM062509/GM/NIGMS NIH HHS/United States
- R01 GM059136/GM/NIGMS NIH HHS/United States
- R56 GM059136/GM/NIGMS NIH HHS/United States
- GM59136/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous