Endogenous expression of Hras(G12V) induces developmental defects and neoplasms with copy number imbalances of the oncogene - PubMed (original) (raw)
. 2009 May 12;106(19):7979-84.
doi: 10.1073/pnas.0900343106. Epub 2009 Apr 29.
Norisato Mitsutake, Krista LaPerle, Nagako Akeno, Pat Zanzonico, Valerie A Longo, Shin Mitsutake, Edna T Kimura, Hartmut Geiger, Eugenio Santos, Hans G Wendel, Aime Franco, Jeffrey A Knauf, James A Fagin
Affiliations
- PMID: 19416908
- PMCID: PMC2674938
- DOI: 10.1073/pnas.0900343106
Endogenous expression of Hras(G12V) induces developmental defects and neoplasms with copy number imbalances of the oncogene
Xu Chen et al. Proc Natl Acad Sci U S A. 2009.
Abstract
We developed mice with germline endogenous expression of oncogenic Hras to study effects on development and mechanisms of tumor initiation. They had high perinatal mortality, abnormal cranial dimensions, defective dental ameloblasts, and nasal septal deviation, consistent with some of the features of human Costello syndrome. These mice developed papillomas and angiosarcomas, which were associated with Hras(G12V) allelic imbalance and augmented Hras signaling. Endogenous expression of Hras(G12V) was also associated with a higher mutation rate in vivo. Tumor initiation by Hras(G12V) likely requires augmentation of signal output, which in papillomas and angiosarcomas is achieved via increased Hras-gene copy number, which may be favored by a higher mutation frequency in cells expressing the oncoprotein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Development of mice with a conditional knock-in activating mutation of Hras. (A) Diagram of Hras targeted allele. (Top) Targeting vector consists of a 5′arm containing the WT Hras gene and a 3′arm containing the mutant Hras gene, which are separated by an _Frt_-flanked Neomycin minigene. (Middle) Targeted allele after crossing with β actin-Flp mice to remove the Neomycin minigene. (Bottom) Targeted allele after crossing with Caggs-Cre mice to remove the WT Hras copy. (B) Southern blot of tail DNA isolated from WT, FR-HrasG12V, or _CC/FR-HrasG12V_± mice cut with XbaI and EcoRV and probed with a genomic fragment containing exons 1–4 of Hras. Wt, wild-type allele; Targ, targeted allele. (C) Sequence trace of products generated by RT-PCR of RNA isolated from MEFs from FR-HrasG12V or CC/FR-HrasG12V embryos. (D) Western blot of activated Hras in MEFs from CC/FR-HrasG12V and control mice. Activated Ras proteins were pulled down with an agarose-conjugated Raf-1 Ras-binding domain, followed by SDS/PAGE gel electrophoresis and immunoblotting with a specific anti-Hras antibody. Western blot of total lysate with Hras IgG is shown below. (E) RT-PCR products of RNA isolated from MEFs from WT or CC/FR-HrasG12V embryos were incubated with or without Gsu I, which digests WT but not mutant Hras cDNA. Total Hras cDNA in the absence of Gsu I was normalized to 100%. The right panel shows that the mutant Hras cDNA is ≈50% of total Hras in CC/FR-HrasG12V MEFs.
Fig. 2.
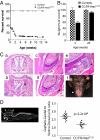
Increased neonatal mortality and cranio-facial deformities in CC/FR-Hras _G12V_± mice. (A) Kaplan-Meier survival plot of CC/FR-HrasG12V and control mice. (B) Decreased body weight in CC/FR-HrasG12V mice at weaning. Weight of CC/FR-HrasG12V mice and control littermates at 3 and 20 weeks of age. Bars represent mean ± SE percent-change in body weight versus controls (n = 8; P = 7.7 ×10−7 at 3 weeks, P = 0.25 at 20 weeks). (C) Coronal sections (4×) across the nasal cavity of a representative WT (a) and CC/FR-HrasG12V (b) mouse. The mutant mouse exhibits marked nasal septal deviation. Low power magnification (20×) of an incisor of a WT (C) and a mutant (D) mouse. The tooth of the _CC/FR-HrasG12V_± mice has an abnormal ameloblast cell lining and defective enamel formation. (e) Higher magnification (40×) demonstrates detachment of the ameloblasts from the adjacent dentin, loss of polarity, and stratification of ameloblasts, as well as areas of enamel sequestration. (f) Mouse with misalignment of incisors and malocclusion. (D) (Left) Whole-body CT scan (sagittal view) illustrating the rectangular region of interest used to determine the overall cephalo-caudal and ventro-dorsal dimensions of the cranial bony structures. (Right) Abnormal cranial dimensions of CC/FR-HrasG12V mice. Mean cephalo-caudal to ventro-dorsal ratio in aged-matched control and CC/FR-HrasG12V mice. All mice studied in this article are HrasG12V heterozygous mice.
Fig. 3.
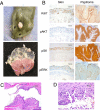
Squamous papilloma and angiosarcoma development in CC/FR-HrasG12V mice is associated with augmented Hras signaling. (A) (Upper) Representative papilloma in a CC/FR-HrasG12V mouse. Papillomas frequently formed in areas exposed to friction. (Lower) Stomach from a 28-week-old CC/FR-HrasG12V mouse showing multiple papillomas in the stomach fundus, but not in the cardia or antrum. (B) Representative IHC staining for Ki67, pAKT, pS6, and pERK1/2 in sections of papilloma and adjacent non-neoplastic skin from CC/FR-HrasG12V mice. (C) H&E-stained sections (10×) of the external auditory canal of a CC/FR-HrasG12V mice with epidermal and sebaceous hyperplasia. (D) Representative H&E section (100×) of angiosarcoma developing in CC/FR-HrasG12V mice.
Fig. 4.
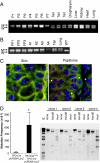
Hras allelic imbalance in papillomas and angiosarcoma from CC/FR-HrasG12V mice. (A) PCR of genomic DNA of papillomas with primers that distinguish mutant from WT Hras alleles (see Fig. 1_A_). W: 622 bp WT allele; M: 666 bp targeted allele because of insertion of loxP site. P1-P7: DNA from papillomas, or indicated nontumoral tissues. (B) PCR of DNA from forestomach papillomas (FP1–FP3), angiosarcomas (A1–A4), or indicated nontumoral tissues. (C) FISH of a representative section from papilloma tissue (P2) using a mouse BAC containing the Hras gene. Papilloma nuclei have 3 to 5 fluorescent signals corresponding to Hras (red), whereas adjacent skin is diploid. Mouse chromosome 7 centromeres are labeled in green. (D) (Left) Increased mutation rate in thyroid cells from FR-HrasG12V/TPO-Cre mice. Plasmids rescued from DNA extracts of thyroid glands of TPO-Cre/pUR288-LacZ or FR-HrasG12V/TPO-Cre/pUR288-LacZ were screened for alterations in LacZ, as described in Methods. Bars represent the mean mutation frequency ± SE of pooled samples from TPO-Cre/pUR288-LacZ (n = 3; 10 thyroids per pool) and FR-HrasG12V/TPO-Cre/pUR288-LacZ (n = 4; 10 thyroids per pool). P < 0.05. (Right) Representative Southern blot of _LacZ_-negative clones. The type of mutation was determined by PCR amplification and restriction digestion of _LacZ_-negative clones. Clone 1 contained an inactivating point mutation of LacZ, whereas the other 3 show distinct restriction profiles consistent with recombination events, insertions, or deletions.
Similar articles
- Mice with an Oncogenic HRAS Mutation are Resistant to High-Fat Diet-Induced Obesity and Exhibit Impaired Hepatic Energy Homeostasis.
Oba D, Inoue SI, Miyagawa-Tomita S, Nakashima Y, Niihori T, Yamaguchi S, Matsubara Y, Aoki Y. Oba D, et al. EBioMedicine. 2018 Jan;27:138-150. doi: 10.1016/j.ebiom.2017.11.029. Epub 2017 Dec 6. EBioMedicine. 2018. PMID: 29254681 Free PMC article. - Mutant Hras(G12V) and Kras(G12D) have overlapping, but non-identical effects on hepatocyte growth and transformation frequency in transgenic mice.
Figueiredo ML, Stein TJ, Jochem A, Sandgren EP. Figueiredo ML, et al. Liver Int. 2012 Apr;32(4):582-91. doi: 10.1111/j.1478-3231.2011.02732.x. Epub 2012 Jan 3. Liver Int. 2012. PMID: 22221894 Free PMC article. - H-Ras and K-Ras Oncoproteins Induce Different Tumor Spectra When Driven by the Same Regulatory Sequences.
Drosten M, Simón-Carrasco L, Hernández-Porras I, Lechuga CG, Blasco MT, Jacob HK, Fabbiano S, Potenza N, Bustelo XR, Guerra C, Barbacid M. Drosten M, et al. Cancer Res. 2017 Feb 1;77(3):707-718. doi: 10.1158/0008-5472.CAN-16-2925. Epub 2016 Nov 21. Cancer Res. 2017. PMID: 27872088 - RAS-mediated oncogenic signaling pathways in human malignancies.
Khan AQ, Kuttikrishnan S, Siveen KS, Prabhu KS, Shanmugakonar M, Al-Naemi HA, Haris M, Dermime S, Uddin S. Khan AQ, et al. Semin Cancer Biol. 2019 Feb;54:1-13. doi: 10.1016/j.semcancer.2018.03.001. Epub 2018 Mar 7. Semin Cancer Biol. 2019. PMID: 29524560 Review. - RAS mutations in human cancers: Roles in precision medicine.
Murugan AK, Grieco M, Tsuchida N. Murugan AK, et al. Semin Cancer Biol. 2019 Dec;59:23-35. doi: 10.1016/j.semcancer.2019.06.007. Epub 2019 Jun 27. Semin Cancer Biol. 2019. PMID: 31255772 Review.
Cited by
- Dominant role of oncogene dosage and absence of tumor suppressor activity in Nras-driven hematopoietic transformation.
Xu J, Haigis KM, Firestone AJ, McNerney ME, Li Q, Davis E, Chen SC, Nakitandwe J, Downing J, Jacks T, Le Beau MM, Shannon K. Xu J, et al. Cancer Discov. 2013 Sep;3(9):993-1001. doi: 10.1158/2159-8290.CD-13-0096. Epub 2013 Jun 3. Cancer Discov. 2013. PMID: 23733505 Free PMC article. - Mouse models of thyroid cancer: A 2015 update.
Kirschner LS, Qamri Z, Kari S, Ashtekar A. Kirschner LS, et al. Mol Cell Endocrinol. 2016 Feb 5;421:18-27. doi: 10.1016/j.mce.2015.06.029. Epub 2015 Jun 27. Mol Cell Endocrinol. 2016. PMID: 26123589 Free PMC article. Review. - Mechanics of a multilayer epithelium instruct tumour architecture and function.
Fiore VF, Krajnc M, Quiroz FG, Levorse J, Pasolli HA, Shvartsman SY, Fuchs E. Fiore VF, et al. Nature. 2020 Sep;585(7825):433-439. doi: 10.1038/s41586-020-2695-9. Epub 2020 Sep 2. Nature. 2020. PMID: 32879493 Free PMC article. - Ras in cancer and developmental diseases.
Fernández-Medarde A, Santos E. Fernández-Medarde A, et al. Genes Cancer. 2011 Mar;2(3):344-58. doi: 10.1177/1947601911411084. Genes Cancer. 2011. PMID: 21779504 Free PMC article. - Briefly bound to activate: transient binding of a second catalytic magnesium activates the structure and dynamics of CDK2 kinase for catalysis.
Bao ZQ, Jacobsen DM, Young MA. Bao ZQ, et al. Structure. 2011 May 11;19(5):675-90. doi: 10.1016/j.str.2011.02.016. Structure. 2011. PMID: 21565702 Free PMC article.
References
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA072597/CA/NCI NIH HHS/United States
- R01 CA050706/CA/NCI NIH HHS/United States
- P30 CA08748/CA/NCI NIH HHS/United States
- T32 DK07313/DK/NIDDK NIH HHS/United States
- CA72597/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R24 CA83084/CA/NCI NIH HHS/United States
- R24 CA083084/CA/NCI NIH HHS/United States
- CA50706/CA/NCI NIH HHS/United States
- T32 DK007313/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous