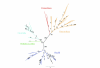Structural analysis of polarizing indels: an emerging consensus on the root of the tree of life - PubMed (original) (raw)
Structural analysis of polarizing indels: an emerging consensus on the root of the tree of life
Ruben E Valas et al. Biol Direct. 2009.
Abstract
Background: The root of the tree of life has been a holy grail ever since Darwin first used the tree as a metaphor for evolution. New methods seek to narrow down the location of the root by excluding it from branches of the tree of life. This is done by finding traits that must be derived, and excluding the root from the taxa those traits cover. However the two most comprehensive attempts at this strategy, performed by Cavalier-Smith and Lake et al., have excluded each other's rootings.
Results: The indel polarizations of Lake et al. rely on high quality alignments between paralogs that diverged before the last universal common ancestor (LUCA). Therefore, sequence alignment artifacts may skew their conclusions. We have reviewed their data using protein structure information where available. Several of the conclusions are quite different when viewed in the light of structure which is conserved over longer evolutionary time scales than sequence. We argue there is no polarization that excludes the root from all Gram-negatives, and that polarizations robustly exclude the root from the Archaea.
Conclusion: We conclude that there is no contradiction between the polarization datasets. The combination of these datasets excludes the root from every possible position except near the Chloroflexi.
Figures
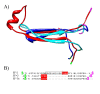
Figure 1
EF-2 contains a derived insert. A) Structural alignment of EF-G from Thermus thermophilus (2BV3 61–89 colored blue), EF-Tu (1EFC 58–86 colored cyan) from Escherichia coli, EF-2 from Saccharomyces cerevisiae (1N0U 67–110 colored red). B) Sequence corresponding to the structural alignment in A. The well conserved glycine and aspartic acid are highlighted green and magenta respectively in both the sequence and structure to show the disordered nature of this region. The 4 positions highlighted in red are aligned in the original alignment, which is why the alignment was critiqued in [22]. The additional insert in Eukaryotes relative to the Archaea is boxed in black.
Figure 2
HisA does not exclude the root from the Eobacteria. A MUSCLE based alignment of all the HisA sequences in Eobacteria. Represenatives from Actinobacteria (Streptomyces coelicolor A3) and other Gram-negatives (Synechocystis sp. PCC 6803) are included to show the indel. All the Eobacterial sequences share the relative deletion with the Firmicutes (Bacillus clausii).
Figure 3
Structural Alignment of MreB/Hsp70. A) A multiple structural alignment of the MreB/Hsp70 C-terminal actin-like ATPase domain. The region around the indel is highlighted as a ribbon diagram. The backbone of the rest of the domain demonstrates high conservation between these structures. The blue chain is MreB from Thermotoga Maritima (1JCF:A 51–86 drawn as ribbon). The red chain is Hsp70 from the Gram-positive bacterium Geobacillus Kaustophilus (2V7Y:A 57–101 drawn as ribbon). The orange chain is Hsp70 from the Gram-negative bacterium Escherichia coli (1DKG:D 56–125 drawn as ribbon). B) The sequences corresponding to the highlighted portion of the structure alignment in A.
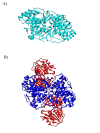
Figure 4
Quaternary Structure of PyrD. A) PyrD 1A from Lactococcus lactis is a homodimer (1JUB colored cyan). B) PyrD from L. lactis 1B is a heterotetramer. The homodimer interface at the center of PyrD 1B (1EP3 colored blue) is similar to the interface in PyrD 1A. PyrD 1B has an additional subunit PyrK (colored red). This implies that PyrD 1B is derived from PyrD 1A.
Figure 5
Maximum likelihood tree of PyrD 1B. Each of the major groups is separated by significant bootstrap values which indicates the distribution of PyrD 1B cannot be due to recent horizontal transfer.
Figure 6
Summary of data. Each circle corresponds to an argument presented above that excludes the root of the tree of life from a particular branch. The Archaea are placed with the Gram-positives, but drawn with a dashed line because we do not wish to argue which Gram-positive group was their ancestor at this time.
Similar articles
- Early evolutionary relationships among known life forms inferred from elongation factor EF-2/EF-G sequences: phylogenetic coherence and structure of the archaeal domain.
Cammarano P, Palm P, Creti R, Ceccarelli E, Sanangelantoni AM, Tiboni O. Cammarano P, et al. J Mol Evol. 1992 May;34(5):396-405. doi: 10.1007/BF00162996. J Mol Evol. 1992. PMID: 1602493 - Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications.
Brown JR, Doolittle WF. Brown JR, et al. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):2441-5. doi: 10.1073/pnas.92.7.2441. Proc Natl Acad Sci U S A. 1995. PMID: 7708661 Free PMC article. - The evidence that the tree of life is not rooted within the Archaea is unreliable: a reply to Skophammer et al.
Di Giulio M. Di Giulio M. Gene. 2007 Jun 1;394(1-2):105-6. doi: 10.1016/j.gene.2007.01.024. Epub 2007 Feb 12. Gene. 2007. PMID: 17395404 - The universal tree of life: an update.
Forterre P. Forterre P. Front Microbiol. 2015 Jul 21;6:717. doi: 10.3389/fmicb.2015.00717. eCollection 2015. Front Microbiol. 2015. PMID: 26257711 Free PMC article. Review. - The natural evolutionary relationships among prokaryotes.
Gupta RS. Gupta RS. Crit Rev Microbiol. 2000;26(2):111-31. doi: 10.1080/10408410091154219. Crit Rev Microbiol. 2000. PMID: 10890353 Review.
Cited by
- Emergence of the chemoautotrophic metabolism in hydrothermal environments and the origin of ancestral bacterial taxa.
Marakushev SA, Belonogova OV. Marakushev SA, et al. Dokl Biochem Biophys. 2011 Jul-Aug;439:161-6. doi: 10.1134/S1607672911040041. Epub 2011 Sep 18. Dokl Biochem Biophys. 2011. PMID: 21928135 No abstract available. - Evolution of mitochondria reconstructed from the energy metabolism of living bacteria.
Degli Esposti M, Chouaia B, Comandatore F, Crotti E, Sassera D, Lievens PM, Daffonchio D, Bandi C. Degli Esposti M, et al. PLoS One. 2014 May 7;9(5):e96566. doi: 10.1371/journal.pone.0096566. eCollection 2014. PLoS One. 2014. PMID: 24804722 Free PMC article. - The divergence and natural selection of autocatalytic primordial metabolic systems.
Marakushev SA, Belonogova OV. Marakushev SA, et al. Orig Life Evol Biosph. 2013 Jun;43(3):263-81. doi: 10.1007/s11084-013-9340-7. Epub 2013 Jul 17. Orig Life Evol Biosph. 2013. PMID: 23860777 - What are the origins and phylogeny of plant hemoglobins?
Vinogradov SN, Hoogewijs D, Arredondo-Peter R. Vinogradov SN, et al. Commun Integr Biol. 2011 Jul;4(4):443-5. doi: 10.4161/cib.4.4.15429. Epub 2011 Jul 1. Commun Integr Biol. 2011. PMID: 21966566 Free PMC article. - Molecular evolution of translin superfamily proteins within the genomes of eubacteria, archaea and eukaryotes.
Gupta GD, Kale A, Kumar V. Gupta GD, et al. J Mol Evol. 2012 Dec;75(5-6):155-67. doi: 10.1007/s00239-012-9534-z. Epub 2012 Nov 28. J Mol Evol. 2012. PMID: 23188094