Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei - PubMed (original) (raw)
Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei
Raquel A Oliveira et al. Nat Cell Biol. 2010 Feb.
Abstract
The metaphase-anaphase transition is orchestrated through proteolysis of numerous proteins by a ubiquitin protein ligase called the anaphase-promoting complex or cyclosome (APC/C). A crucial aspect of this process is sister chromatid separation, which is thought to be mediated by separase, a thiol protease activated by the APC/C. Separase cleaves cohesin, a ring-shaped complex that entraps sister DNAs. It is a matter of debate whether cohesin-independent forces also contribute to sister chromatid cohesion. Using 4D live-cell imaging of Drosophila melanogaster syncytial embryos blocked in metaphase (via APC/C inhibition), we show that artificial cohesin cleavage is sufficient to trigger chromosome disjunction. This is nevertheless insufficient for correct chromosome segregation. Kinetochore-microtubule attachments are rapidly destabilized by the loss of tension caused by cohesin cleavage in the presence of high Cdk1 (cyclin-dependent kinase 1) activity, as occurs when the APC/C cannot destroy mitotic cyclins. Metaphase chromosomes undergo a bona fide anaphase when cohesin cleavage is combined with Cdk1 inhibition. We conclude that only two key events, opening of cohesin rings and downregulation of Cdk1, are sufficient to drive proper segregation of chromosomes in anaphase.
Figures
Figure 1. Injection of Mad2 L13Q or UbcH10C114S prevents anaphase onset in Drosophila syncytial embryos
Embryos expressing His2A-mRFP1 were (a) not injected, (b) injected with 25 mg/ml of active Mad2 (Mad2L13Q) or (c) injected with 30 mg/ml of a dominant negative form of the human E2 ubiquitin-conjugating enzyme (UbcH10C114S); scale bars are 10 μm; bottom rows show higher magnifications (3x) of a single nuclear division. In all experiments performed (n≥25 embryos) both Mad2L13Q and UbcH10C114S induced a stable metaphase arrest.
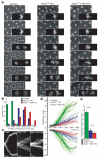
Figure 2. Cleavage of cohesin is sufficient to trigger sister chromatid disjunction
Embryos containing TEV-cleavable Rad21 (Rad21TEV) as their sole source of Rad21 were either unperturbed (control) or arrested in metaphase by injection of Mad2L13Q, followed by injection of either TEV protease alone (Mad2L13Q-TEV) or ICRF-193 and TEV protease (Mad2L13Q-ICRF-TEV). a) Images show selected frames of His2A-mRFP1 expressing embryos; times (min:sec) are relative to the last metaphase figure (control) or time of TEV injection; scale bars are 10 μm; insets show higher magnification (2.5x) of a single nuclear division; b) asynchrony of sister chromatid disjunction measured by the duration of anaphase onset, calculated as the time between separation of the first and last kinetochore pairs of a single metaphase (n=30 metaphases for each experimental condition); c) representative kymographs showing centromere (Cid-EGFP) separation; each kymograph shows all centromeres from a single metaphase; vertical scale bars are 2 μm and horizontal scale bars correspond to 30 sec; d) kymograph analysis of averaged centromere movement. Thin lines represent the average movement of all centromeres from individual experiments (n=30) and thick lines show the average of all experiments; e) average segregation speeds for each experimental condition; error bars show standard deviation; *** P<0.0001 and ** P<0.001 (two-tailed t-test).
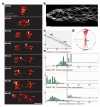
Figure 3. Loss of cohesin leads to unstable kinetochore-microtubule attachments
Rad21TEV embryos were injected with TEV protease to trigger sister centromere disjunction in Mad2L13Q-induced metaphase-arrested embryos. a) Selected stills from a movie showing His2Av-mRFP1 (red) and Cid-EFGP (green); arrows follow the trajectory of a single centromere after the initial pole-ward movement; times (min:sec) are relative to TEV injection; scale bar is 5 μm; b) representative kymograph showing centromere behaviour for 15 minutes after TEV-induced segregation. Vertical scale bars are 5 μm and horizontal scale bars correspond to 1 minute; c) example of a single centromere trajectory for 15 minutes after TEV-mediated centromere disjunction; pauses are in red and runs in blue (away from starting point) and green (towards starting point); grey line represents the segregation axis; d) directions of centromere movement (measured by the angle of trajectory) in Mad2-TEV experiments plotted on rose diagrams to show overall bias in run directions towards the axis of segregation (0°-180°); e) frequency (%) of maximum speed per run in control and Mad2-TEV embryos; top two panels show the maximum speeds per run observed during the initial segregation phase (until 2 minutes after the initial separation of sister centromeres); bottom panel shows later chromosome movements observed in Mad2-TEV experiments (from 2 minutes after separation onwards); insets show speeds >10 μm/min enlarged (5x) on the y axis; single centromere trajectories were analysed from 80 individual centromeres (out of 5 independent experiments); note that while during the segregation phase the chromosome movements in TEV-induced pseudo-anaphases are consistently slower than in controls, later chromosome movements are still overall slower but exhibit infrequent very fast movements (>12 μm/min).
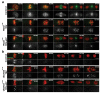
Figure 4. Localization of Aurora B and BubRI after TEV-induced sister chromatid disjunction
Early embryos surviving on Rad21TEV and expressing His2A-mRFP1 (red) were injected with mRNA encoding Aurora B-EGFP (a) or EGFP-BubRI (b) (shown in green); a) upper panel - Aurora B localization in a control embryo undergoing an unperturbed mitosis; middle panel - Aurora B localization in an metaphase arrested embryo (Mad2L13Q) injected with TEV protease; lower panel - localization of Aurora B in a Mad2L13Q-induced metaphase embryo that has been injected simultaneously with TEV protease and the Cdk inhibitor human p27; b) upper panel - BubRI localization in a control embryo undergoing an unperturbed mitosis; middle panel - BubRI localization in a metaphase-arrested embryos (UbcH10S114S) with TEV protease; lower panel - localization of BubRI in a UbcH10C114S-induced metaphase embryo that has been injected simultaneously with TEV protease and p27; in all figures, times (min:sec) are relative to the last metaphase (control) or to TEV injection; scale bars are 5 μm.
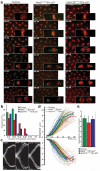
Figure 5. Cleavage of cohesin and Cdk inhibition trigger formation of daughter nuclei
Embryos surviving on Rad21TEV were either uninjected (control) or arrested in metaphase with Mad2L13Q or UbcH10C114S and subsequently co-injected with TEV protease and the CDK inhibitor human p27. a) Images show selected frames of embryos expressing His2A-mRFP1 (red) and Cid-EGFP (green); times (min:sec) are relative to the last metaphase figure (control) or to the time of TEV-p27 injection; scale bars are 10 μm; insets show higher magnification (2.5x) of a single nuclear division; b) asynchrony of sister chromatid disjunction measured by the duration of anaphase onset, calculated as the time between separation of the first and the last kinetochore pairs of a single metaphase (n=30 metaphases for each experimental condition); c) representative kymographs showing centromere (Cid-EGFP) separation; each kymograph shows all centromeres from a single metaphase; horizontal scale bars are 2 μm and vertical scale bars correspond to 30 sec; d) kymograph analysis of averaged centromere movement. Thin lines represent the average movement of all centromeres from individual experiments (n=30) and thick lines show the average of all experiments; e) average segregation speeds for each experimental condition; error bars show standard deviation.
Similar articles
- Splitting the nucleus: what's wrong with the tripartite ring model?
Nasmyth K, Oliveira RA. Nasmyth K, et al. Cold Spring Harb Symp Quant Biol. 2010;75:375-88. doi: 10.1101/sqb.2010.75.019. Epub 2011 Jan 5. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21209385 - Essential roles for cohesin in kinetochore and spindle function in Xenopus egg extracts.
Kenney RD, Heald R. Kenney RD, et al. J Cell Sci. 2006 Dec 15;119(Pt 24):5057-66. doi: 10.1242/jcs.03277. J Cell Sci. 2006. PMID: 17158911 - Cdk1 phosphorylation sites on Cdc27 are required for correct chromosomal localisation and APC/C function in syncytial Drosophila embryos.
Huang JY, Morley G, Li D, Whitaker M. Huang JY, et al. J Cell Sci. 2007 Jun 15;120(Pt 12):1990-7. doi: 10.1242/jcs.006833. Epub 2007 May 22. J Cell Sci. 2007. PMID: 17519285 Free PMC article. - Sharpening the anaphase switch.
Meadows JC, Millar JB. Meadows JC, et al. Biochem Soc Trans. 2015 Feb;43(1):19-22. doi: 10.1042/BST20140250. Biochem Soc Trans. 2015. PMID: 25619242 Review. - How do so few control so many?
Nasmyth K. Nasmyth K. Cell. 2005 Mar 25;120(6):739-46. doi: 10.1016/j.cell.2005.03.006. Cell. 2005. PMID: 15797376 Review.
Cited by
- Deprotection of centromeric cohesin at meiosis II requires APC/C activity but not kinetochore tension.
Mengoli V, Jonak K, Lyzak O, Lamb M, Lister LM, Lodge C, Rojas J, Zagoriy I, Herbert M, Zachariae W. Mengoli V, et al. EMBO J. 2021 Apr 1;40(7):e106812. doi: 10.15252/embj.2020106812. Epub 2021 Mar 1. EMBO J. 2021. PMID: 33644894 Free PMC article. - A single Drosophila embryo extract for the study of mitosis ex vivo.
Telley IA, Gáspár I, Ephrussi A, Surrey T. Telley IA, et al. Nat Protoc. 2013 Feb;8(2):310-24. doi: 10.1038/nprot.2013.003. Epub 2013 Jan 17. Nat Protoc. 2013. PMID: 23329004 - Condensin confers the longitudinal rigidity of chromosomes.
Houlard M, Godwin J, Metson J, Lee J, Hirano T, Nasmyth K. Houlard M, et al. Nat Cell Biol. 2015 Jun;17(6):771-81. doi: 10.1038/ncb3167. Epub 2015 May 11. Nat Cell Biol. 2015. PMID: 25961503 Free PMC article. - TORC2 is required to maintain genome stability during S phase in fission yeast.
Schonbrun M, Kolesnikov M, Kupiec M, Weisman R. Schonbrun M, et al. J Biol Chem. 2013 Jul 5;288(27):19649-60. doi: 10.1074/jbc.M113.464974. Epub 2013 May 23. J Biol Chem. 2013. PMID: 23703609 Free PMC article. - Sister chromatids separate during anaphase in a three-stage program as directed by interaxis bridges.
Chu L, Zhang Z, Mukhina M, Zickler D, Kleckner N. Chu L, et al. Proc Natl Acad Sci U S A. 2022 Mar 8;119(10):e2123363119. doi: 10.1073/pnas.2123363119. Epub 2022 Mar 2. Proc Natl Acad Sci U S A. 2022. PMID: 35235450 Free PMC article.
References
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. - PubMed
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. - PubMed
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. - PubMed
- Yanagida M. Clearing the way for mitosis: is cohesin a target? Nat Rev Mol Cell Biol. 2009;10:489–496. - PubMed
- Diaz-Martinez LA, Gimenez-Abian JF, Clarke DJ. Chromosome cohesion - rings, knots, orcs and fellowship. J Cell Sci. 2008;121:2107–2114. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous