TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration - PubMed (original) (raw)
Review
. 2010 Apr 15;19(R1):R46-64.
doi: 10.1093/hmg/ddq137. Epub 2010 Apr 15.
Affiliations
- PMID: 20400460
- PMCID: PMC3167692
- DOI: 10.1093/hmg/ddq137
Review
TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration
Clotilde Lagier-Tourenne et al. Hum Mol Genet. 2010.
Abstract
Amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) are neurodegenerative diseases with clinical and pathological overlap. Landmark discoveries of mutations in the transactive response DNA-binding protein (TDP-43) and fused in sarcoma/translocated in liposarcoma (FUS/TLS) as causative of ALS and FTLD, combined with the abnormal aggregation of these proteins, have initiated a shifting paradigm for the underlying pathogenesis of multiple neurodegenerative diseases. TDP-43 and FUS/TLS are both RNA/DNA-binding proteins with striking structural and functional similarities. Their association with ALS and other neurodegenerative diseases is redirecting research efforts toward understanding the role of RNA processing regulation in neurodegeneration.
Figures
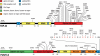
Figure 1.
TDP-43 and FUS/TLS mutations in ALS and FTLD patients. Thirty-eight dominant mutations have been identified in TDP-43 in sporadic and familial ALS patients and in rare FTLD patients, with most lying in the C-terminal glycine-rich region. All are missense mutations, except for the truncating mutation TDP-43Y374X (upper panel). Thirty mutations have been identified in FUS/TLS in familial and sporadic ALS cases and in rare FTLD patients (R514S and G515S were found in cis). Most mutations are clustered in the last 17 amino acids and in the glycine-rich region (lower panel). NLS, nuclear localization signal; NES, nuclear export signal. Domains have been defined according to
and
http://www.cbs.dtu.dk/services/NetNES
.
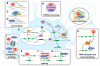
Figure 2.
Proposed physiological roles of TDP-43 and FUS/TLS. (A) Summary of major steps in RNA processing from transcription to translation or degradation. (B) TDP-43 binds single-stranded TG-rich elements in promoter regions thereby blocking transcription of the downstream gene [shown for TAR DNA of HIV (38) and mouse SP-10 gene (132,150)]. (C) FUS/TLS associates with TBP within the TFIID complex suggesting that it participates in the general transcriptional machinery (–159). (D) In response to DNA damage, FUS/TLS is recruited in the promoter region of cyclin D1 (CCND1) by sense and antisense non-coding RNAs (ncRNAs) and represses CCND1 transcription (145). (E) TDP-43 binds a UG track in intronic regions preceding alternatively spliced exons and enhances their exclusion [shown for CFTR (38,40,96,130,131,160) and apolipoprotein A-II (161)]. (F) FUS/TLS was identified as a part of the spliceosome (–170) and (G) was shown to promote exon inclusion in H-ras mRNA, through indirect binding to structural regulatory elements located on the downstream intron (177). (H) Both proteins were found in a complex with Drosha, suggesting that they may be involved in miRNA processing (183). (I) Both TDP-43 and FUS/TLS shuttle between the nucleus and the cytosol (134,142) and (J) are incorporated in SGs where they form complexes with mRNAs and other RNA binding proteins (,,,–204). (K) TDP-43 and FUS/TLS are both involved in the transport of mRNAs to dendritic spines and/or the axonal terminal where they may facilitate local translation (–189,191). Examples of such cargo transcripts are the low molecular weight NFL for TDP-43 (51,208) and the actin-stabilizing protein Nd1-L for FUS/TLS (147).
Similar articles
- Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins.
Ratti A, Buratti E. Ratti A, et al. J Neurochem. 2016 Aug;138 Suppl 1:95-111. doi: 10.1111/jnc.13625. Epub 2016 Jun 15. J Neurochem. 2016. PMID: 27015757 Review. - Molecular basis of amyotrophic lateral sclerosis.
Liscic RM, Breljak D. Liscic RM, et al. Prog Neuropsychopharmacol Biol Psychiatry. 2011 Mar 30;35(2):370-2. doi: 10.1016/j.pnpbp.2010.07.017. Epub 2010 Jul 23. Prog Neuropsychopharmacol Biol Psychiatry. 2011. PMID: 20655970 Review. - FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations.
Neumann M, Bentmann E, Dormann D, Jawaid A, DeJesus-Hernandez M, Ansorge O, Roeber S, Kretzschmar HA, Munoz DG, Kusaka H, Yokota O, Ang LC, Bilbao J, Rademakers R, Haass C, Mackenzie IR. Neumann M, et al. Brain. 2011 Sep;134(Pt 9):2595-609. doi: 10.1093/brain/awr201. Epub 2011 Aug 19. Brain. 2011. PMID: 21856723 Free PMC article. - TDP-43 and FUS/TLS: cellular functions and implications for neurodegeneration.
Fiesel FC, Kahle PJ. Fiesel FC, et al. FEBS J. 2011 Oct;278(19):3550-68. doi: 10.1111/j.1742-4658.2011.08258.x. Epub 2011 Aug 24. FEBS J. 2011. PMID: 21777389 Review. - How do the RNA-binding proteins TDP-43 and FUS relate to amyotrophic lateral sclerosis and frontotemporal degeneration, and to each other?
Baloh RH. Baloh RH. Curr Opin Neurol. 2012 Dec;25(6):701-7. doi: 10.1097/WCO.0b013e32835a269b. Curr Opin Neurol. 2012. PMID: 23041957 Review.
Cited by
- Caenorhabditis elegans RNA-processing protein TDP-1 regulates protein homeostasis and life span.
Zhang T, Hwang HY, Hao H, Talbot C Jr, Wang J. Zhang T, et al. J Biol Chem. 2012 Mar 9;287(11):8371-82. doi: 10.1074/jbc.M111.311977. Epub 2012 Jan 9. J Biol Chem. 2012. PMID: 22232551 Free PMC article. - Knockdown of the Drosophila fused in sarcoma (FUS) homologue causes deficient locomotive behavior and shortening of motoneuron terminal branches.
Sasayama H, Shimamura M, Tokuda T, Azuma Y, Yoshida T, Mizuno T, Nakagawa M, Fujikake N, Nagai Y, Yamaguchi M. Sasayama H, et al. PLoS One. 2012;7(6):e39483. doi: 10.1371/journal.pone.0039483. Epub 2012 Jun 19. PLoS One. 2012. PMID: 22724023 Free PMC article. - Exome sequencing identifies FUS mutations as a cause of essential tremor.
Merner ND, Girard SL, Catoire H, Bourassa CV, Belzil VV, Rivière JB, Hince P, Levert A, Dionne-Laporte A, Spiegelman D, Noreau A, Diab S, Szuto A, Fournier H, Raelson J, Belouchi M, Panisset M, Cossette P, Dupré N, Bernard G, Chouinard S, Dion PA, Rouleau GA. Merner ND, et al. Am J Hum Genet. 2012 Aug 10;91(2):313-9. doi: 10.1016/j.ajhg.2012.07.002. Epub 2012 Aug 2. Am J Hum Genet. 2012. PMID: 22863194 Free PMC article. - RNA processing in neurological tissue: development, aging and disease.
Szeto RA, Tran T, Truong J, Negraes PD, Trujillo CA. Szeto RA, et al. Semin Cell Dev Biol. 2021 Jun;114:57-67. doi: 10.1016/j.semcdb.2020.09.004. Epub 2020 Oct 16. Semin Cell Dev Biol. 2021. PMID: 33077405 Free PMC article. Review. - Characterization of β-domains in C-terminal fragments of TDP-43 by scanning tunneling microscopy.
Xu M, Zhu L, Liu J, Yang Y, Wu JY, Wang C. Xu M, et al. J Struct Biol. 2013 Jan;181(1):11-6. doi: 10.1016/j.jsb.2012.10.011. Epub 2012 Nov 5. J Struct Biol. 2013. PMID: 23138004 Free PMC article.
References
- Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J.P., Deng H.X., et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi:10.1038/362059a0. - DOI - PubMed
- Boillee S., Vande Velde C., Cleveland D.W. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi:10.1016/j.neuron.2006.09.018. - DOI - PubMed
- Pasinelli P., Brown R.H. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat. Rev. Neurosci. 2006;7:710–723. doi:10.1038/nrn1971. - DOI - PubMed
- Valdmanis P.N., Rouleau G.A. Genetics of familial amyotrophic lateral sclerosis. Neurology. 2008;70:144–152. doi:10.1212/01.wnl.0000296811.19811.db. - DOI - PubMed
- Ilieva H., Polymenidou M., Cleveland D.W. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J. Cell. Biol. 2009;187:761–772. doi:10.1083/jcb.200908164. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous