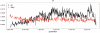PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals - PubMed (original) (raw)
PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals
Robert Kofler et al. PLoS One. 2011.
Abstract
Recent statistical analyses suggest that sequencing of pooled samples provides a cost effective approach to determine genome-wide population genetic parameters. Here we introduce PoPoolation, a toolbox specifically designed for the population genetic analysis of sequence data from pooled individuals. PoPoolation calculates estimates of θ(Watterson), θ(π), and Tajima's D that account for the bias introduced by pooling and sequencing errors, as well as divergence between species. Results of genome-wide analyses can be graphically displayed in a sliding window plot. PoPoolation is written in Perl and R and it builds on commonly used data formats. Its source code can be downloaded from http://code.google.com/p/popoolation/. Furthermore, we evaluate the influence of mapping algorithms, sequencing errors, and read coverage on the accuracy of population genetic parameter estimates from pooled data.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Outline of a population genetic analysis from pooled sequence data.
Sequencer figure from
Figure 2. Graphical output of polymorphism and divergence estimates using PoPoolation.
Sliding window analysis of θ π of a Portuguese D. melanogaster population on chromosome 3R (black line). The red line shows divergence (dxy) between D. melanogaster and D. simulans using the same window size and step size as for θ π. Note that dxy is scaled by 1/10. Both lines are based on non-overlapping windows of 50 kb.
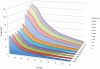
Figure 3. Sequencing errors in relation to coverage, minor allele count, and sequence quality.
PhiX sequences (74 bp) generated with an Illumina GAIIx sequencer were analyzed for sequencing error rate (number of mutated bases after quality filtering). The gray bar indicates the presence of a polymorphic site in the PhiX sequence, which results in a minimum sequencing error rate.
Figure 4. Improvement of the alignment for diverged regions using the PE-SW remap algorithm.
IGV screenshot of the mapping of pooled sequence reads in a highly divergent region of D. melanogaster. The upper panel shows an alignment of the PE reads without the PE-SW remap and the lower panel shows the same region with the PE-SW remap.
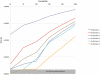
Figure 5. The influence of coverage and window size on the accuracy of the estimated θ π.
The accuracy was measured as the mean standardized difference between θ π estimated for a given window size and its expectation.
Similar articles
- PoPoolation DB: a user-friendly web-based database for the retrieval of natural polymorphisms in Drosophila.
Pandey RV, Kofler R, Orozco-terWengel P, Nolte V, Schlötterer C. Pandey RV, et al. BMC Genet. 2011 Mar 2;12:27. doi: 10.1186/1471-2156-12-27. BMC Genet. 2011. PMID: 21366916 Free PMC article. - SNP calling by sequencing pooled samples.
Raineri E, Ferretti L, Esteve-Codina A, Nevado B, Heath S, Pérez-Enciso M. Raineri E, et al. BMC Bioinformatics. 2012 Sep 20;13:239. doi: 10.1186/1471-2105-13-239. BMC Bioinformatics. 2012. PMID: 22992255 Free PMC article. - The next generation of molecular markers from massively parallel sequencing of pooled DNA samples.
Futschik A, Schlötterer C. Futschik A, et al. Genetics. 2010 Sep;186(1):207-18. doi: 10.1534/genetics.110.114397. Epub 2010 May 10. Genetics. 2010. PMID: 20457880 Free PMC article. - Model-based quality assessment and base-calling for second-generation sequencing data.
Bravo HC, Irizarry RA. Bravo HC, et al. Biometrics. 2010 Sep;66(3):665-74. doi: 10.1111/j.1541-0420.2009.01353.x. Biometrics. 2010. PMID: 19912177 Free PMC article. Review. - Statistical challenges associated with detecting copy number variations with next-generation sequencing.
Teo SM, Pawitan Y, Ku CS, Chia KS, Salim A. Teo SM, et al. Bioinformatics. 2012 Nov 1;28(21):2711-8. doi: 10.1093/bioinformatics/bts535. Epub 2012 Aug 31. Bioinformatics. 2012. PMID: 22942022 Review.
Cited by
- Genomics of sex allocation in the parasitoid wasp Nasonia vitripennis.
Pannebakker BA, Cook N, van den Heuvel J, van de Zande L, Shuker DM. Pannebakker BA, et al. BMC Genomics. 2020 Jul 20;21(1):499. doi: 10.1186/s12864-020-06904-4. BMC Genomics. 2020. PMID: 32689940 Free PMC article. - LDx: estimation of linkage disequilibrium from high-throughput pooled resequencing data.
Feder AF, Petrov DA, Bergland AO. Feder AF, et al. PLoS One. 2012;7(11):e48588. doi: 10.1371/journal.pone.0048588. Epub 2012 Nov 9. PLoS One. 2012. PMID: 23152785 Free PMC article. - Dissecting the invasion history of Spotted-Wing Drosophila (Drosophila suzukii) in Portugal using genomic data.
Sario S, Marques JP, Farelo L, Afonso S, Santos C, Melo-Ferreira J. Sario S, et al. BMC Genomics. 2024 Aug 29;25(1):813. doi: 10.1186/s12864-024-10739-8. BMC Genomics. 2024. PMID: 39210249 Free PMC article. - The distribution of fitness effects among synonymous mutations in a gene under directional selection.
Lebeuf-Taylor E, McCloskey N, Bailey SF, Hinz A, Kassen R. Lebeuf-Taylor E, et al. Elife. 2019 Jul 19;8:e45952. doi: 10.7554/eLife.45952. Elife. 2019. PMID: 31322500 Free PMC article. - Contribution of epigenetic variation to adaptation in Arabidopsis.
Schmid MW, Heichinger C, Coman Schmid D, Guthörl D, Gagliardini V, Bruggmann R, Aluri S, Aquino C, Schmid B, Turnbull LA, Grossniklaus U. Schmid MW, et al. Nat Commun. 2018 Oct 25;9(1):4446. doi: 10.1038/s41467-018-06932-5. Nat Commun. 2018. PMID: 30361538 Free PMC article.
References
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat Genet. 2010. - PubMed
- Rubin CJ, Zody MC, Eriksson J, Meadows JR, Sherwood E, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–591. - PubMed
- Quinlan AR, Stewart DA, Stromberg MP, Marth GT. Pyrobayes: an improved base caller for SNP discovery in pyrosequences. Nat Methods. 2008;5:179–181. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous