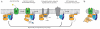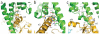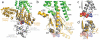Crystal structure of the β2 adrenergic receptor-Gs protein complex - PubMed (original) (raw)
. 2011 Jul 19;477(7366):549-55.
doi: 10.1038/nature10361.
Brian T DeVree, Yaozhong Zou, Andrew C Kruse, Ka Young Chung, Tong Sun Kobilka, Foon Sun Thian, Pil Seok Chae, Els Pardon, Diane Calinski, Jesper M Mathiesen, Syed T A Shah, Joseph A Lyons, Martin Caffrey, Samuel H Gellman, Jan Steyaert, Georgios Skiniotis, William I Weis, Roger K Sunahara, Brian K Kobilka
Affiliations
- PMID: 21772288
- PMCID: PMC3184188
- DOI: 10.1038/nature10361
Crystal structure of the β2 adrenergic receptor-Gs protein complex
Søren G F Rasmussen et al. Nature. 2011.
Abstract
G protein-coupled receptors (GPCRs) are responsible for the majority of cellular responses to hormones and neurotransmitters as well as the senses of sight, olfaction and taste. The paradigm of GPCR signalling is the activation of a heterotrimeric GTP binding protein (G protein) by an agonist-occupied receptor. The β(2) adrenergic receptor (β(2)AR) activation of Gs, the stimulatory G protein for adenylyl cyclase, has long been a model system for GPCR signalling. Here we present the crystal structure of the active state ternary complex composed of agonist-occupied monomeric β(2)AR and nucleotide-free Gs heterotrimer. The principal interactions between the β(2)AR and Gs involve the amino- and carboxy-terminal α-helices of Gs, with conformational changes propagating to the nucleotide-binding pocket. The largest conformational changes in the β(2)AR include a 14 Å outward movement at the cytoplasmic end of transmembrane segment 6 (TM6) and an α-helical extension of the cytoplasmic end of TM5. The most surprising observation is a major displacement of the α-helical domain of Gαs relative to the Ras-like GTPase domain. This crystal structure represents the first high-resolution view of transmembrane signalling by a GPCR.
© 2011 Macmillan Publishers Limited. All rights reserved
Figures
Figure 1. G protein cycle for the β2AR-Gs complex
a, Extracellular agonist binding to the β2AR leads to conformational rearrangements of the cytoplasmic ends of transmembrane segments that enable the Gs heterotrimer (α, β, and γ) to bind the receptor. GDP is released from the α subunit upon formation of β2AR-Gs complex. The GTP binds to the nucleotide-free α subunit resulting in dissociation of the α and βγ subunits from the receptor. The subunits regulate their respective effector proteins adenylyl cyclase (AC) and Ca2+ channels. The Gs heterotrimer reassembles from α and βγ subunits following hydrolysis of GTP to GDP in the α subunit. b, The purified nucleotide-free β2AR-Gs protein complex maintained in detergent micelles. The Gαs subunit consists of two domains, the Ras domain (αRas) and the α-helical domain (αAH). Both are involved in nucleotide binding. In the nucleotide-free state, the αAH domain has a variable position relative the αRas domain.
Figure 2. Overall structure of the β2AR Gs complex
a, Lattice packing of the complex shows alternating layers of receptor and G protein within the crystal. Abundant contacts are formed among proteins within the aqueous layers. b, The overall structure of the asymmetric unit contents shows the β2AR (green) bound to an agonist (yellow spheres) and engaged in extensive interactions with Gαs (orange). Gαs together with Gβ (cyan) and Gγ (purple) constitute the heterotrimeric G protein Gs. A Gs binding nanobody (red) binds the G protein between the α and β subunits. The nanobody (Nb35) facilitates crystallization, as does T4 lysozyme (magenta) fused to the amino terminus of the β2AR. c, The biological complex omitting crystallization aids, showing its location and orientation within a cell membrane.
Figure 3. Comparison of active and inactive β2AR structures
a, Side and cytoplasmic views of the β2AR-Gs structure (green) compared to the inactive carazolol-bound β2 AR structure (blue). Significant structural changes are seen for the intracellular domains of TM5 and TM6. TM5 is extended by two helical turns while TM6 is moved outward by 14 Å as measured at the α-carbons of Glu268 (yellow arrow) in the two structures. b, β2AR-Gs compared with the nanobody-stabilized active state β2AR-Nb80 structure (orange). c, The positions of residues in the E/DRY and NPxxY motifs and other key residues of the β2AR-Gs and β2AR-Nb80 structures. All residues occupy very similar positions except Arg131 which in the β2AR-Nb80 structure interacts with the nanobody. d, View from the cytoplasmic side of residues shown in (c).
Figure 4. Receptor-G protein interactions
a, b The α5-helix of Gαs docks into a cavity formed on the intracellular side of the receptor by the opening of transmembrane helices 5 and 6. a. Within the transmembrane core, the interactions are primarily non-polar. An exception involves packing of Tyr391 of the α5-helix against Arg131 of the conserved DRY sequence in TM3 (see also Figure S3). Arg131 also packs against Tyr of the conserved NPxxY sequence in TM7. b. As α5-helix exits the receptor it forms a network of polar interactions with TM5 and TM3. c, Receptor residues Thr68 and Asp130 interact with the ICL2 helix of the β2AR via Tyr141, positioning the helix so that Phe139 of the receptor docks into a hydrophobic pocket on the G protein surface, thereby structurally linking receptor-G protein interactions with the highly conserved DRY motif of the β2AR.
Figure 5. Conformational changes in Gαs
a, A comparison of Gαs in the β2AR-Gs complex (orange) with the GTPγS-bound Gαs (grey) (PDB ID: 1AZT). GTPγS is shown as spheres. The helical domain of Gαs (GαsAH) exhibits a dramatic displacement relative to its position in the GTPγS-bound state. b, The α5-helix of Gαs is rotated and displaced toward the β2AR, perturbing the β6-α5 loop which otherwise forms part of the GTPγS binding pocket. c, The β1-α1 loop (P-loop) and β6-α5 loop of Gαs interact with the phosphates and purine ring, respectively, of GTPγS in the GTPγS-Gαs structure. d, The β1-α1 and β6-α5 loops are rearranged in the nucleotide-free β2AR-Gs structure.
Figure 6. Possible sequence of β2AR-Gs complex formation
a, b, Comparison of the β2AR-Gs structure (green and gold) with metarhodopsin II (PDB ID: 3PQR) (purple) bound with the carboxyl-terminal peptide of transducin (blue). TM7 has been omitted in panel a to better visualize the G proteins. c, Cartoon of the β2AR-Gαs peptide fusion construct used in the binding experiments (d). d, Competition binding experiments between [3H]-DHA and full agonist isoproterenol. Top panel shows binding data (reproduced from Rasmussen et al., 2011) on β2AR reconstituted in HDL particles with and without Gs heterotrimer. The fraction of β2AR in the Ki high state for the β2AR with Gs is 0.55. Bottom panel shows binding to β2AR and a β2AR-Gαs peptide fusion expressed in _Sf_9 cell membranes. The fraction of β2AR in the Ki high state for the β2AR-Gαs peptide fusion is 0.68. e, The initial interaction of agonist bound β2AR and GαsRas may involve an orientation of the carboxyl-terminus of GαsRas similar to that of the carboxyl-terminal peptide of transducin in the structure of metarhodopsin II. f, The final position of GαsRas on the β2AR as observed in the β2AR-Gs complex.
Comment in
- Structural biology: snapshot of a signalling complex.
Schwartz TW, Sakmar TP. Schwartz TW, et al. Nature. 2011 Sep 28;477(7366):540-1. doi: 10.1038/477540a. Nature. 2011. PMID: 21956322 No abstract available.
Similar articles
- Conformational changes in the G protein Gs induced by the β2 adrenergic receptor.
Chung KY, Rasmussen SG, Liu T, Li S, DeVree BT, Chae PS, Calinski D, Kobilka BK, Woods VL Jr, Sunahara RK. Chung KY, et al. Nature. 2011 Sep 28;477(7366):611-5. doi: 10.1038/nature10488. Nature. 2011. PMID: 21956331 Free PMC article. - Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex.
Westfield GH, Rasmussen SG, Su M, Dutta S, DeVree BT, Chung KY, Calinski D, Velez-Ruiz G, Oleskie AN, Pardon E, Chae PS, Liu T, Li S, Woods VL Jr, Steyaert J, Kobilka BK, Sunahara RK, Skiniotis G. Westfield GH, et al. Proc Natl Acad Sci U S A. 2011 Sep 20;108(38):16086-91. doi: 10.1073/pnas.1113645108. Epub 2011 Sep 13. Proc Natl Acad Sci U S A. 2011. PMID: 21914848 Free PMC article. - Single-molecule analysis of ligand efficacy in β2AR-G-protein activation.
Gregorio GG, Masureel M, Hilger D, Terry DS, Juette M, Zhao H, Zhou Z, Perez-Aguilar JM, Hauge M, Mathiasen S, Javitch JA, Weinstein H, Kobilka BK, Blanchard SC. Gregorio GG, et al. Nature. 2017 Jul 6;547(7661):68-73. doi: 10.1038/nature22354. Epub 2017 Jun 7. Nature. 2017. PMID: 28607487 Free PMC article. - Structural features of β2 adrenergic receptor: crystal structures and beyond.
Bang I, Choi HJ. Bang I, et al. Mol Cells. 2015;38(2):105-11. doi: 10.14348/molcells.2015.2301. Epub 2014 Dec 24. Mol Cells. 2015. PMID: 25537861 Free PMC article. Review. - Structural insights into agonist-induced activation of G-protein-coupled receptors.
Deupi X, Standfuss J. Deupi X, et al. Curr Opin Struct Biol. 2011 Aug;21(4):541-51. doi: 10.1016/j.sbi.2011.06.002. Epub 2011 Jun 30. Curr Opin Struct Biol. 2011. PMID: 21723721 Review.
Cited by
- The role of ligands on the equilibria between functional states of a G protein-coupled receptor.
Kim TH, Chung KY, Manglik A, Hansen AL, Dror RO, Mildorf TJ, Shaw DE, Kobilka BK, Prosser RS. Kim TH, et al. J Am Chem Soc. 2013 Jun 26;135(25):9465-74. doi: 10.1021/ja404305k. Epub 2013 Jun 14. J Am Chem Soc. 2013. PMID: 23721409 Free PMC article. - Combined docking and machine learning identify key molecular determinants of ligand pharmacological activity on β2 adrenoceptor.
Jiménez-Rosés M, Morgan BA, Jimenez Sigstad M, Tran TDZ, Srivastava R, Bunsuz A, Borrega-Román L, Hompluem P, Cullum SA, Harwood CR, Koers EJ, Sykes DA, Styles IB, Veprintsev DB. Jiménez-Rosés M, et al. Pharmacol Res Perspect. 2022 Oct;10(5):e00994. doi: 10.1002/prp2.994. Pharmacol Res Perspect. 2022. PMID: 36029004 Free PMC article. - Complementary biosensors reveal different G-protein signaling modes triggered by GPCRs and non-receptor activators.
Garcia-Marcos M. Garcia-Marcos M. Elife. 2021 Mar 31;10:e65620. doi: 10.7554/eLife.65620. Elife. 2021. PMID: 33787494 Free PMC article. - Structural basis for chemokine receptor CCR6 activation by the endogenous protein ligand CCL20.
Wasilko DJ, Johnson ZL, Ammirati M, Che Y, Griffor MC, Han S, Wu H. Wasilko DJ, et al. Nat Commun. 2020 Jun 15;11(1):3031. doi: 10.1038/s41467-020-16820-6. Nat Commun. 2020. PMID: 32541785 Free PMC article. - Advances in receptor conformation research: the quest for functionally selective conformations focusing on the β2-adrenoceptor.
Woo AY, Song Y, Zhu W, Xiao RP. Woo AY, et al. Br J Pharmacol. 2015 Dec;172(23):5477-88. doi: 10.1111/bph.13049. Epub 2015 Feb 27. Br J Pharmacol. 2015. PMID: 25537131 Free PMC article. Review.
References
- Dixon RA, et al. Cloning of the gene and cDNA for mammalian beta-adrenergic receptor and homology with rhodopsin. Nature. 1986;321:75–79. doi:10.1038/321075a0. - PubMed
- Rasmussen SG, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi:nature06325 [pii] 10.1038/nature06325. - PubMed
- Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi:1150609 [pii] 10.1126/science.1150609. - PubMed
- Lefkowitz RJ. Seven transmembrane receptors: something old, something new. Acta Physiol (Oxf) 2007;190:9–19. doi:APS1693 [pii] 10.1111/j.1365-201X.2007.01693.x. - PubMed
- Brandt DR, Asano T, Pedersen SE, Ross EM. Reconstitution of catecholamine-stimulated guanosinetriphosphatase activity. Biochemistry. 1983;22:4357–4362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM075915/GM/NIGMS NIH HHS/United States
- P50GM073210/GM/NIGMS NIH HHS/United States
- R01 GM068603-03/GM/NIGMS NIH HHS/United States
- P60DK-20572/DK/NIDDK NIH HHS/United States
- T32-GM008270/GM/NIGMS NIH HHS/United States
- R01 GM068603-04/GM/NIGMS NIH HHS/United States
- R01 GM068603-05/GM/NIGMS NIH HHS/United States
- GM75915/GM/NIGMS NIH HHS/United States
- U54GM094599/GM/NIGMS NIH HHS/United States
- P50 GM073210/GM/NIGMS NIH HHS/United States
- NS028471/NS/NINDS NIH HHS/United States
- GM083118/GM/NIGMS NIH HHS/United States
- U54 GM094599/GM/NIGMS NIH HHS/United States
- R01 GM083118/GM/NIGMS NIH HHS/United States
- P01 GM075913/GM/NIGMS NIH HHS/United States
- P60 DK020572/DK/NIDDK NIH HHS/United States
- R01 GM068603-01/GM/NIGMS NIH HHS/United States
- R01 NS028471/NS/NINDS NIH HHS/United States
- R01 GM068603-02/GM/NIGMS NIH HHS/United States
- P01 GM75913/GM/NIGMS NIH HHS/United States
- R01 GM056169/GM/NIGMS NIH HHS/United States
- R37 NS028471/NS/NINDS NIH HHS/United States
- GM56169/GM/NIGMS NIH HHS/United States
- R01 GM068603/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials