Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells - PubMed (original) (raw)
. 2011 Oct 12;478(7369):391-4.
doi: 10.1038/nature10424.
S Tamir Rashid, Helene Strick-Marchand, Ignacio Varela, Pei-Qi Liu, David E Paschon, Elena Miranda, Adriana Ordóñez, Nicholas R F Hannan, Foad J Rouhani, Sylvie Darche, Graeme Alexander, Stefan J Marciniak, Noemi Fusaki, Mamoru Hasegawa, Michael C Holmes, James P Di Santo, David A Lomas, Allan Bradley, Ludovic Vallier
Affiliations
- PMID: 21993621
- PMCID: PMC3198846
- DOI: 10.1038/nature10424
Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells
Kosuke Yusa et al. Nature. 2011.
Abstract
Human induced pluripotent stem cells (iPSCs) represent a unique opportunity for regenerative medicine because they offer the prospect of generating unlimited quantities of cells for autologous transplantation, with potential application in treatments for a broad range of disorders. However, the use of human iPSCs in the context of genetically inherited human disease will require the correction of disease-causing mutations in a manner that is fully compatible with clinical applications. The methods currently available, such as homologous recombination, lack the necessary efficiency and also leave residual sequences in the targeted genome. Therefore, the development of new approaches to edit the mammalian genome is a prerequisite to delivering the clinical promise of human iPSCs. Here we show that a combination of zinc finger nucleases (ZFNs) and piggyBac technology in human iPSCs can achieve biallelic correction of a point mutation (Glu342Lys) in the α(1)-antitrypsin (A1AT, also known as SERPINA1) gene that is responsible for α(1)-antitrypsin deficiency. Genetic correction of human iPSCs restored the structure and function of A1AT in subsequently derived liver cells in vitro and in vivo. This approach is significantly more efficient than any other gene-targeting technology that is currently available and crucially prevents contamination of the host genome with residual non-human sequences. Our results provide the first proof of principle, to our knowledge, for the potential of combining human iPSCs with genetic correction to generate clinically relevant cells for autologous cell-based therapies.
Figures
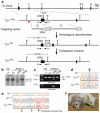
Figure 1. Correction of the G290T mutation in the Tyr gene in mIPSCs
a, The strategy for precise genome modification using the piggyBac transposon. Top line, structure of the Tyr gene; red line, 5′ external probe for Southern blot analysis; open arrow, piggyBac transposon carrying a PGK-puro_Δ_tk cassette; P1, P2 and P3, PCR primers; B, _Bam_HI; E, _Eco_NI. b, c, Southern blot (b) and PCR analyses (c) showing insertion (c/PB) and excision (c/Rev) of the piggyBac transposon. ES, mouse ESCs as a control. d, e, Sequence analyses revealed correction of the G290T mutation (d) and seamless excision of the piggyBac transposon (e). Note that two silent mutations (A and T indicated by arrowheads) introduced near the TTAA site were also detected. f, A chimeric mouse generated by injecting corrected Tyr c/Rev mIPSCs (left) displays black coat color. Right, a non-injected albino mouse.
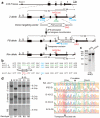
Figure 2. Correction of the Z mutation in A1ATD-hIPSCs
a, The strategy for precise genome modification using ZFNs and the piggyBac transposon. Top line, structure of the A1AT gene; blue lines, Southern blot probes; thin and thick boxes, non-coding and coding exons, respectively; open arrow, piggyBac transposon; B, _Bam_HI; A, _Afl_III. b, Sequences of wild-type (Reference), Z, PB, and Rev alleles. Amino acid position 342 (blue), recognition sites for ZFNs (green), piggyBac excision site (red) are shown. Sequence changes in Rev allele from Z allele were indicated by asterisks. c, Surveyor nuclease assay showing the cleavage of Z mutation in ZFNs-transfected K562 cells. Non-transfected cells were used as a control. d, Southern blot analysis showing bi-allelic piggyBac insertion (B-16) and bi-allelic excision (B-16-C2, -C3 and -C6) during correction of the A1ATD-hIPSCs line B. Genomic DNA was digested by _Bam_HI (5′ and PB probes) or _Alf_III (3′ probe). Genotype: ZZ, homozygous for Z allele; PP, homozygous for insertion of piggyBac; RR, homozygous for reverted allele. e, Sequence analysis showing correction of Z mutation in 3 corrected hIPSC lines. Wild-type sequence (top line) and A1ATD-hIPSC (second line).
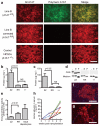
Figure 3. Functional analysis of restored A1AT in c-hIPSCs-derived hepatocyte-like cells
a, Immunofluorescence showing the absence of polymeric A1AT protein in hepatocyte-like cells generated from c-hIPSCs. All forms of A1AT (left panels) and misfolded polymeric A1AT (middle panels). b, c, ELISA to assess the intracellular (b) and secreted (c) levels of polymeric A1AT protein in hepatocyte-like cells derived from A1ATD-hIPSCs (ZZ), c-hIPSCs (RR) and control hIPSCs (++). d, Endoglycosidase H (E) and peptide:_N_-glycosidase (P) digestion of A1AT immunoprecipitated from uncorrected (ZZ), corrected (RR) and control (++) hIPSC-derived hepatocyte-like cells (upper panels) and corresponding culture medium (lower panels). e, Chymotrypsin ELISA showing that corrected cells (RR) have A1AT enzymatic inhibitory activity that is superior to uncorrected cells (ZZ) and close to adult hepatocytes. f, g, Immunofluorescence of transplanted liver sections detecting human albumin (f) and A1AT (g). DNA was counterstained with DAPI. h, ELISA read-out of human albumin in the mouse serum longitudinally followed for each mouse. Asterisk, the mouse was subjected to histology analysis. Scale bars, 100 μm. Data in b, c and e are shown as mean ± s.d. (_n_=3). Student’s _t_-test was performed. NS, not significant.
Comment in
- A ZFN/piggyBac step closer to autologous liver cell therapy.
Mattis AN, Willenbring H. Mattis AN, et al. Hepatology. 2012 Jun;55(6):2033-5. doi: 10.1002/hep.25715. Hepatology. 2012. PMID: 22422378 No abstract available.
Similar articles
- Sustained knockdown of a disease-causing gene in patient-specific induced pluripotent stem cells using lentiviral vector-based gene therapy.
Eggenschwiler R, Loya K, Wu G, Sharma AD, Sgodda M, Zychlinski D, Herr C, Steinemann D, Teckman J, Bals R, Ott M, Schambach A, Schöler HR, Cantz T. Eggenschwiler R, et al. Stem Cells Transl Med. 2013 Sep;2(9):641-54. doi: 10.5966/sctm.2013-0017. Epub 2013 Aug 7. Stem Cells Transl Med. 2013. PMID: 23926210 Free PMC article. - hiPSC hepatocyte model demonstrates the role of unfolded protein response and inflammatory networks in α1-antitrypsin deficiency.
Segeritz CP, Rashid ST, de Brito MC, Serra MP, Ordonez A, Morell CM, Kaserman JE, Madrigal P, Hannan NRF, Gatto L, Tan L, Wilson AA, Lilley K, Marciniak SJ, Gooptu B, Lomas DA, Vallier L. Segeritz CP, et al. J Hepatol. 2018 Oct;69(4):851-860. doi: 10.1016/j.jhep.2018.05.028. Epub 2018 Jun 5. J Hepatol. 2018. PMID: 29879455 Free PMC article. - Adenine base editing reduces misfolded protein accumulation and toxicity in alpha-1 antitrypsin deficient patient iPSC-hepatocytes.
Werder RB, Kaserman JE, Packer MS, Lindstrom-Vautrin J, Villacorta-Martin C, Young LE, Aratyn-Schaus Y, Gregoire F, Wilson AA. Werder RB, et al. Mol Ther. 2021 Nov 3;29(11):3219-3229. doi: 10.1016/j.ymthe.2021.06.021. Epub 2021 Jul 2. Mol Ther. 2021. PMID: 34217893 Free PMC article. - Gene correction in patient-specific iPSCs for therapy development and disease modeling.
Jang YY, Ye Z. Jang YY, et al. Hum Genet. 2016 Sep;135(9):1041-58. doi: 10.1007/s00439-016-1691-5. Epub 2016 Jun 2. Hum Genet. 2016. PMID: 27256364 Free PMC article. Review. - Alpha-1 antitrypsin deficiency: a conformational disease associated with lung and liver manifestations.
Greene CM, Miller SD, Carroll T, McLean C, O'Mahony M, Lawless MW, O'Neill SJ, Taggart CC, McElvaney NG. Greene CM, et al. J Inherit Metab Dis. 2008 Feb;31(1):21-34. doi: 10.1007/s10545-007-0748-y. Epub 2008 Jan 16. J Inherit Metab Dis. 2008. PMID: 18193338 Review.
Cited by
- Bioengineered Organoids Offer New Possibilities for Liver Cancer Studies: A Review of Key Milestones and Challenges.
Jabri A, Khan J, Taftafa B, Alsharif M, Mhannayeh A, Chinnappan R, Alzhrani A, Kazmi S, Mir MS, Alsaud AW, Yaqinuddin A, Assiri AM, AlKattan K, Vashist YK, Broering DC, Mir TA. Jabri A, et al. Bioengineering (Basel). 2024 Apr 1;11(4):346. doi: 10.3390/bioengineering11040346. Bioengineering (Basel). 2024. PMID: 38671768 Free PMC article. Review. - Computational Tools to Assist in Analyzing Effects of the SERPINA1 Gene Variation on Alpha-1 Antitrypsin (AAT).
Mróz J, Pelc M, Mitusińska K, Chorostowska-Wynimko J, Jezela-Stanek A. Mróz J, et al. Genes (Basel). 2024 Mar 6;15(3):340. doi: 10.3390/genes15030340. Genes (Basel). 2024. PMID: 38540399 Free PMC article. Review. - Precise genetic engineering with piggyBac transposon in plants.
Nishizawa-Yokoi A, Toki S. Nishizawa-Yokoi A, et al. Plant Biotechnol (Tokyo). 2023 Dec 25;40(4):255-262. doi: 10.5511/plantbiotechnology.23.0525a. Plant Biotechnol (Tokyo). 2023. PMID: 38434112 Free PMC article. - Stem cell modeling of nervous system tumors.
Furnari FB, Anastasaki C, Bian S, Fine HA, Koga T, Le LQ, Rodriguez FJ, Gutmann DH. Furnari FB, et al. Dis Model Mech. 2024 Feb 1;17(2):dmm050533. doi: 10.1242/dmm.050533. Epub 2024 Feb 14. Dis Model Mech. 2024. PMID: 38353122 Free PMC article. Review. - Efficient gene editing in induced pluripotent stem cells enabled by an inducible adenine base editor with tunable expression.
Nandy K, Babu D, Rani S, Joshi G, Ijee S, George A, Palani D, Premkumar C, Rajesh P, Vijayanand S, David E, Murugesan M, Velayudhan SR. Nandy K, et al. Sci Rep. 2023 Dec 11;13(1):21953. doi: 10.1038/s41598-023-42174-2. Sci Rep. 2023. PMID: 38081875 Free PMC article.
References
- Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. - PubMed
- Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–141. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0701448/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- G1000847/MRC_/Medical Research Council/United Kingdom
- 077187/WT_/Wellcome Trust/United Kingdom
- G0901786/MRC_/Medical Research Council/United Kingdom
- G0800784/MRC_/Medical Research Council/United Kingdom
- G0601840/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous