P(I) Release Limits the Intrinsic and RNA-Stimulated ATPase Cycles of DEAD-Box Protein 5 (Dbp5) - PubMed (original) (raw)
P(I) Release Limits the Intrinsic and RNA-Stimulated ATPase Cycles of DEAD-Box Protein 5 (Dbp5)
Emily V Wong et al. J Mol Biol. 2016.
Abstract
mRNA export from the nucleus depends on the ATPase activity of the DEAD-box protein Dbp5/DDX19. Although Dbp5 has measurable ATPase activity alone, several regulatory factors (e.g., RNA, nucleoporin proteins, and the endogenous small molecule InsP6) modulate catalytic activity in vitro and in vivo to facilitate mRNA export. An analysis of the intrinsic and regulator-activated Dbp5 ATPase cycle is necessary to define how these factors control Dbp5 and mRNA export. Here, we report a kinetic and equilibrium analysis of the Saccharomyces cerevisiae Dbp5 ATPase cycle, including the influence of RNA on Dbp5 activity. These data show that ATP binds Dbp5 weakly in rapid equilibrium with a binding affinity (KT~4 mM) comparable to the KM for steady-state cycling, while ADP binds an order of magnitude more tightly (KD~0.4 mM). The overall intrinsic steady-state cycling rate constant (kcat) is limited by slow, near-irreversible ATP hydrolysis and even slower subsequent phosphate release. RNA increases kcat and rate-limiting Pi release 20-fold, although Pi release continues to limit steady-state cycling in the presence of RNA, in conjunction with RNA binding. Together, this work identifies RNA binding and Pi release as important biochemical transitions within the Dbp5 ATPase cycle and provides a framework for investigating the means by which Dbp5 and mRNA export is modulated by regulatory factors.
Keywords: RNA helicase; kinetics; mRNA export; mantATP; thermodynamics.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
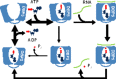
Graphical abstract
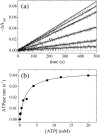
Fig. 1
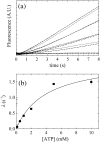
Steady-state ATPase activity of Dbp5. (a) Time courses of absorbance change at 340 nm assayed with the NADH-coupled assay after mixing 660 nM Dbp5 with (lower to upper curves) 0, 0.25, 0.5, 1, 2, 8, or 20 mM ATP. The continuous lines through the data represent the best fits to linear functions, yielding the steady-state ATPase rates from the slopes. Broken lines represent kinetic simulations of Scheme 1 and the corresponding rate and equilibrium constants obtained from global fitting with KinTek Explorer in Table 2. (b) [ATP] dependence of the Dbp5 steady-state ATPase rate. The continuous line through the data represents the best fit to a rectangular hyperbola, yielding the maximum velocity per enzyme (_k_cat) from the amplitude and the _K_M,ATP from the [ATP] at half-maximum velocity (Table 1). Uncertainty bars represent standard errors in the fits and are within the data points.
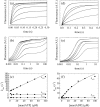
Fig. 2
Mant-Nucleotide binding to Dbp5. (a) Time courses of fluorescence change after mixing 0.5 μM Dbp5 with (lower to upper) 0, 2.5, 5, 10, 20, 30, or 50 μM mantATP. The smooth lines through the data represent the best fits to three exponentials. (b) Full time courses from (a) shown on a log scale to 200 s. (c) [mantATP] dependence of the observed rate constants associated with mantATP binding. Continuous lines represent the best linear fits to the data. Uncertainty bars representing the standard errors of the best fits are within the data points. (d–f) Analogous mantADP binding data fitted to two exponentials shown on linear (d) and log (e) scales and with rate constants fitted to hyperbolic or linear functions (f).
Fig. 3
Interactions between mant-ADP·BeF3 and Dbp5. (a) The mant fluorophore buries adjacent residues (Val181, M110, and Phe112) of ∆90Dbp5 via hydrophobic interactions (< 4.0 Å). The mant moiety hydrogen bonds (broken lines) with waters at the solvent-exposed surface. (b) Displacement of a network of hydrogen-bonded water molecules by the mant fluorophore. ∆90Dbp5-ADP·BeF3 (blue) and the associated water molecules (red) of 3PEY superimposed to ∆90Dbp5-mant-ADP·BeF3 and the associated water molecules (gray). π Stacking of the adenine is maintained. rmsd = 0.1106 Å.
Fig. 4
Unlabeled nucleotide binding affinity measured by competition with mant nucleotides. (a) Time courses of fluorescence change after mixing 0.5 μM Dbp5 with 50 μM mantADP and 0, 0.175, 0.75, 2, 5, or 10 mM ADP (upper to lower). Continuous lines through the data represent best fits to two exponentials. (b) [nucleotide] Dependence of the fastest observed rate constant (_k_1,obs) after mixing 0.5 μM Dbp5 with a solution of 50 μM mantADP supplemented with 0, 0.1, 0.3, 0.6, 1, 2.5, 5, or 11 mM ATP (black squares). Black circles represent the fastest observed rate constant of mantADP binding on mixing 0.5 μM Dbp5 with a solution of 50 μM mantADP supplemented with 0.005, 0.015, 0.03, 0.0625, 0.175, 0.75, 2, 5, or 10 mM ADP. Continuous lines represent best fits to Eq. (13).
Fig. 5
Transient and steady-state Pi release during Dbp5 ATPase cycling. (a) Time courses of fluorescence change after mixing 0.5 μM Dbp5 with 5 μM PiBiP containing (lower to upper curves) 0, 0.5, 1, 2, or 5 mM ATP. Fits to Eq. (14) are shown as broken lines through the data. Simulated data (using rate and equilibrium constants obtained from KinTek Explorer global fitting; Table 2) are shown as dotted lines through the data. (b) [ATP] dependence of the observed Pi release lag phase rate constant (λ). Continuous line through the data points represents best fit to a rectangular hyperbola [Eq. (15)].
Fig. 6
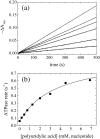
RNA-stimulated steady-state ATPase activity of Dbp5. Time courses of absorbance change at 340 nm assayed with the NADH-coupled assay after mixing 100 nM Dbp5 and 20 mM ATP with (lower to upper curves) 0, 0.45, 1.25, 2, 3, or 4.5 mM polyuridylic acid (concentration refers to total nucleotides). The continuous lines through the data represent the best fits to linear functions, yielding the steady-state ATPase rates from the slopes. (b) [ATP] dependence of the RNA-stimulated Dbp5 steady-state ATPase rate. The continuous line through the data represents the best fit to Eq. (11), yielding the maximum velocity per enzyme (_k_cat) from the amplitude and the _K_M,RNA from the [ATP] at half-maximum velocity (Table 1). Uncertainty bars represent standard errors in the fits and are within the data points.
Fig. 7
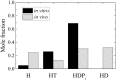
Steady-state distribution of Dbp5 ATPase cycle intermediates. In vitro conditions are 20 mM ATP, 0 mM ADP, and 0 mM Pi. In vivo conditions are 2.1 mM ATP , 470 μM ADP , 2.5 mM Pi.
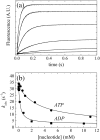
Fig. 8
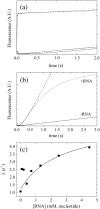
Transient RNA-stimulated Pi release during Dbp5 ATPase cycling. (a) Time courses of fluorescence change after mixing 5 μM Dbp5 with 4 mM ATP and aging for (lower to upper) 0.05, 6, or 25 s prior to rapidly mixing with 5 μM PiBiP. Broken lines are best fits to (lower to upper) Eq. (14), linear equation, or bimolecular binding plus a linear phase, respectively. (b) Time courses of fluorescence change after mixing 5 μM Dbp5 with 4 mM ATP, aging for 6 s, and then rapidly mixing with 0 or 10 mM RNA in the presence of 5 μM PiBiP. Fits to Eq. (16) are shown as broken lines through the data. (c) [RNA] dependence of the observed Pi release lag phase rate constant (λ) where pre-equilibrated Dbp5 and RNA are rapidly mixed with ATP and PiBiP. Continuous line through the data points represents best fit to a rectangular hyperbola [Eq. (15)], yielding a maximum observed rate constant of 5.5 ± 2 s− 1 upon extrapolation of the fit to saturating RNA. The [RNA] at half maximal is ~ 3 mM under these nonsaturating ATP concentrations.
Similar articles
- The nucleoporin Gle1 activates DEAD-box protein 5 (Dbp5) by promoting ATP binding and accelerating rate limiting phosphate release.
Gray S, Cao W, Montpetit B, De La Cruz EM. Gray S, et al. Nucleic Acids Res. 2022 Apr 22;50(7):3998-4011. doi: 10.1093/nar/gkac164. Nucleic Acids Res. 2022. PMID: 35286399 Free PMC article. - A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export.
Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. Montpetit B, et al. Nature. 2011 Apr 14;472(7342):238-42. doi: 10.1038/nature09862. Epub 2011 Mar 27. Nature. 2011. PMID: 21441902 Free PMC article. - Nup159 Weakens Gle1 Binding to Dbp5 But Does Not Accelerate ADP Release.
Wong EV, Gray S, Cao W, Montpetit R, Montpetit B, De La Cruz EM. Wong EV, et al. J Mol Biol. 2018 Jul 6;430(14):2080-2095. doi: 10.1016/j.jmb.2018.05.025. Epub 2018 May 19. J Mol Biol. 2018. PMID: 29782832 Free PMC article. - Dbp5 - from nuclear export to translation.
Tieg B, Krebber H. Tieg B, et al. Biochim Biophys Acta. 2013 Aug;1829(8):791-8. doi: 10.1016/j.bbagrm.2012.10.010. Epub 2012 Nov 2. Biochim Biophys Acta. 2013. PMID: 23128325 Review. - Emerging molecular functions and novel roles for the DEAD-box protein Dbp5/DDX19 in gene expression.
Arul Nambi Rajan A, Montpetit B. Arul Nambi Rajan A, et al. Cell Mol Life Sci. 2021 Mar;78(5):2019-2030. doi: 10.1007/s00018-020-03680-y. Epub 2020 Nov 17. Cell Mol Life Sci. 2021. PMID: 33205304 Free PMC article. Review.
Cited by
- An aromatic-rich loop couples DNA binding and ATP hydrolysis in the PriA DNA helicase.
Windgassen TA, Keck JL. Windgassen TA, et al. Nucleic Acids Res. 2016 Nov 16;44(20):9745-9757. doi: 10.1093/nar/gkw690. Epub 2016 Aug 2. Nucleic Acids Res. 2016. PMID: 27484483 Free PMC article. - Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Schmitt A, Hamann F, Neumann P, Ficner R. Schmitt A, et al. Acta Crystallogr D Struct Biol. 2018 Jul 1;74(Pt 7):643-654. doi: 10.1107/S2059798318006356. Epub 2018 Jun 8. Acta Crystallogr D Struct Biol. 2018. PMID: 29968674 Free PMC article. - Gle1 is required for tRNA to stimulate Dbp5 ATPase activity in vitro and promote Dbp5-mediated tRNA export in vivo in Saccharomyces cerevisiae.
Arul Nambi Rajan A, Asada R, Montpetit B. Arul Nambi Rajan A, et al. Elife. 2024 Jan 8;12:RP89835. doi: 10.7554/eLife.89835. Elife. 2024. PMID: 38189406 Free PMC article. - Motif-VI loop acts as a nucleotide valve in the West Nile Virus NS3 Helicase.
Roy P, Walter Z, Berish L, Ramage H, McCullagh M. Roy P, et al. Nucleic Acids Res. 2024 Jul 22;52(13):7447-7464. doi: 10.1093/nar/gkae500. Nucleic Acids Res. 2024. PMID: 38884215 Free PMC article. - Gle1 is required for tRNA to stimulate Dbp5 ATPase activity in vitro and to promote Dbp5 mediated tRNA export in vivo.
Rajan AAN, Asada R, Montpetit B. Rajan AAN, et al. bioRxiv [Preprint]. 2023 Nov 9:2023.06.29.547072. doi: 10.1101/2023.06.29.547072. bioRxiv. 2023. PMID: 37425677 Free PMC article. Updated. Preprint.
References
- Linder P., Lasko P.F., Ashburner M., Leroy P., Nielsen P.J., Nishi K. Birth of the D-E-A-D box. Nature. 1989;337:121–122. - PubMed
- Rocak S., Linder P. DEAD-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 2004;5:232–241. - PubMed
- Cordin O., Banroques J., Tanner N.K., Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. - PubMed
- Fairman M.E., Maroney P.A., Wang W., Bowers H.A., Gollnick P., Nilsen T.W. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous