Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic Ma-Pi 2 diet - PubMed (original) (raw)
doi: 10.1017/S0007114516001045. Epub 2016 May 6.
Elena Biagi 1, Matteo Soverini 1, Clarissa Consolandi 2, Sara Quercia 1, Marco Severgnini 2, Clelia Peano 2, Silvia Turroni 1, Simone Rampelli 1, Paolo Pozzilli 3, Mario Pianesi 4, Francesco Fallucca 5, Patrizia Brigidi 1
Affiliations
- PMID: 27151248
- PMCID: PMC4894062
- DOI: 10.1017/S0007114516001045
Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic Ma-Pi 2 diet
Marco Candela et al. Br J Nutr. 2016 Jul.
Abstract
The gut microbiota exerts a role in type 2 diabetes (T2D), and deviations from a mutualistic ecosystem layout are considered a key environmental factor contributing to the disease. Thus, the possibility of improving metabolic control in T2D by correcting gut microbiome dysbioses through diet has been evaluated. Here, we explore the potential of two different energy-restricted dietary approaches - the fibre-rich macrobiotic Ma-Pi 2 diet or a control diet recommended by Italian professional societies for T2D treatment - to correct gut microbiota dysbioses in T2D patients. In a previous 21-d open-label MADIAB trial, fifty-six overweight T2D patients were randomised to the Ma-Pi 2 or the control diet. For the present study, stools were collected before and after intervention from a subset of forty MADIAB participants, allowing us to characterise the gut microbiota by 16S rRNA sequencing and imputed metagenomics. To highlight microbiota dysbioses in T2D, the gut microbiota of thirteen normal-weight healthy controls were characterised. According to our findings, both diets were effective in modulating gut microbiome dysbioses in T2D, resulting in an increase of the ecosystem diversity and supporting the recovery of a balanced community of health-promoting SCFA producers, such as Faecalibacterium, Roseburia, Lachnospira, Bacteroides and Akkermansia. The Ma-Pi 2 diet, but not the control diet, was also effective in counteracting the increase of possible pro-inflammatory groups, such as Collinsella and Streptococcus, in the gut ecosystem, showing the potential to reverse pro-inflammatory dysbioses in T2D, and possibly explaining the greater efficacy in improving the metabolic control.
Keywords: CTR control diet; Dysbiosis; FBG fasting blood glucose; Fibre-rich diets; GM gut microbiota; Gut microbiota; KO KEGG Orthology; Macrobiotic diets; PCoA principal coordinates analysis; T2D type 2 diabetes; Type 2 diabetes.
Figures
Fig. 1
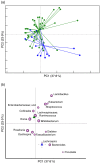
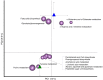
Comparison of the gut microbiota compositional structure between overweight type 2 diabetes (T2D) patients at baseline and healthy controls. (a) Principal coordinates analysis (PCoA) based on weighted UniFrac distances shows separation between forty overweight T2D patients at T0 and thirteen normal-weight healthy controls. , Healthy controls;
, Healthy controls;  , T2D patients. P<0·001; permutation test with pseudo_F_ ratios. (b) Superimposition of microbial genera on the PCoA plot in order to identify the genera involved in this separation. Sphere width is proportional to the mean relative abundance of the genus across all samples. The two components explain 37·8 and 23·9 % of the variance, respectively.
, T2D patients. P<0·001; permutation test with pseudo_F_ ratios. (b) Superimposition of microbial genera on the PCoA plot in order to identify the genera involved in this separation. Sphere width is proportional to the mean relative abundance of the genus across all samples. The two components explain 37·8 and 23·9 % of the variance, respectively.  ,
, , Centroids for each group with indication of standard errors on each coordinate axis; uncl, unclassified.
, Centroids for each group with indication of standard errors on each coordinate axis; uncl, unclassified.
Fig. 2
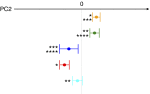
Variation of the weighted UniFrac PC2 coordinates between the study groups. , Ma-Pi 2 diet group at T0;
, Ma-Pi 2 diet group at T0; , control (CTR) diet group at T0;
, control (CTR) diet group at T0; , healthy controls;
, healthy controls;  , Ma-Pi 2 diet group at T1;
, Ma-Pi 2 diet group at T1;  , CTR diet group at T1. For each group, average (±
, CTR diet group at T1. For each group, average (±
sem
, error bar) PC2 coordinates are shown. The significance of the differences between the PC2 coordinates of the groups is indicated as follows: *P<0·001 (Ma-Pi 2 diet group at T0 v. T1), ** _P_=0·01 (CTR diet group at T0 v. T1), ***_P_=0·01 (Ma-Pi 2 diet group at T0 v. healthy controls), **** _P_=0·03 (CTR diet group at T0 _v._healthy controls); Wilcoxon’s signed rank-sum test.
Fig. 3
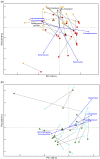
Comparison of the gut microbiota compositional structure of type 2 diabetes (T2D) patients before and after the nutritional interventions. (a) Principal coordinates analysis (PCoA) based on weighted UniFrac distances for T2D subjects following the Ma-Pi 2 diet (n 21) shows separation between T0 ( ) and T1 (
) and T1 ( ). The two components explain 35·0 and 24·8 % of the variance, respectively. _P_=0·01; permutation test with pseudo F ratios. (b) PCoA based on weighted UniFrac distances for T2D subjects following the control (CTR) diet (n 19).
). The two components explain 35·0 and 24·8 % of the variance, respectively. _P_=0·01; permutation test with pseudo F ratios. (b) PCoA based on weighted UniFrac distances for T2D subjects following the control (CTR) diet (n 19). , T0;
, T0;  , T1. The two components explain 32·0 and 20·2 % of the variance, respectively._P_=0·04; permutation test with pseudo F ratios. Lines connect T0 and T1 samples from the same patient.
, T1. The two components explain 32·0 and 20·2 % of the variance, respectively._P_=0·04; permutation test with pseudo F ratios. Lines connect T0 and T1 samples from the same patient.  ,
, responding bacterial genera and biochemical parameters, respectively;
responding bacterial genera and biochemical parameters, respectively;  , direction of significant correlations;
, direction of significant correlations;  ,
,  ,
,  ,
, , centroids for each time point.
, centroids for each time point.
Fig. 4
Impact of dietary interventions on the taxonomic structure of the core microbiota in type 2 diabetes (T2D) patients. Heat maps were calculated for both Ma-Pi 2 and control (CTR) diet, based on the log-ratio of the median genus abundance in T2D patients (T0 and T1 samples) and healthy controls. The colour code and segment line reveal the deviation in terms of fold change from the median profile of healthy subjects ( ). T2D patients following the Ma-Pi 2 diet (n 21); T2D patients following the CTR diet (n 19); healthy controls (n 13).
). T2D patients following the Ma-Pi 2 diet (n 21); T2D patients following the CTR diet (n 19); healthy controls (n 13).
Fig. 5
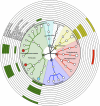
Impact of dietary components on the gut microbiota taxonomic structure. Cladogram obtained with the GraPhlAn tool, showing the family-level gut microbiota profile of T1 samples from both intervention groups with a phylum-based colour code ( , Firmicutes;
, Firmicutes;  , Bacteroidetes;
, Bacteroidetes;  , Proteobacteria;
, Proteobacteria;  , Actinobacteria;
, Actinobacteria;  , Verrucomicrobia;
, Verrucomicrobia;  , Synergistetes). Families with relative abundance of at least 0·5 % in at least two samples are plotted. Larger circles identify bacterial families having a positive correlation with at least one dietary component; the names of these families are reported. Filled circles identify bacterial families that showed significantly higher abundance in T1 samples of the Ma-Pi 2 diet group (
, Synergistetes). Families with relative abundance of at least 0·5 % in at least two samples are plotted. Larger circles identify bacterial families having a positive correlation with at least one dietary component; the names of these families are reported. Filled circles identify bacterial families that showed significantly higher abundance in T1 samples of the Ma-Pi 2 diet group ( ) or the CTR diet group (
) or the CTR diet group ( ). Bacterial family-food component correlations are indicated by filled boxes in the external rings of the plot, referring to the list of dietary components. T2D patients following the Ma-Pi 2 diet (n 21); T2D patients following the CTR diet (_n_19).
). Bacterial family-food component correlations are indicated by filled boxes in the external rings of the plot, referring to the list of dietary components. T2D patients following the Ma-Pi 2 diet (n 21); T2D patients following the CTR diet (_n_19).
Fig. 6
Functional dysbioses of the gut microbiome in type 2 diabetes (T2D) patients. Metabolic pathways were superimposed on the principal component analysis plot based on Euclidean distances, and the pathways responsible for the separation are shown. , Healthy controls (_n_13);
, Healthy controls (_n_13);  , T2D patients (n 40). Sphere width is proportional to the mean relative abundance of the function across all samples. The two components explain 14·0 and 9·2 % of the variance, respectively. P_=0·03; permutation test with pseudo_F ratios.
, T2D patients (n 40). Sphere width is proportional to the mean relative abundance of the function across all samples. The two components explain 14·0 and 9·2 % of the variance, respectively. P_=0·03; permutation test with pseudo_F ratios.  ,
,  , Centroids for each group with indication of standard errors on each coordinate axis.
, Centroids for each group with indication of standard errors on each coordinate axis.
Fig. 7
Impact of Ma-Pi 2 dietary intervention on the functional configuration of the gut microbiome in type 2 diabetes (T2D) patients. Metabolic pathways were superimposed on the principal component analysis plot based on Euclidean distances in T2D patients before (T0,  ) and after (T1,
) and after (T1,  ) the Ma-Pi 2 diet (n 21). The two components explain 22·3 and 14·9 % of the variance, respectively. _P_=0·007; permutation test with pseudo F ratios.
) the Ma-Pi 2 diet (n 21). The two components explain 22·3 and 14·9 % of the variance, respectively. _P_=0·007; permutation test with pseudo F ratios.  ,
,  , Centroids for each group with indication of standard errors on each coordinate axis.
, Centroids for each group with indication of standard errors on each coordinate axis.
References
- Xu Y (2013) Prevalence and control of diabetes in Chinese adults. JAMA 310, 948. - PubMed
- Zhang P, Zhang X, Brown J, et al. (2010) Global healthcare expenditure on diabetes for 2010 and 2030. Diabetes Res Clin Pract 87, 293–301. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous