Ataxin-2: From RNA Control to Human Health and Disease - PubMed (original) (raw)
Review
Ataxin-2: From RNA Control to Human Health and Disease
Lauren A Ostrowski et al. Genes (Basel). 2017.
Abstract
RNA-binding proteins play fundamental roles in the regulation of molecular processes critical to cellular and organismal homeostasis. Recent studies have identified the RNA-binding protein Ataxin-2 as a genetic determinant or risk factor for various diseases including spinocerebellar ataxia type II (SCA2) and amyotrophic lateral sclerosis (ALS), amongst others. Here, we first discuss the increasingly wide-ranging molecular functions of Ataxin-2, from the regulation of RNA stability and translation to the repression of deleterious accumulation of the RNA-DNA hybrid-harbouring R-loop structures. We also highlight the broader physiological roles of Ataxin-2 such as in the regulation of cellular metabolism and circadian rhythms. Finally, we discuss insight from clinically focused studies to shed light on the impact of molecular and physiological roles of Ataxin-2 in various human diseases. We anticipate that deciphering the fundamental functions of Ataxin-2 will uncover unique approaches to help cure or control debilitating and lethal human diseases.
Keywords: ALS; RNA metabolism; RNA-DNA hybrids; SCA2; stress granules.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Figure 1
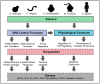
Structure of Ataxin-2 and its conserved domains across model species. The RNA-binding domains of Ataxin-2, Like-Sm (LSm) and LSm-associated (LSmAD), are located at the N-terminal region and are conserved across species. The C-terminal region of Ataxin-2 harbours a PAM2 domain, which interacts with the poly(A)-binding protein (PABP1). A polyglutamine (polyQ) tract is also located in the N-terminal of most mammalian species. For species with multiple Q residues upstream of the LSm/LSmAD domains (S. pombe, D. rerio, M. musculus), the Q represented was chosen based on amino acid sequence similarities with the regions flanking the polyQ site in human ATXN2. Conserved proline-rich domains are also depicted (consensus sequences: PPAXPTXXSP and PPSRPSRPPS). ATSS = alternative translational start site.
Figure 2
Functions of Ataxin-2 proteins under stressed and non-stressed conditions. Studies from a variety of model organisms demonstrate a role for Ataxin-2 proteins in the regulation of mRNA polyadentylation, stability, translation, R-loops, circadian rhythms, branched-chain amino acid (BCAA) metabolism, mTOR activity, stress granule (SG) assembly, and P-body morphology. Dashed lines represent indirect regulation by Ataxin-2. Reference numbers for papers are shown.
Figure 3
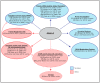
Multifaceted functions of Ataxin-2 and links to disease. Ataxin-2 functions to regulate several stages of RNA processing, with roles in physiological pathways. These functions include promoting mRNA stability and translation, as well as the regulation of R-loop and stress granule formation. These functions contribute to the control of metabolic pathways such as TOR and circadian rhythmicity. Deregulation of any of these processes can give rise to a wide range of cellular dysfunction, which can promote disease. SCA2, spinocerebellar ataxia type II; ALS, amyotrophic lateral sclerosis; PD, Parkinson’s disease; SCA1, spinocerebellar ataxia type I; MJD, Machado-Joseph’s disease; POAG, primary open angle glaucoma.
Similar articles
- R-loops highlight the nucleus in ALS.
Salvi JS, Mekhail K. Salvi JS, et al. Nucleus. 2015;6(1):23-9. doi: 10.1080/19491034.2015.1004952. Nucleus. 2015. PMID: 25587791 Free PMC article. Review. - Taking a risk: a therapeutic focus on ataxin-2 in amyotrophic lateral sclerosis?
van den Heuvel DM, Harschnitz O, van den Berg LH, Pasterkamp RJ. van den Heuvel DM, et al. Trends Mol Med. 2014 Jan;20(1):25-35. doi: 10.1016/j.molmed.2013.09.001. Epub 2013 Oct 16. Trends Mol Med. 2014. PMID: 24140266 Review. - FTLD-ALS of TDP-43 type and SCA2 in a family with a full ataxin-2 polyglutamine expansion.
Bäumer D, East SZ, Tseu B, Zeman A, Hilton D, Talbot K, Ansorge O. Bäumer D, et al. Acta Neuropathol. 2014 Oct;128(4):597-604. doi: 10.1007/s00401-014-1277-z. Epub 2014 Apr 10. Acta Neuropathol. 2014. PMID: 24718895 - Genetic ablation of ataxin-2 increases several global translation factors in their transcript abundance but decreases translation rate.
Fittschen M, Lastres-Becker I, Halbach MV, Damrath E, Gispert S, Azizov M, Walter M, Müller S, Auburger G. Fittschen M, et al. Neurogenetics. 2015 Jul;16(3):181-92. doi: 10.1007/s10048-015-0441-5. Epub 2015 Feb 27. Neurogenetics. 2015. PMID: 25721894 Free PMC article.
Cited by
- Ataxin-2: a powerful RNA-binding protein.
Li L, Wang M, Huang L, Zheng X, Wang L, Miao H. Li L, et al. Discov Oncol. 2024 Jul 22;15(1):298. doi: 10.1007/s12672-024-01158-y. Discov Oncol. 2024. PMID: 39039334 Free PMC article. Review. - RNA Deregulation in Amyotrophic Lateral Sclerosis: The Noncoding Perspective.
Laneve P, Tollis P, Caffarelli E. Laneve P, et al. Int J Mol Sci. 2021 Sep 24;22(19):10285. doi: 10.3390/ijms221910285. Int J Mol Sci. 2021. PMID: 34638636 Free PMC article. Review. - The disease-associated proteins Drosophila Nab2 and Ataxin-2 interact with shared RNAs and coregulate neuronal morphology.
Rounds JC, Corgiat EB, Ye C, Behnke JA, Kelly SM, Corbett AH, Moberg KH. Rounds JC, et al. Genetics. 2022 Jan 4;220(1):iyab175. doi: 10.1093/genetics/iyab175. Genetics. 2022. PMID: 34791182 Free PMC article. - The Importance of Offering Exome or Genome Sequencing in Adult Neuromuscular Clinics.
Dratch L, Bardakjian TM, Johnson K, Babaian N, Gonzalez-Alegre P, Elman L, Quinn C, Guo MH, Scherer SS, Amado DA. Dratch L, et al. Biology (Basel). 2024 Feb 2;13(2):93. doi: 10.3390/biology13020093. Biology (Basel). 2024. PMID: 38392311 Free PMC article. - Updates on Disease Mechanisms and Therapeutics for Amyotrophic Lateral Sclerosis.
Nguyen L. Nguyen L. Cells. 2024 May 21;13(11):888. doi: 10.3390/cells13110888. Cells. 2024. PMID: 38891021 Free PMC article. Review.
References
- Imbert G., Saudou F., Yvert G., Devys D., Trottier Y., Garnier J.M., Weber C., Mandel J.L., Cancel G., Abbas N., et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat. Genet. 1996;14:285–291. doi: 10.1038/ng1196-285. - DOI - PubMed
- Pulst S.M., Nechiporuk A., Nechiporuk T., Gispert S., Chen X.N., Lopes-Cendes I., Pearlman S., Starkman S., Orozco-Diaz G., Lunkes A., et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat. Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. - DOI - PubMed
- Sanpei K., Takano H., Igarashi S., Sato T., Oyake M., Sasaki H., Wakisaka A., Tashiro K., Ishida Y., Ikeuchi T., et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat. Genet. 1996;14:277–284. doi: 10.1038/ng1196-277. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous