Peptidoglycan-Associated Cyclic Lipopeptide Disrupts Viral Infectivity - PubMed (original) (raw)
. 2019 Oct 29;93(22):e01282-19.
doi: 10.1128/JVI.01282-19. Print 2019 Nov 15.
Adam Hage 1, Birte Kalveram 2, Megan Mears 2, Jessica A Plante 3 1, Sergio E Rodriguez 1, Zhixia Ding 2 4, Xuemei Luo 5 6, Dennis Bente 1, Shelton S Bradrick 5, Alexander N Freiberg 2, Vsevolod Popov 2 4, Ricardo Rajsbaum 1 7, Shannan Rossi 1 2, William K Russell 5 6, Vineet D Menachery 8 7
Affiliations
- PMID: 31462558
- PMCID: PMC6819921
- DOI: 10.1128/JVI.01282-19
Peptidoglycan-Associated Cyclic Lipopeptide Disrupts Viral Infectivity
Bryan A Johnson et al. J Virol. 2019.
Abstract
Enteric viruses exploit bacterial components, including lipopolysaccharides (LPS) and peptidoglycan (PG), to facilitate infection in humans. Because of their origin in the bat enteric system, we wondered if severe acute respiratory syndrome coronavirus (SARS-CoV) or Middle East respiratory syndrome CoV (MERS-CoV) also use bacterial components to modulate infectivity. To test this question, we incubated CoVs with LPS and PG and evaluated infectivity, finding no change following LPS treatment. However, PG from Bacillus subtilis reduced infection >10,000-fold, while PG from other bacterial species failed to recapitulate this. Treatment with an alcohol solvent transferred inhibitory activity to the wash, and mass spectrometry revealed surfactin, a cyclic lipopeptide antibiotic, as the inhibitory compound. This antibiotic had robust dose- and temperature-dependent inhibition of CoV infectivity. Mechanistic studies indicated that surfactin disrupts CoV virion integrity, and surfactin treatment of the virus inoculum ablated infection in vivo Finally, similar cyclic lipopeptides had no effect on CoV infectivity, and the inhibitory effect of surfactin extended broadly to enveloped viruses, including influenza, Ebola, Zika, Nipah, chikungunya, Una, Mayaro, Dugbe, and Crimean-Congo hemorrhagic fever viruses. Overall, our results indicate that peptidoglycan-associated surfactin has broad viricidal activity and suggest that bacteria by-products may negatively modulate virus infection.IMPORTANCE In this article, we consider a role for bacteria in shaping coronavirus infection. Taking cues from studies of enteric viruses, we initially investigated how bacterial surface components might improve CoV infection. Instead, we found that peptidoglycan-associated surfactin is a potent viricidal compound that disrupts virion integrity with broad activity against enveloped viruses. Our results indicate that interactions with commensal bacterial may improve or disrupt viral infections, highlighting the importance of understanding these microbial interactions and their implications for viral pathogenesis and treatment.
Keywords: MERS-CoV; SARS-CoV; coronavirus; cyclic lipopeptide; microbiome; surfactin.
Copyright © 2019 American Society for Microbiology.
Figures
FIG 1
Peptidoglycan from Bacillus subtilis reduces coronavirus infectivity. (A) Bacterial envelope components such as LPS are bound to CoVs, increasing their thermostability (right) relative to that of untreated samples (left). (B) Relative infectivity of HCoV-229E (n = 4) and MERS-CoV (n = 5) after treatment with PBS alone (black), 1 mg/ml LPS from E. coli (gray), or 1 mg/ml PG from B. subtilis (green) following 2 h of incubation at 37°C. (C) HCoV-299E (circles) and MERS-CoV (triangles) infectivity after treatment for 2 h at 37°C with peptidoglycan from the indicated bacterial species (n = 3). (D) HCoV-229E and (E) MERS-CoV after treatment with 1 mg/ml PG from B. subtilis at room temperature (RT), 32°C, and 37°C (n = 3). For all dot plots, the centered bar represents the group mean, while the error bars represent standard deviation (SD). P values are based on the two-tailed Student’s t test, indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FIG 2
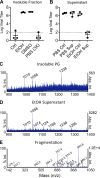
Identification of surfactin from B. subtilis peptidoglycan. (A and B) PG from B. subtilis in a PBS solution was clarified, washed with the indicated solvents, and clarified again. Supernatants were decanted and retained, while the insoluble fractions were resuspended in PBS. The (A) insoluble fraction and (B) supernatants were then used to treat HCoV-229E, and relative infectivity was determined (n = 3). (C, D, and E) Mass spectrometry was performed on PG (C) and ethanol wash (D). The peak corresponding to the molecular mass of 1,058 kDa in the ethanol wash was then further fragmented (E) to determine the identity of the molecule. Representative spectra are shown (n = 3). For all dot plots, the centered bar represents the group mean and error bars the SD.
FIG 3
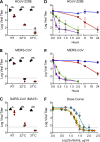
Characterization of CoV inhibition by surfactin. (A) HCoV-229E, (B) MERS-CoV, and (C) SARS-CoV (MA15) were treated with PBS alone (black) or 100 μg/ml surfactin (red) and at room temperature (RT), 32°C, and 37°C, and infectivity was determined (n = 3). HCoV-229E (D) and MERS-CoV (E) were treated for the indicated time at 4°C (blue), RT (red), 32°C (green), or 37°C (purple), and infectivity was determined (n = 3). (F) HCoV-229E (blue), MERS-CoV (orange), and SARS-CoV MA15 (green) were diluted in concentrations ranging from 0.5 to 200 μg/ml, and viral infectivity was determined (n = 3). For dot plots, each point represents the titer from an independent experiment, while the group mean is indicated by a line. Each point on the line graph represents the group mean. All error bars represent SD. The two-tailed Student’s t test was used to determine P values, indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
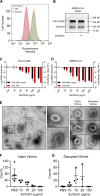
FIG 4
Surfactin disrupts CoV structural integrity. (A) HUH7 cells were first treated with either PBS or 100 μg/ml surfactin solution. Cells were then incubated with recombinant MERS-CoV S1-Fc protein and then stained with an anti-human IgG-FITC secondary in order to examine DPP4 levels. Unstained (red), PBS treated (blue), and surfactin treated (gold). Shown is a representative figure from 3 independent experiments. (B) MERS-CoV were treated with 100 μg/ml surfactin or PBS, as indicated, and then immunoblotted for MERS-CoV spike. Shown is a representative figure from 3 independent experiments. (C) HCoV-229E and (D) MERS-CoV were treated with 0 (ctrl) 10, 20, 30, 100, and 200 μg/ml of surfactin, as indicated. Viral infectivity was then determined (red) or samples were then treated with RNase I, RNA was extracted, and viral genome copy number determined by RT-qPCR (black). Bar graph bars represent the group mean for each experiment, and error bars represent SD (n = 3). (E to G) PBS- or 100 μg/ml surfactin-treated HCoV-229E samples were negatively stained and examined by TEM, and intact and disrupted virions were counted. A representative micrograph shown in panel E, with intact (i) and disrupted (ii) virions (left). Additional examples of intact and disrupted virions are also shown (right). Total counts of intact (F) and disrupted (G) virions are shown at various concentrations, with no less than 3 independent experiments having been performed for each concentration of surfactin. Horizontal lines represent group mean, while error bars represent SD. A two-tailed Student’s t test determined significance. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
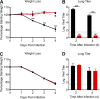
FIG 5
In vivo characterization of surfactin treatment on SARS-CoV infection. (A and B) BALB/c mice were then intranasally infected with 104 PFU of PBS- (black) or 100 μg/ml surfactin (red)-treated SARS-CoV MA15 and (A) monitored for weight loss over 4 days. Dotted lines and triangles represent mock-infected animals receiving PBS alone (black) or surfactin alone (red). (B) Lung tissue was harvested and viral titer determined at days 2 and 4. n = 4 for all infected groups, n = 2 for mock groups. (C and D) BALB/c mice were pretreated intranasally with 50 μl of either PBS (black) or surfactin in PBS (red). Eighteen h later, BALB/c mice were infected with 104 PFU of SARS-CoV (MA15) and (C) monitored for weight loss over 4 days. (D) Lung titer determined 2 (n = 5) and 4 days postinfection (n = 10). Dots on line graphs and bars on bar graphs represent the group mean. ND, no titers were detected. All error bars represent SD. P values were calculated using the two-tailed Student’s t test, with *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
FIG 6
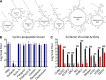
Surfactin, but not other cyclic lipopeptides, broadly reduces the infectivity of enveloped viruses. (A) Biochemical models of each of the seven cyclic lipopeptides tested. The number of amino acids present in the cyclic ring is shown in parentheses. (B) HCoV-229E (blue) and MERS-CoV (gray) were treated with PBS or 100 μg/ml of the indicated cyclic lipopeptides in PBS and incubated for 2 h at 37°C. Viral infectivity was then determined (n = 3). (C) The indicated viruses were diluted in PBS (black) or 100 μg/ml of surfactin (red), incubated for 2 h at 37°C, and viral infectivity was determined (n = 3). Viruses are as follows: coxsackievirus (CVB3), Dugbe (DUGV), Crimean-Congo hemorrhagic fever virus (CCHFV), Zika virus (ZIKV), Nipah virus (NiV), chikungunya virus (CHIKV), Una virus, Mayaro virus, influenza A strains H1N1 and H3N2, and Ebola virus (EBOV). Bar graph bars represent the group means (n = 3). Error bars represent SD. ND, no titers were detected. The Student’s t test was used to calculate P values, with *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Similar articles
- TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection.
Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. Iwata-Yoshikawa N, et al. J Virol. 2019 Mar 5;93(6):e01815-18. doi: 10.1128/JVI.01815-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626688 Free PMC article. - Ca2+ Ions Promote Fusion of Middle East Respiratory Syndrome Coronavirus with Host Cells and Increase Infectivity.
Straus MR, Tang T, Lai AL, Flegel A, Bidon M, Freed JH, Daniel S, Whittaker GR. Straus MR, et al. J Virol. 2020 Jun 16;94(13):e00426-20. doi: 10.1128/JVI.00426-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32295925 Free PMC article. - Glycopeptide Antibiotics Potently Inhibit Cathepsin L in the Late Endosome/Lysosome and Block the Entry of Ebola Virus, Middle East Respiratory Syndrome Coronavirus (MERS-CoV), and Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV).
Zhou N, Pan T, Zhang J, Li Q, Zhang X, Bai C, Huang F, Peng T, Zhang J, Liu C, Tao L, Zhang H. Zhou N, et al. J Biol Chem. 2016 Apr 22;291(17):9218-32. doi: 10.1074/jbc.M116.716100. Epub 2016 Mar 7. J Biol Chem. 2016. PMID: 26953343 Free PMC article. - Middle East Respiratory Syndrome Virus Pathogenesis.
Singh SK. Singh SK. Semin Respir Crit Care Med. 2016 Aug;37(4):572-7. doi: 10.1055/s-0036-1584796. Epub 2016 Aug 3. Semin Respir Crit Care Med. 2016. PMID: 27486737 Free PMC article. Review. - SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium.
Sims AC, Burkett SE, Yount B, Pickles RJ. Sims AC, et al. Virus Res. 2008 Apr;133(1):33-44. doi: 10.1016/j.virusres.2007.03.013. Epub 2007 Apr 23. Virus Res. 2008. PMID: 17451829 Free PMC article. Review.
Cited by
- Antibiotics daptomycin interacts with S protein of SARS-CoV-2 to promote cell invasion of Omicron (B1.1.529) pseudovirus.
Cao X, Huang L, Tang M, Liang Y, Liu X, Hou H, Liang S. Cao X, et al. Virulence. 2024 Dec;15(1):2339703. doi: 10.1080/21505594.2024.2339703. Epub 2024 Apr 24. Virulence. 2024. PMID: 38576396 Free PMC article. - Gut Microbiome Disruption Following SARS-CoV-2: A Review.
Righi E, Dalla Vecchia I, Auerbach N, Morra M, Górska A, Sciammarella C, Lambertenghi L, Gentilotti E, Mirandola M, Tacconelli E, Sartor A. Righi E, et al. Microorganisms. 2024 Jan 9;12(1):131. doi: 10.3390/microorganisms12010131. Microorganisms. 2024. PMID: 38257958 Free PMC article. Review. - Alkyl Derivatives of Perylene Photosensitizing Antivirals: Towards Understanding the Influence of Lipophilicity.
Mikhnovets IE, Holoubek J, Panina IS, Kotouček J, Gvozdev DA, Chumakov SP, Krasilnikov MS, Zhitlov MY, Gulyak EL, Chistov AA, Nikitin TD, Korshun VA, Efremov RG, Alferova VA, Růžek D, Eyer L, Ustinov AV. Mikhnovets IE, et al. Int J Mol Sci. 2023 Nov 18;24(22):16483. doi: 10.3390/ijms242216483. Int J Mol Sci. 2023. PMID: 38003673 Free PMC article. - The Synthetic Peptide GA-Hecate and Its Analogs Inhibit Multiple Steps of the Chikungunya Virus Infection Cycle In Vitro.
Ayusso GM, da Silva Sanches PR, Carvalho T, Santos IA, Martins DOS, Lima MLD, da Conceição PJP, Bittar C, Merits A, Cilli EM, Jardim ACG, Rahal P, Calmon MF. Ayusso GM, et al. Pharmaceuticals (Basel). 2023 Sep 30;16(10):1389. doi: 10.3390/ph16101389. Pharmaceuticals (Basel). 2023. PMID: 37895860 Free PMC article. - Gastrointestinal Manifestations of SARS-CoV-2: Transmission, Pathogenesis, Immunomodulation, Microflora Dysbiosis, and Clinical Implications.
Durairajan SSK, Singh AK, Saravanan UB, Namachivayam M, Radhakrishnan M, Huang JD, Dhodapkar R, Zhang H. Durairajan SSK, et al. Viruses. 2023 May 24;15(6):1231. doi: 10.3390/v15061231. Viruses. 2023. PMID: 37376531 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R21 AI132479/AI/NIAID NIH HHS/United States
- R01 AI134907/AI/NIAID NIH HHS/United States
- R00 AG049092/AG/NIA NIH HHS/United States
- R21 AI126012/AI/NIAID NIH HHS/United States
- T32 AI007526/AI/NIAID NIH HHS/United States
- R24 AI120942/AI/NIAID NIH HHS/United States
- U19 AI100625/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous