The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase - PubMed (original) (raw)
. 2020 Jun;92(6):693-697.
doi: 10.1002/jmv.25761. Epub 2020 Mar 18.
Affiliations
- PMID: 32167173
- PMCID: PMC7228302
- DOI: 10.1002/jmv.25761
The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase
Jrhau Lung et al. J Med Virol. 2020 Jun.
Erratum in
- Corrigendum.
Lung J, Lin YS, Yang YH, Chou YL, Shu LH, Cheng YC, Te Liu H, Wu CY. Lung J, et al. J Med Virol. 2020 Oct;92(10):2248. doi: 10.1002/jmv.26176. Epub 2020 Jul 22. J Med Virol. 2020. PMID: 33411369 Free PMC article. No abstract available.
Abstract
An outbreak of coronavirus disease 2019 (COVID-19) occurred in Wuhan and it has rapidly spread to almost all parts of the world. For coronaviruses, RNA-dependent RNA polymerase (RdRp) is an important polymerase that catalyzes the replication of RNA from RNA template and is an attractive therapeutic target. In this study, we screened these chemical structures from traditional Chinese medicinal compounds proven to show antiviral activity in severe acute respiratory syndrome coronavirus (SARS-CoV) and the similar chemical structures through a molecular docking study to target RdRp of SARS-CoV-2, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV). We found that theaflavin has a lower idock score in the catalytic pocket of RdRp in SARS-CoV-2 (-9.11 kcal/mol), SARS-CoV (-8.03 kcal/mol), and MERS-CoV (-8.26 kcal/mol) from idock. To confirm the result, we discovered that theaflavin has lower binding energy of -8.8 kcal/mol when it docks in the catalytic pocket of SARS-CoV-2 RdRp by using the Blind Docking server. Regarding contact modes, hydrophobic interactions contribute significantly in binding and additional hydrogen bonds were found between theaflavin and RdRp. Moreover, one π-cation interaction was formed between theaflavin and Arg553 from the Blind Docking server. Our results suggest that theaflavin could be a potential SARS-CoV-2 RdRp inhibitor for further study.
Keywords: RNA-dependent RNA polymerase; SARS-CoV-2; theaflavin; traditional Chinese medicinal compounds.
© 2020 Wiley Periodicals, Inc.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures
Figure 1
(A) Sequence alignment for the amino acids of RdRp between SARS‐CoV‐2, SARS‐CoV, and MERS‐CoV. (B) A modeled structure of SARS‐CoV‐2 RdRp, SARS‐CoV RdRp, and MERS‐CoV RdRp and Grid box size for binding site using MODELLER. The active site (Val557) and grid box size (light yellow) were showed for binding site. MERS, Middle East respiratory syndrome; RdRp, RNA‐dependent RNA polymerase; SARS‐CoV, severe acute respiratory syndrome coronavirus
Figure 2
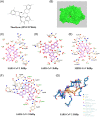
(A) The structure of theaflavin (ZINC3978446). (B) Red and green molecules, respectively, represent crystallographic and predicted pose for theaflavin. (C‐E) The contact model between theaflavin and SARS‐CoV‐2 RdRp (C), SARS‐CoV RdRp (D), and MERS‐CoV RdRp (E) are shown in two‐dimensional (2D) interaction diagram by idock. (F) The contact model between theaflavin and SARS‐CoV‐2 RdRp is shown in the 2D interaction diagram by the Blind Dock server. Their relative distances between amino acid residues and theaflavin are analyzed and illustrated by LigPlot+. Carbon, oxygen, nitrogen, and fluoride molecules are marked as white, red, blue, and green circles, respectively. Covalent bonds in theaflavin and amino acid residues of RdRp are labeled in purple and orange solid lines, respectively. The light blue dot lines label the distance (in Å) of hydrogen bonds formed between the functional moieties of theaflavin and amino acid residues. Hydrophobic interactions between theaflavin and RdRp are depicted by the name of involving amino acid residues, which are labeled with dark green with dark red eyelashes pointing to the involved functional moiety of theaflavin. (G) The hydrogen bonds and π‐cation interaction established by theaflavin with the closest residues are showed through Protein‐Ligand Interaction Profiler (PLIP). MERS, Middle East respiratory syndrome; RdRp, RNA‐dependent RNA polymerase; SARS‐CoV, severe acute respiratory syndrome coronavirus
Similar articles
- Anti-HCV, nucleotide inhibitors, repurposing against COVID-19.
Elfiky AA. Elfiky AA. Life Sci. 2020 May 1;248:117477. doi: 10.1016/j.lfs.2020.117477. Epub 2020 Feb 28. Life Sci. 2020. PMID: 32119961 Free PMC article. - Theaflavin-3'-O-gallate a Black-tea Constituent Blocked SARS CoV-2 RNA dependant RNA Polymerase Active-site with Better Docking Results than Remdesivir.
Banerjee A, Kanwar M, Maiti S. Banerjee A, et al. Drug Res (Stuttg). 2021 Oct;71(8):462-472. doi: 10.1055/a-1467-5828. Epub 2021 Sep 13. Drug Res (Stuttg). 2021. PMID: 34517419 - Intermolecular interaction among Remdesivir, RNA and RNA-dependent RNA polymerase of SARS-CoV-2 analyzed by fragment molecular orbital calculation.
Kato K, Honma T, Fukuzawa K. Kato K, et al. J Mol Graph Model. 2020 Nov;100:107695. doi: 10.1016/j.jmgm.2020.107695. Epub 2020 Jul 15. J Mol Graph Model. 2020. PMID: 32702590 Free PMC article. - Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.
Akaji K, Konno H. Akaji K, et al. Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review. - RNA-Dependent RNA Polymerase as a Target for COVID-19 Drug Discovery.
Zhu W, Chen CZ, Gorshkov K, Xu M, Lo DC, Zheng W. Zhu W, et al. SLAS Discov. 2020 Dec;25(10):1141-1151. doi: 10.1177/2472555220942123. Epub 2020 Jul 13. SLAS Discov. 2020. PMID: 32660307 Free PMC article. Review.
Cited by
- Systematic Studies on the Anti-SARS-CoV-2 Mechanisms of Tea Polyphenol-Related Natural Products.
Li CW, Chao TL, Lai CL, Lin CC, Pan MY, Cheng CL, Kuo CJ, Wang LH, Chang SY, Liang PH. Li CW, et al. ACS Omega. 2024 May 17;9(22):23984-23997. doi: 10.1021/acsomega.4c02392. eCollection 2024 Jun 4. ACS Omega. 2024. PMID: 38854515 Free PMC article. - Natural compounds may contribute in preventing SARS-CoV-2 infection: a narrative review.
Bizzoca ME, Leuci S, Mignogna MD, Muzio EL, Caponio VCA, Muzio LL. Bizzoca ME, et al. Food Sci Hum Wellness. 2022 Sep;11(5):1134-1142. doi: 10.1016/j.fshw.2022.04.005. Epub 2022 Jun 2. Food Sci Hum Wellness. 2022. PMID: 38621001 Free PMC article. Review. - Natural medicinal plant products as an immune-boosters: A possible role to lessen the impact of Covid-19.
Islam SS, Midya S, Sinha S, Saadi SMAI. Islam SS, et al. Case Stud Chem Environ Eng. 2021 Dec;4:100105. doi: 10.1016/j.cscee.2021.100105. Epub 2021 May 5. Case Stud Chem Environ Eng. 2021. PMID: 38620656 Free PMC article. - The Potential of Usnic-Acid-Based Thiazolo-Thiophenes as Inhibitors of the Main Protease of SARS-CoV-2 Viruses.
Yarovaya OI, Filimonov AS, Baev DS, Borisevich SS, Zaykovskaya AV, Chirkova VY, Marenina MK, Meshkova YV, Belenkaya SV, Shcherbakov DN, Gureev MA, Luzina OA, Pyankov OV, Salakhutdinov NF, Khvostov MV. Yarovaya OI, et al. Viruses. 2024 Jan 31;16(2):215. doi: 10.3390/v16020215. Viruses. 2024. PMID: 38399993 Free PMC article. - Application of artificial intelligence (AI) to control COVID-19 pandemic: Current status and future prospects.
Ashique S, Mishra N, Mohanto S, Garg A, Taghizadeh-Hesary F, Gowda BHJ, Chellappan DK. Ashique S, et al. Heliyon. 2024 Feb 9;10(4):e25754. doi: 10.1016/j.heliyon.2024.e25754. eCollection 2024 Feb 29. Heliyon. 2024. PMID: 38370192 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous