Subtraction hybridization identifies a transformation progression-associated gene PEG-3 with sequence homology to a growth arrest and DNA damage-inducible gene - PubMed (original) (raw)
Subtraction hybridization identifies a transformation progression-associated gene PEG-3 with sequence homology to a growth arrest and DNA damage-inducible gene
Z Z Su et al. Proc Natl Acad Sci U S A. 1997.
Free PMC article
Erratum in
- Proc Natl Acad Sci U S A 1997 Oct 28;94(22):12241
Abstract
Cancer is a progressive multigenic disorder characterized by defined changes in the transformed phenotype that culminates in metastatic disease. Determining the molecular basis of progression should lead to new opportunities for improved diagnostic and therapeutic modalities. Through the use of subtraction hybridization, a gene associated with transformation progression in virus- and oncogene-transformed rat embryo cells, progression elevated gene-3 (PEG-3), has been cloned. PEG-3 shares significant nucleotide and amino acid sequence homology with the hamster growth arrest and DNA damage-inducible gene gadd34 and a homologous murine gene, MyD116, that is induced during induction of terminal differentiation by interleukin-6 in murine myeloid leukemia cells. PEG-3 expression is elevated in rodent cells displaying a progressed-transformed phenotype and in rodent cells transformed by various oncogenes, including Ha-ras, v-src, mutant type 5 adenovirus (Ad5), and human papilloma virus type 18. The PEG-3 gene is transcriptionally activated in rodent cells, as is gadd34 and MyD116, after treatment with DNA damaging agents, including methyl methanesulfonate and gamma-irradiation. In contrast, only PEG-3 is transcriptionally active in rodent cells displaying a progressed phenotype. Although transfection of PEG-3 into normal and Ad5-transformed cells only marginally suppresses colony formation, stable overexpression of PEG-3 in Ad5-transformed rat embryo cells elicits the progression phenotype. These results indicate that PEG-3 is a new member of the gadd and MyD gene family with similar yet distinct properties and this gene may directly contribute to the transformation progression phenotype. Moreover, these studies support the hypothesis that constitutive expression of a DNA damage response may mediate cancer progression.
Figures
Figure 1
PEG-3 expression in Ad5-transformed RE cells displaying different stages of transformation progression. Fifteen micrograms of cellular RNA isolated from the indicated cell types were electrophoresed, transferred to nylon membranes, and hybridized with an ≈700 bp 3′ region of the PEG-3 gene (Upper) and then stripped and probed with GAPDH (Lower).
Figure 2
PEG-3 expression in γ-irradiated and oncogene-transformed CREF cells. The experimental procedure was as described in the legend to Fig. 1. CREF cells were γ-irradiated with 10 Gy and RNA was isolated 4 and 24 hr later.
Figure 3
Predicted amino acid sequences of the PEG-3, gadd34, and MyD116 proteins. Sequences shared by the three genes are shaded. PEG-3 encodes a putative protein of 457 aa (_M_r of ≈50 kDa), the _gadd34_gene encodes a putative protein of 589 aa (M_r of ≈65 kDa), and the_MyD116 gene encodes a putative protein of 657 aa (_M_r of ≈72 kDa).
Figure 4
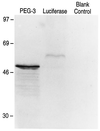
In vitro translation of the_PEG-3_ gene. Lanes: Luciferase, _in vitro_translation of the luciferase gene (≈61 kDa) (positive control); Blank Control, contains the same reaction mixture without mRNA (negative control); PEG-3, contains the translated products of this cDNA. Rainbow protein standards (Amersham) were used to determine the sizes of the in vitro translated products.
Figure 5
Transcription of the PEG-3,gadd34, and MyD116 genes as a function of DNA damage and transformation progression. Nuclear run-on assays were performed to determine comparative rates of transcription. Nuclei were isolated from CREF cells treated with methyl methanesulfonate (MMS) (100 μg/ml for 2 hr followed by growth for 4 hr in complete medium) or γ-irradiation (10 Gy followed by 2 hr growth in complete medium). DNA probes include PEG-3, MyD116,gadd34, GAPDH, and pBR322.
Figure 6
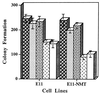
Effect of transfection with PEG-3, mda-7, and p21 (mda-6) on colony formation of E11 and E11-NMT cells in monolayer culture. Target cells were transfected with 10 μg of a Zeocin vector (pZeoSV), the PEG-3 gene cloned in pZeoSV (PEG-3), the pREP4 vector, the mda-7 gene cloned in pREP4 (mda-7), and the mda-6 (p21) gene cloned in pREP4 (p21 (mda-6). Data represent the average number of Zeocin- or hygromycin (pREP4 transfection)-resistant colonies ± SD for four plates seeded at 1 × 105 cells per 6-cm plate. , Zeocin vector; ░⃞, PEG-3;
, Zeocin vector; ░⃞, PEG-3;  , pREP4 vector; ▩, mda-7; □, p21 (mda-6).
, pREP4 vector; ▩, mda-7; □, p21 (mda-6).
Figure 7
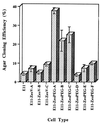
Effect of stable PEG-3 expression on anchorage-independent growth of E11 cells. Agar cloning efficiency of E11, Zeocin-resistant pZeoV (vector)-transfected E11, and Zeocin-resistant pZeoPEG-transfected E11 cells. Average number of colonies developing in four replicate plates ± SD.
Figure 8
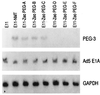
Expression of PEG-3, Ad5 E1A, and GAPDH RNA in pZeoPEG-transfected E11 cells. The experimental procedure was as described in the legend to Fig. 1. Blots were probed sequentially with PEG-3, Ad5 E1A, and GAPDH. The E11-ZeoPEG clones are the same clones analyzed for anchorage independence in Fig. 7.
Similar articles
- PEA3 sites within the progression elevated gene-3 (PEG-3) promoter and mitogen-activated protein kinase contribute to differential PEG-3 expression in Ha-ras and v-raf oncogene transformed rat embryo cells.
Su Z, Shi Y, Friedman R, Qiao L, McKinstry R, Hinman D, Dent P, Fisher PB. Su Z, et al. Nucleic Acids Res. 2001 Apr 15;29(8):1661-71. doi: 10.1093/nar/29.8.1661. Nucleic Acids Res. 2001. PMID: 11292838 Free PMC article. - Potential molecular mechanism for rodent tumorigenesis: mutational generation of Progression Elevated Gene-3 (PEG-3).
Su ZZ, Emdad L, Sarkar D, Randolph A, Valerie K, Yacoub A, Dent P, Fisher PB. Su ZZ, et al. Oncogene. 2005 Mar 24;24(13):2247-55. doi: 10.1038/sj.onc.1208420. Oncogene. 2005. PMID: 15674324 - Differentiation primary response genes and proto-oncogenes as positive and negative regulators of terminal hematopoietic cell differentiation.
Liebermann DA, Hoffman B. Liebermann DA, et al. Stem Cells. 1994 Jul;12(4):352-69. doi: 10.1002/stem.5530120402. Stem Cells. 1994. PMID: 7951003 Review. - Myeloid differentiation (MyD)/growth arrest DNA damage (GADD) genes in tumor suppression, immunity and inflammation.
Liebermann DA, Hoffman B. Liebermann DA, et al. Leukemia. 2002 Apr;16(4):527-41. doi: 10.1038/sj.leu.2402477. Leukemia. 2002. PMID: 11960329 Review.
Cited by
- PEA3 sites within the progression elevated gene-3 (PEG-3) promoter and mitogen-activated protein kinase contribute to differential PEG-3 expression in Ha-ras and v-raf oncogene transformed rat embryo cells.
Su Z, Shi Y, Friedman R, Qiao L, McKinstry R, Hinman D, Dent P, Fisher PB. Su Z, et al. Nucleic Acids Res. 2001 Apr 15;29(8):1661-71. doi: 10.1093/nar/29.8.1661. Nucleic Acids Res. 2001. PMID: 11292838 Free PMC article. - Conversion of a Non-Cancer-Selective Promoter into a Cancer-Selective Promoter.
Bhoopathi P, Pradhan AK, Kumar A, Maji S, Mannangatti P, Deng X, Bandyopadhyay D, Sarkar D, Wang XY, Landry JW, Das SK, Emdad L, Fisher PB. Bhoopathi P, et al. Cancers (Basel). 2022 Mar 15;14(6):1497. doi: 10.3390/cancers14061497. Cancers (Basel). 2022. PMID: 35326649 Free PMC article. - Cancer imaging: Gene transcription-based imaging and therapeutic systems.
Bhang HE, Pomper MG. Bhang HE, et al. Int J Biochem Cell Biol. 2012 May;44(5):684-9. doi: 10.1016/j.biocel.2012.02.001. Epub 2012 Feb 10. Int J Biochem Cell Biol. 2012. PMID: 22349219 Free PMC article. Review. - MRI detection of the malignant transformation of stem cells through reporter gene expression driven by a tumor-specific promoter.
Sun J, Huang J, Bao G, Zheng H, Wang C, Wei J, Fu Y, Qiu J, Liao Y, Cai J. Sun J, et al. Stem Cell Res Ther. 2021 May 12;12(1):284. doi: 10.1186/s13287-021-02359-w. Stem Cell Res Ther. 2021. PMID: 33980305 Free PMC article. - mda-7 (IL-24) Mediates selective apoptosis in human melanoma cells by inducing the coordinated overexpression of the GADD family of genes by means of p38 MAPK.
Sarkar D, Su ZZ, Lebedeva IV, Sauane M, Gopalkrishnan RV, Valerie K, Dent P, Fisher PB. Sarkar D, et al. Proc Natl Acad Sci U S A. 2002 Jul 23;99(15):10054-9. doi: 10.1073/pnas.152327199. Epub 2002 Jul 11. Proc Natl Acad Sci U S A. 2002. PMID: 12114539 Free PMC article.
References
- Fisher P B. In: Tumor Promotion and Cocarcinogenesis in Vitro: Mechanisms of Tumor Promotion. Slaga T J, editor. Boca Raton, FL: CRC; 1984. pp. 57–123.
- Bishop J M. Cell. 1991;64:235–248. - PubMed
- Vogelstein B, Kinzler K W. Trends Genet. 1991;9:138–141. - PubMed
- Levine A J. Annu Rev Biochem. 1993;62:623–651. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous