Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA - PubMed (original) (raw)
Comparative Study
Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA
U A Ochsner et al. J Bacteriol. 1999 Feb.
Abstract
A novel outer membrane lipoprotein in Pseudomonas aeruginosa is encoded by the omlA gene, which was identified immediately upstream of the fur (ferric uptake regulator) gene. The omlA and fur genes were divergently transcribed and had overlapping promoter regions. The proximal fur P2 promoter and the omlA promoter shared a 5-bp DNA motif for their -10 promoter elements. The distal fur P1 promoter was located within the omlA coding sequence, and the omlA and fur T1 mRNAs overlapped by 154 nucleotides. Optimal expression of both fur and omlA required roughly 200 bp of DNA upstream of the promoter regions, suggesting the presence of cis-acting transcriptional activation elements located within the omlA and fur genes, respectively. The levels of Fur and OmlA proteins had no influence on omlA or fur expression, excluding any trans-acting cross-regulation between fur and omlA. Expression of omlA was constitutive regardless of growth phase, oxygen tension, iron concentration, pH, and temperature. OmlA contained a signal sequence typical of bacterial lipoproteins, with a cysteine as a putative cleavage and lipid attachment site. Inhibition of signal peptidase II by globomycin resulted in failure to process OmlA, thus giving strong evidence that OmlA is a lipoprotein. Cell fractionation followed by Western blot analysis indicated that all OmlA protein is localized in the outer membrane. Mature OmlA was an acidic (pI = 4. 5) protein of 17.3 kDa and had close to 40% amino acid sequence identity to SmpA (small protein A) of Escherichia coli, Vibrio cholerae, and Haemophilus influenzae, a protein of unknown function. All P. aeruginosa strains tested as well as Pseudomonas fluorescens were found to produce OmlA. A mutant strain with impaired production of OmlA but no change in the expression of the overlapping fur gene was constructed. The omlA mutant was hypersusceptible to anionic detergents such as sodium dodecyl sulfate and deoxycholate, and it showed increased susceptibility to various antibiotics, including nalidixic acid, rifampin, novobiocin, and chloramphenicol. A structural role of OmlA in maintaining the cell envelope integrity is proposed.
Figures
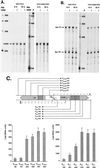
FIG. 1
(A) P. aeruginosa ORF1-ORF2-omlA-fur region. The transcripts are indicated by arrows above the corresponding genes, and an experimentally confirmed transcriptional terminator is shown between ORF2 and omlA. Also shown are the homologous regions of E. coli and V. cholerae, with the corresponding genes indicated by thick dashed arrows. (B) DNA sequence of the omlA gene. The corresponding amino acid sequence of OmlA is given below the DNA coding region. Also shown is the start of the divergent fur gene on the opposite DNA strand, with the amino-terminal protein sequence given above the 5′ fur coding sequence. Promoter elements for omlA and fur are indicated by brackets, dashed arrows represent transcriptional start sites for omlA, fur T1, and fur T2, and a transcriptional terminator is shown by head-to-head arrows. Also included are translational signals such as Shine-Dalgarno sequences (S/D) and the translational start sites for both omlA and fur. (C) Amino acid sequence alignment of OmlA from P. aeruginosa and P. fluorescens with SmpA from H. influenzae (GenBank no. 1175311), V. cholerae (GenBank no. U39068), and E. coli (GenBank no. D90888).
FIG. 2
(A) RNase protection analysis of omlA expression. A 444-nucleotide riboprobe spanning the region from 343 to −101 of the omlA sequence was hybridized to total RNA isolated from P. aeruginosa PAO1 grown for 6 or 10 h aerobically or microaerobically under low-iron (−) or high-iron (+) iron conditions. The protected omlA mRNA is indicated with an arrow. (B) RNase protection analysis of fur expression. The 444-nucleotide riboprobe covered from −101 to 343 of the omlA sequence, and the protected transcripts T1 and T2 are indicated by arrows. (C) Expression of omlA and fur. The map of the fur-omlA locus shows the omlA promoter PA, the fur promoters P1 and P2, the relevant transcripts (dashed arrows), and regions for cis activation (hatched boxes). Promoter fragments (e.g., PomlA, Pfur) of increasing size as indicated by arrows were translationally fused to the lacZ reporter gene. The β-galactosidase activities were measured in triplicate cultures grown for 4 h in D-TSB medium.
FIG. 3
(A) Locations of single base pair mutations in CS and in its revertants, CSR#0, CSR#1, and CSR#5. The relevant promoter elements, transcripts, and translational motifs within the omlA-fur intergenic region are indicated. S/D, Shine-Dalgarno site. (B) RNase protection of omlA (left) and fur (right) transcripts in PAO1, CS, CSR#0, and CSR#1. The RNA was isolated after 8 h of growth in high-iron D-TSB at 25°C. (C) Western blot analyses of OmlA (top) and Fur (bottom) proteins in CS, CSR#0, CSR#1, CSR#5, and wild-type PAO1. The cells were grown for 10 h in high-iron D-TSB at 25°C, and whole-cell extract samples were prepared and normalized for cell densities.
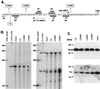
FIG. 4
(A) Autoradiography of overexpressed and radiolabeled OmlA. The omlA gene was expressed in a T7 system in E. coli/pET-omlA in the absence or presence of globomycin. Also shown is the pET23a vector control. (B through G) Western blot analysis of P. aeruginosa cell fractions, standardized GST-OmlA, and total protein from 1.35 × 107 PAO1 cells and of whole-cell extracts prepared from different P. aeruginosa wild-type strains, from P. fluorescens, from omlA mutants, and from a complemented omlA strain carrying pOML24. (H) Outer membrane protein profiles of wild-type PAO1 and omlA mutants on a Coomassie-stained 15% acrylamide SDS gel.
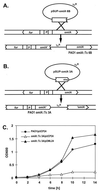
FIG. 5
(A) Construction of PAO1 omlA::Tc mutant 6B. The integration of the plasmid creates a promoterless omlA gene without affecting the divergent fur gene. (B) Construction of PAO1 omlA::Tc mutant 3A. The single crossover at omlA generates a truncated omlA gene due to an early stop codon. (C) Growth inhibition of 3A in M9 medium containing 0.1% SDS. Growth curves are shown for PAO1/pUCP24 (wild type with a control plasmid), 3A/pUCP24 (omlA mutant with a control plasmid), and 3A/pOML24 (omlA mutant genetically complemented with the omlA gene on a multicopy plasmid).
Similar articles
- The omlA gene is involved in multidrug resistance and its expression is inhibited by coumarins in Xanthomonas campestris pv. phaseoli.
Fuangthong M, Sallabhan R, Atichartpongkul S, Rangkadilok N, Sriprang R, Satayavivad J, Mongkolsuk S. Fuangthong M, et al. Arch Microbiol. 2008 Mar;189(3):211-8. doi: 10.1007/s00203-007-0310-1. Epub 2007 Oct 24. Arch Microbiol. 2008. PMID: 17957353 - The Burkholderia cepacia fur gene: co-localization with omlA and absence of regulation by iron.
Lowe CA, Asghar AH, Shalom G, Shaw JG, Thomas MS. Lowe CA, et al. Microbiology (Reading). 2001 May;147(Pt 5):1303-1314. doi: 10.1099/00221287-147-5-1303. Microbiology (Reading). 2001. PMID: 11320133 - Cloning, sequencing, and transcriptional regulation of viuA, the gene encoding the ferric vibriobactin receptor of Vibrio cholerae.
Butterton JR, Stoebner JA, Payne SM, Calderwood SB. Butterton JR, et al. J Bacteriol. 1992 Jun;174(11):3729-38. doi: 10.1128/jb.174.11.3729-3738.1992. J Bacteriol. 1992. PMID: 1317381 Free PMC article. - Cloning and sequence analysis of a gene (pchR) encoding an AraC family activator of pyochelin and ferripyochelin receptor synthesis in Pseudomonas aeruginosa.
Heinrichs DE, Poole K. Heinrichs DE, et al. J Bacteriol. 1993 Sep;175(18):5882-9. doi: 10.1128/jb.175.18.5882-5889.1993. J Bacteriol. 1993. PMID: 8397186 Free PMC article. - Genome-wide analysis and literature-based survey of lipoproteins in Pseudomonas aeruginosa.
Remans K, Vercammen K, Bodilis J, Cornelis P. Remans K, et al. Microbiology (Reading). 2010 Sep;156(Pt 9):2597-2607. doi: 10.1099/mic.0.040659-0. Epub 2010 Jul 8. Microbiology (Reading). 2010. PMID: 20616104 Review.
Cited by
- Cell Envelope Stress Response in Pseudomonas aeruginosa.
Chevalier S, Bouffartigues E, Tortuel D, David A, Tahrioui A, Labbé C, Barreau M, Tareau AS, Louis M, Lesouhaitier O, Cornelis P. Chevalier S, et al. Adv Exp Med Biol. 2022;1386:147-184. doi: 10.1007/978-3-031-08491-1_6. Adv Exp Med Biol. 2022. PMID: 36258072 - Tracing phylogenomic events leading to diversity of Haemophilus influenzae and the emergence of Brazilian Purpuric Fever (BPF)-associated clones.
Papazisi L, Ratnayake S, Remortel BG, Bock GR, Liang W, Saeed AI, Liu J, Fleischmann RD, Kilian M, Peterson SN. Papazisi L, et al. Genomics. 2010 Nov;96(5):290-302. doi: 10.1016/j.ygeno.2010.07.005. Epub 2010 Jul 30. Genomics. 2010. PMID: 20654709 Free PMC article. - Genetic and physiological characterization of ohr, encoding a protein involved in organic hydroperoxide resistance in Pseudomonas aeruginosa.
Ochsner UA, Hassett DJ, Vasil ML. Ochsner UA, et al. J Bacteriol. 2001 Jan;183(2):773-8. doi: 10.1128/JB.183.2.773-778.2001. J Bacteriol. 2001. PMID: 11133975 Free PMC article. - Identification, isolation, and analysis of a gene cluster involved in iron acquisition by Pseudomonas mendocina ymp.
Awaya JD, Dubois JL. Awaya JD, et al. Biometals. 2008 Jun;21(3):353-66. doi: 10.1007/s10534-007-9124-5. Epub 2007 Dec 6. Biometals. 2008. PMID: 18058194 Free PMC article. - The peptidoglycan-associated lipoprotein OprL helps protect a Pseudomonas aeruginosa mutant devoid of the transactivator OxyR from hydrogen peroxide-mediated killing during planktonic and biofilm culture.
Panmanee W, Gomez F, Witte D, Pancholi V, Britigan BE, Hassett DJ. Panmanee W, et al. J Bacteriol. 2008 May;190(10):3658-69. doi: 10.1128/JB.00022-08. Epub 2008 Feb 29. J Bacteriol. 2008. PMID: 18310335 Free PMC article.
References
- Barton H A, Johnson Z, Cox C D, Vasil A I, Vasil M L. Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol Microbiol. 1996;21:1001–1017. - PubMed
- Bertrand K P, Postle K, Wray L V, Jr, Reznikoff W S. Overlapping divergent promoters control expression of Tn10 tetracycline resistance. Gene. 1983;23:149–156. - PubMed
- Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous