HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype (original) (raw)
Abstract
Some rare HIV-1-infected individuals, referred to as HIV controllers (HIC), have persistently undetectable plasma viral load in the absence of therapy. This control of HIV-1 replication has been associated with a strong, multifunctional specific CD8+ T cell response. However, no direct link between this immune response and the control of viremia has so far been provided. We investigated parameters of specific CD8+ T cell response and in vitro susceptibility to HIV-1 infection in 11 HIC. We found high frequencies of HIV-specific CD8+ T cells. Interestingly, these cells expressed the activation marker HLA-DR but not CD38. This unique phenotype differentiates HIV-specific CD8+ T cells from HIC and noncontroller subjects and likely reflects a high potential to expand upon exposure to antigen and a capacity to exert effector functions. Accordingly, although CD4+ T cells from HIC were fully susceptible to HIV-1 superinfection, their CD8+ T cells effectively suppressed HIV-1 infection. Remarkably, this potent anti-HIV activity was observed without prior stimulation of CD8+ T cells. This activity was not mediated by secreted inhibitory factors but was due to the elimination of infected CD4+ T cells and was observed only with autologous CD4+ T cells, indicating an HLA-restricted cytotoxic mechanism. This constitutive antiviral capacity of CD8+ T cells could account for the control of viral replication in HIC.
Keywords: HIV suppression, CD8+ T cells, HLA-DR, CD38
Most untreated HIV-1-infected individuals have continuous viral replication and ultimately progress to AIDS. However, a rare subpopulation of HIV-infected patients spontaneously control viral replication for long periods in the absence of treatment (1–5). These individuals, referred to here as HIV controllers (HIC), are characterized by undetectable plasma HIV-1 RNA. Some HIC have been found to be infected by replication-incompetent viruses (6). However, a potent immune response to HIV-1 is thought to be pivotal in these patients (3, 7, 8).
Actually, HIC generally exhibit a strong CD8+ T cell specific response and high frequencies of HIV-specific CD8+ T cells despite very low levels of viral antigens (3, 4, 7). Furthermore, (i) HIV-specific CD8+ T cells from HIC are qualitatively different from those of progressors (8, 9); (ii) some HLA-Bw4 haplotypes (e.g., B27 and B57) are overrepresented in HIC (10, 11), suggesting an important role of class I-restricted CD8+ T cells; and (iii) multiepitopic and de novo CD8+ T cell responses are associated with suppression of viremia despite cytotoxic T lymphocyte escape mutations (12).
However, the mechanisms by which CD8+ T cells restrain HIV-1 infection in HIC are still unclear. The remarkable spontaneous viral control in HIC offers a unique model in which to shed some light on efficient in vivo mechanisms of CD8+ T cell-mediated HIV-1 control. We characterized the parameters of the CD8+ T cell response in 11 HIC from a previously described group (5), providing evidence that their HIV-specific CD8+ T cells possess a unique phenotype. In addition, we demonstrated an extraordinary capacity of their CD8+ T cells ex vivo to suppress HIV-1 infection.
Results
Study Population.
The characteristics of the 11 HIC are reported in Table 1. HLA alleles B57 and B27, associated with protection from progression to AIDS (10, 11), were overrepresented (seven and four subjects possessed these alleles, respectively).
Table 1.
Characteristics of the 11 HIC in whom plasma HIV RNA load had been undetectable for >10 years in the absence of antiretroviral treatment
| Patients | Sex/age | HIV diagnosis | CD4 count | HIV RNA | HIV DNA | HLA class I |
|---|---|---|---|---|---|---|
| A1 | F/49 | 1988 | 982/1,010 | <50/<50 | 3 | A02/—/B27/B57 |
| A2 | M/48 | 1985 | 753/786 | <50/<50 | 7 | A03/A23/B7/B57 |
| A3 | F/42 | 1993 | 876/1,159 | <50/<50 | Negative | A03/A30/B15/B57 |
| A4 | M/47 | 1987 | 592/749 | <50/<50 | 80 | A02/A32/B27/B40 |
| A5 | M/42 | 1991 | 728/740 | <50/<50 | 65 | A02/A26/B38/B44 |
| A6 | M/69 | 1985 | 706/524 | <50/150 | 40 | A02/—/B27/B57 |
| A7 | M/38 | 1983 | 609/580 | <50/<50 | 15 | A02/—/B44/B57 |
| A9 | M/44 | 1992 | 844/522 | <50/<50 | 60 | A02/A29/B27/B57 |
| A10 | F/48 | 1985 | 1,033/1,036 | <50/<50 | 47 | A03/A11/B44/B78 |
| B2 | F/40 | 1987 | 928/860 | <50/180 | 170 | A02/A26/B44/B64 |
| B5 | M/57 | 1989 | 799/800 | <50/<50 | 33 | A02/A68/B1402/B57 |
HIC Exhibit Large Numbers of HIV-Specific CD8+ T Cells with Variable Maturation Profiles.
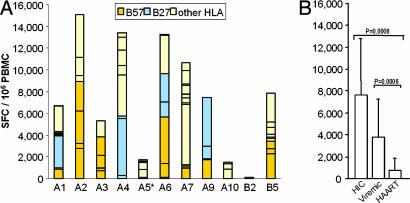
High frequencies of HIV-specific CD8+ T cells [7,546 ± 5,121 spot-forming cells (SFC) per 106 peripheral blood mononuclear cells (PBMC)] with a broad response (7 ± 3 peptides recognized) were observed in most HIC (Fig. 1A). Subjects bearing either the HLA-B57 or -B27 allele had the highest frequencies of HIV-specific CD8+ T cells. These frequencies were not significantly different from those of viremic controls (3,719 ± 3,455 SFC per 106 PBMC) but significantly higher than those of highly active antiretroviral therapy (HAART) subjects (758 ± 1,025 SFC per 106 PBMC) (Fig. 1B).
Fig. 1.
Frequencies of HIV-specific IFN-γ-secreting CD8+ T cells in HIC, viremic, and HAART subjects. (A) For all subjects (except A5) an average of 41 ± 9 individual peptides were tested, depending on the results of HLA typing. Each bar corresponds to the sum of SFC per 106 PBMC obtained with peptides described to be restricted by HLA-B57 (orange), HLA-B27 (blue), or other HLA antigens (yellow). ∗, For subject A5, whose HLA-typing was lacking at the time of the study, 12 pools of optimal peptides were used instead of individual peptides. The bar for A5 corresponds to the sum of SFC per 106 PBMC obtained with these pools. (B) Comparison of SFC (mean ± SD) in PBMC from HIC, viremic, and HAART subjects. Statistical differences in SFC between groups are indicated above the bars.
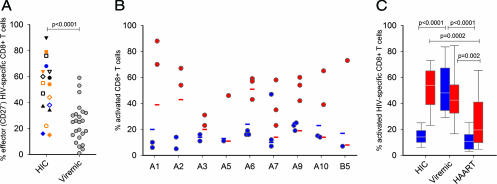
The maturation status of HIV-specific CD8+ T cells was analyzed by measuring the membrane expression of CD45RO and the differentiation marker CD27. This combination readily identifies effector cells as being CD27− [CD27−/CD45RO+ defining the classical effector cells and CD27−/CD45RO− (i.e., CD45RA+) defining the terminally differentiated CD8+ T cells (13)]. Overall, higher percentages of CD27− (effector) HIV-specific CD8+ T cells were observed in HIC than in viremic subjects (Fig. 2A). However, the profiles were extremely heterogeneous within HIC [Fig. 2A and supporting information (SI) Fig. 5], and only a minority of cells expressed high levels of perforin (2.3 ± 1.6%) whatever their maturation profile.
Fig. 2.
Differentiation and activation status of HIV-specific CD8+ T cells from HIC and comparison with viremic and HAART subjects. (A) Individual HIV-specific CD8+ CD27- T cell frequencies on CD8+ tetramer+ cells (left) in nine HIC (each symbol represents one individual, one to three specificities were tested per subject) and viremic subjects. For HIC, results obtained with B*2705-gag 263–272 and B*5701-gag 162–172 tetramers are represented in blue and orange, respectively. (B) Frequencies of CD38 (blue) and HLA-DR (red) expression on total CD8+ T cells (horizontal bars) and HIV-specific CD8+ T cells (circles) in nine HIC. (C) Comparison of CD38 (blue) and HLA-DR (red) expression (median, 25th–75th and 10th–90th percentiles) on HIV-specific CD8+ T cells in HIC, viremic, and HAART groups. Statistical differences between groups are indicated above the bars.
HIV-Specific HIC CD8+ T Cells Possess an Unusual Activation Profile.
We examined the activation status of CD8+ T cells by evaluating HLA-DR and CD38 expression. HIV-specific CD8+ T cells showed a low expression of CD38 (15 ± 9%), similar to that observed among the total CD8+ T cell population (19 ± 5%) (Fig. 2B). In contrast, HIV-specific CD8+ T cells expressed a high level of HLA-DR (52 ± 17%), higher than that observed among total CD8+ T cells (25 ± 16%, P = 0.001) (Fig. 2B). These results may suggest an activation of HIV-specific CD8+ T cells, which is unexpected in subjects with undetectable plasma RNA levels and puzzling in the absence of high CD38 expression (SI Fig. 6_A_). Actually, HIV-specific CD8+ T cells from viremic subjects exhibited high expression of both CD38 and HLA-DR (51 ± 20% and 46 ± 21%, respectively), whereas those from HAART subjects had low expressions of these two markers (11 ± 8% and 26 ± 20% for CD38 and HLA-DR, respectively) (Fig. 2C and SI Fig. 6_A_). In addition to activation, expression of HLA-DR has been associated with proliferation (14). HIV-specific CD8+ T cells from HIC had a high proliferation capacity (on average, 60 ± 31% of HIV-specific CD8+ T cells proliferated) that was positively correlated to the expression of HLA-DR (P = 0.03, Spearman's rank correlation) (data not shown). This profile may reflect the capacity to proliferate rather than ongoing proliferation in vivo, because Ki-67 expression was low on ex vivo HIV-specific CD8+ T cells (data not shown). Consistent with a high proliferative potential, in proliferation assays all HIV-specific proliferating CD8+ T cells were HLA-DR+ (SI Fig. 6_B_). Finally, we observed a mutually exclusive expression of HLA-DR and the CD57 marker on HIV-specific CD8+ T cells (SI Fig. 6_C_ and data not shown).
Ex Vivo Unstimulated HIC CD8+ T Cells Control HIV-1 Infection.
Seeking a direct link between the particular CD8+ T cell response in HIC and the control of viremia, we next evaluated the capacity of circulating HIC CD8+ T cells to suppress, in the absence of exogenous stimulation, HIV-1 infection in autologous CD4+ T cells.
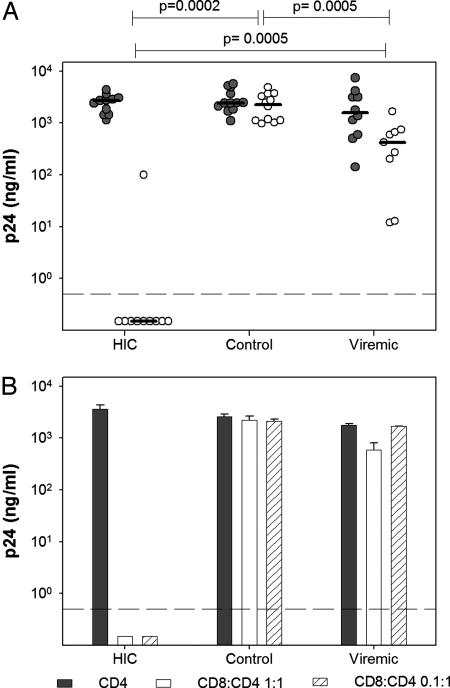
Upon superinfection, purified CD4+ T cells from HIC supported viral replication at levels similar to those observed in healthy controls (2,520 ± 997 and 2,729 ± 1,248 ng/ml p24 at the peak of viral replication in the 11 HIC and 12 healthy controls, respectively) (Fig. 3A). As expected (15), the levels of viral replication in CD4+ T cells from 10 untreated viremic HIV-1-infected individuals were highly heterogeneous (2,331 ± 2,204 ng/ml p24) (Fig. 3A).
Fig. 3.
Control of HIV-1 infection by ex vivo HIC CD8+ T cells. HIV-1 in vitro infection assays were done with cells from HIC, uninfected donors (control), or HIV viremic individuals [median plasma viral load was 25,350 RNA copies per milliliter (range 7,800–321,000)] (viremic). (A) PHA-activated CD4+ T cells were infected with the replicative HIV-1 BaL in the absence (gray) or presence (white) of autologous unstimulated CD8+ T cells (1:1 ratio). Circles represent the average (n = 3 independent infections) peak p24 values for each studied individual. Horizontal lines indicate median values for each group. Statistical differences in CD8+ T cell-mediated inhibition between groups are indicated above the graph. (B) PHA-activated CD4+ T cells (filled bars) and cocultures of autologous unstimulated CD8+ T cells and PHA-activated CD4+ T cells at ratios of 1:1 (open bars) and 0.1:1 (hatched bars) from five HIC, five uninfected blood donors (control), and nine HIV viremic individuals (viremic) were infected with HIV-1 BaL. One representative experiment (HIC A9) is shown. Bars represent peak levels of p24 in supernatants (mean ± SD, n = 3). Values below the dashed lines were at background level.
When cocultures of CD4+ T cells and autologous unstimulated CD8+ T cells at a 1:1 ratio were infected with HIV-1, viral replication was undetectable in 9 of the 10 HIC tested and strongly reduced in the remaining subject, B2 (4.0 ± 1.1 log p24 decrease; CD8:CD4 vs. CD4; n = 10) (Fig. 3A). HIC B2 interestingly also showed the lowest HIV-specific CD8+ T cell response (see Fig. 1A, 130 SFC per 106 PBMC; 1 peptide recognized). In contrast, unstimulated CD8+ T cells from healthy donors were devoid of antiviral activity (0.1 ± 0.2 log p24 decrease; CD8:CD4 vs. CD4; n = 12) and unstimulated CD8+ T cells from viremic individuals were never able to efficiently control HIV-1 superinfection in autologous CD4+ T cells (0.9 ± 0.6 log p24 decrease; CD8:CD4+ vs. CD4; n = 9) (Fig. 3A). Furthermore, the antiviral activity of CD8+ T cells from viremic individuals was rapidly lost when CD8+ T cells were diluted, whereas HIC CD8+ T cells were still fully effective at a ratio of 0.1 CD8+ T cell to 1 CD4+ T cell (Fig. 3B).
This remarkable anti-HIV-1 activity of unstimulated HIC CD8+ T cells was observed with a wide range of infectious doses (BaL HIV-1 multiplicity of infection = 10−2.8 to 10−0.8) (data not shown) and with both laboratory-adapted HIV strains and primary isolates, regardless of clade and tropism [HIV-1 NL4.3 (X4); 132W (dual tropic), subtype E; and DH12 (dual tropic) and BX08 (R5), subtype B] (data not shown). Experiments performed without exogenous IL-2 (that was regularly present in the coculture medium) showed, as expected, a reduced viral replication in infected CD4+ T cells, but the antiviral capacity of HIC unstimulated CD8+ T cells did not depend on the presence of the cytokine (SI Fig. 7).
Soluble Factors Are Not Responsible for CD8+ T Cell-Mediated HIV-1 Control in HIC.
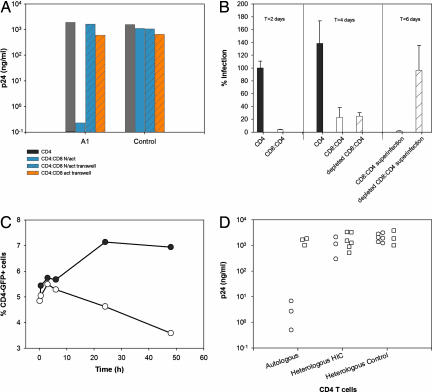
To clarify the mechanism(s) responsible for the control of HIV-1 infection in the cells from HIC, we assessed the role of soluble factors (16, 17). In HIC, the control of HIV-1 replication was totally lost when unstimulated CD8+ T cells were separated from autologous CD4+ T cells by semipermeable membranes (Fig. 4A). By contrast, when mitogen-activated CD8+ T cells were separated from autologous CD4+ T cells by semipermeable membranes, a (modest) reduction in viral replication mediated by soluble factors was observed in HIC and frequently in healthy donors (Fig. 4A). Therefore HIC CD8+ T cells, although able to secrete inhibitory soluble factors upon in vitro stimulation, do not exert their potent anti-HIV-1 activity ex vivo through this mechanism.
Fig. 4.
Mechanism of HIC CD8+ T cell-mediated control of HIV-1 infection. (A) CD4+ T cells from HIC and from uninfected donors were infected with replicative HIV-1 BaL. CD4+ T cells cultured alone are shown as a reference (gray bars). Autologous unstimulated CD8+ T cells were added directly to the CD4+ T cell culture (blue solid bars) or to Transwell inserts that were placed in the CD4+ T cell-containing well (blue hatched bars). Autologous PHA-stimulated CD8+ T cells were added to Transwell (orange hatched bars). The ratio of CD8+ to CD4+ T cells was 1:1. One representative experiment with HIC subject A1 and one uninfected control is shown. Results are peak p24 levels. (B) CD4+ T cells from HIC subject A4 were infected with a single-round HIV-1 BaL pseudotype bearing the luciferase gene alone (filled bars) or in coculture (1:1 ratio) with unstimulated autologous CD8+ T cells (open bars). (Left) Results (mean ± SD, n = 3) are the percentage of infection relative to luciferase activity detected in lysates of infected CD4+ T cells 48 h after infection. (Center) At this time point, CD8+ T cells from half the coculture were depleted and viral replication was analyzed 2 days later. (Right) Both the CD8:CD4 coculture (open bar) and the repurified CD4+ T cells (hatched bar) were then rechallenged with the HIV-1 BaL pseudotype, and luciferase activity was measured 48 h later. (C) CD4+ T cells from HIC were infected with a HIV-1 vesicular stomatitis virus glycoprotein pseudotype bearing the GFP reporter gene (filled circles). Forty-eight hours later, autologous unstimulated CD8+ T cells were added to half of the CD4+ T cells at a 1:1 ratio (open circles). At the indicated time points, an aliquot of each cell suspension was labeled with anti-CD4 antibodies and the quantity of double-positive CD4+GFP+ was assessed by flow cytometry. One representative experiment with HIC subject A7 is shown. (D) Unstimulated CD8+ T cells from HIC A1, A3, and B5 (circles) and uninfected blood donors (squares) were cocultured (1:1 ratio) with autologous or heterologous HIC CD4+ T cells (A1:A3, A3:A1, and B5:A7: CD8:CD4 T cell cultures) or uninfected controls CD4+ T cells and infected with replicative HIV-1 BaL. The results (average of three independent infections) show peak p24 levels.
CD8+ T Cells from HIC Eliminate the HIV-1-Infected CD4+ T Cells.
To evaluate whether the observed control of HIV-1 infection was achieved through the physical elimination of infected CD4+ T cells by HIC CD8+ T cells, we removed CD8+ T cells from superinfected autologous cocultures. The removal of the CD8+ T cells would be expected to rescue HIV-1 replication in the repurified CD4+ T cells only if HIV-1 control was caused by a CD8-mediated intracellular block, such as the postintegration inhibition induced by CD8+ T cell antiviral factor (18).
Single-round infections with pseudotyped HIV-1 particles were used to avoid confusing contribution of subsequent viral replication. A marked control of HIV-1 infection was observed at 48 h when HIC unstimulated CD8+ T cells were cocultured with autologous CD4+ T cells (Fig. 4B Left). The coculture was then split, and CD8+ T cells were depleted from one of the two resulting cocultures. Two days later, a comparable strong reduction of HIV-1 infection was observed in both cultures (Fig. 4B Center). However, when the two cultures were rechallenged with the HIV-1 pseudotype, the CD8-depleted culture was readily superinfected, whereas control of infection was still observed in the nondepleted coculture (Fig. 4B Right). These results are compatible with a CD8+ T cell-mediated elimination of infected CD4+ T cells. Consistent with these results, the addition of HIC CD8+ T cells to autologous CD4+ T cells that had been infected for 2 days with GFP reporter HIV-1 particles provoked a reduction in the number of infected GFP-positive CD4+ T cells (Fig. 4C).
Anti-HIV-1 Activity of HIC CD8+ T Cells Requires Contact with Autologous CD4+ T Cells.
The preceding results strongly support the theory that control of HIV-1 infection was mediated by a cytotoxic mechanism. To find out whether the HIC CD8+ T cell antiviral activity was MHC-restricted, we assessed the capacity of unstimulated HIC CD8+ T cells to control HIV-1 infection in autologous and heterologous CD4+ T cells. The same HIC CD8+ T cells that were able to control HIV-1 in vitro when cocultured with autologous CD4+ T cells, were ineffective when cocultured with heterologous CD4+ T cells both from HIC and from healthy controls (Fig. 4D). These results support an MHC-restricted mechanism of suppression.
Discussion
Here we provide evidence that HIV-specific CD8+ T cells from HIC are characterized by a unique CD38low/HLA-DRhigh phenotype, which likely reflects a capacity to proliferate upon antigenic stimulation and exert effector functions. Importantly, we also report that circulating HIC CD8+ T cells are able to efficiently control HIV-1 infection ex vivo without further stimulation, suggesting that this antiviral activity is functional in vivo. This CD8+ T cell-mediated control is independent of the secretion of antiviral molecules, requires contact with their matching CD4+ T cells, and is caused by the elimination of infected CD4+ T cells, likely due to HIV-specific CD8+ T cells. This spontaneous capacity of CD8+ T cells to clear the virus through the killing of infected cells is particular to HIC.
In keeping with the previous studies in a comparable group of patients (7, 8), we observed that most HIC possessed high frequencies of IFN-γ-secreting HIV-specific CD8+ T cells that, overall, did not differ from those seen in chronically viremic patients and were strikingly higher than those observed in HAART-treated nonviremic patients (Fig. 1) (7, 8, 19, 20). These HIV-specific CD8+ T cells had a broad repertoire, particularly in HLA-B27- and/or HLA-B57-positive individuals.
The high proliferative potential observed in HIC has been previously proposed (8) together with high functionality (9) as hallmarks of a high-quality HIV-specific CD8 T+ cell response in HIC. However, no phenotypical differences had been found that might distinguish HIC from other patients and be associated with an effective immune response. We evaluated the differentiation status of HIC HIV-specific CD8+ T cells as the skewed maturation of these cells may partly explain the lack of effective control of HIV replication in most HIV-infected patients (21–25). We observed that HIV-specific CD8+ T cells were indeed more differentiated in HIC than in HIV-viremic patients (21, 26). Nevertheless, variable profiles were observed within and among the HIC, and, therefore, optimal differentiation of their specific CD8+ T cells does not appear as a distinctive element to explain the effective control of infection.
The activation phenotype of HIV-specific CD8+ T cells in HIC had not been previously defined. We and others (26–28) have shown a positive correlation between the level of viral replication and the activation status of HIV-specific CD8+ T cells. Accordingly, and in contrast to viremic controls, CD38 expression by HIV-specific CD8+ T cells from HIC was very low in this study. Conversely, HLA-DR expression was remarkably high, different from HAART patients, and much higher than that observed on the total CD8+ T cell population. An increased HLA-DR expression associated with a low CD38 expression had been reported on the global CD8+ T cell population in asymptomatic HIV-infected patients with stable CD4+ T cell counts (29). In HIC, we observed that the discordance between the expression of CD38 and HLA-DR was much more pronounced on HIV-specific CD8+ T cells, which is puzzling in the context of high and long-lasting viral control. The low expression of CD38 may reflect the lack of general immune activation, and the expression of HLA-DR may actually characterize T cells with high proliferative potential. This hypothesis is supported by several data: (i) Although we found little evidence of in vivo cycling or proliferation, because very few HIV-specific CD8+ T cells expressed Ki67, HIV-specific CD8+ T cells from HIC had potent in vitro proliferative capacity upon exposure to antigen; (ii) most HLA-DR+ HIV-specific CD8+ T cells did not express the senescence marker CD57, the lack of expression of which has been clearly linked to proliferative capacity (30); (iii) finally, we observed a correlation between HLA-DR expression and the percentage of proliferating cells among HIV-specific CD8+ T cells.
Although the control of HIV-1 replication in HIC has been associated with a strong, multifunctional, specific CD8 T cell response, the anti-HIV activity of HIC CD8+ T cells had not been thoroughly addressed and a direct link between the immune response and the control of viremia was lacking. First, we show that HIC CD4+ T cells are highly susceptible to HIV-1 in vitro infection, thus discarding the idea that an intrinsic resistance of CD4+ T cells to HIV-1 could contribute to HIC status. Second, we found that HIC purified CD8+ T cells were able to efficiently control HIV-1 infection in vitro of autologous CD4+ T cells, even if 10-times more diluted than the CD4+ T cells. Our results are particularly relevant because our experiments were conducted with ex vivo CD8+ T cells in the absence of mitogen activation, showing that CD8+ T cells circulating in the blood of HIC are suitably prepared to control HIV-1 infection. The remarkable spontaneous anti-HIV capacity was a homogeneous feature among CD8+ T cells from HIC, except for patient B2, for whom other factors may contribute to the control of HIV infection. Differences in the CD8+ T cell antiviral activity between HIC and viremic individuals were striking because none of the 9 viremic controls tested suppressed the viral replication as opposed to 9 of the 10 HIC tested.
Although CD8+ T cells in HIV patients have been reported to be able to suppress HIV infection through the secretion of soluble factors (reviewed in ref. 17) or HLA-class I restricted cytolysis (31–33), the relative weight of these mechanisms is controversial. We demonstrate that the ability of HIC CD8+ T cells to control HIV-1 infection ex vivo is not linked to the secretion of soluble factors and required contact with infected CD4+ T cells. The contact of CD8+ T cells with CD4+ T cells did not induce the production of a restriction factor in the latter but rather the elimination of the infected cells. This antiviral activity was effective only on autologous CD4+ T cells. Altogether, our results point to HLA-restricted cytotoxicity as the mechanism associated to the in vivo control of infection achieved by HIC.
In conclusion, we provide direct evidence that circulating CD8+ T cells from HIC are spontaneously able to control HIV-1 infection, a striking difference with cells from viremic subjects. Our study strongly suggests a pivotal role of HIV-specific CD8+ T cells in the viral control of these individuals. These cells have a unique phenotype that may correspond to activated status in vivo but more likely reflects a propensity to expand rapidly upon antigen exposure. This capacity is probably linked to an intact ability to secrete IL-2 and other cytokines (9, 34) and may explain the maintenance of high frequencies of HIV-specific CD8+ T cells in these subjects. The generation of this optimal CD8+ T cell profile is a major research focus. Further studies should provide new insights into the precise mechanisms of HIV control and may serve to a better design of future vaccination or immune-based therapies. Already, both the expression of HLA-DR and CD38 on HIV-specific CD8+ T cells and their capacity to suppress ex vivo HIV-1 might be used as surrogate markers to evaluate the efficiency of induced cytotoxic T lymphocyte responses in vaccine trials.
Materials and Methods
Study Subjects.
Eleven patients infected by HIV-1 for >10 years who had never received antiretroviral treatment and in whom >90% of plasma HIV RNA load tests gave values <400 copies per milliliter (HIC) were studied here: nine have been described elsewhere (5), and two were newly recruited. All subjects were infected with HIV-1 group M, clade B. HLA-typing was performed by genotype analysis (I. Theodorou, Institut National de la Santé et de la Recherche Médicale, Paris, France).
Among HIV-infected patients recruited and monitored at Le Centre Hospitalier Universitaire (Kremlin-Bicêtre, France), 42 who had an exhaustive evaluation of their HIV-specific CD8+ T cells served as controls; 24 were untreated viremic patients [HIV plasma RNA >7,500 copies per milliliter] and 18 were HAART-treated, virologically controlled patients [HIV plasma RNA <200 copies per milliliter]. Twelve healthy controls were blood donors from the Etablissement Français du Sang (Paris, France). All subjects gave their written informed consent.
Isolation of Primary Cells.
CD4+ and CD8+ cells were purified (>99%) from freshly isolated PBMC by positive selection with antibody-coated magnetic beads (Miltenyi Biotec, Paris, France). CD4+ cells were stimulated for 3 days with phytohaemagglutinin (PHA) at 1 μg/ml in the presence of IL-2 (Chiron, Suresnes, France) at 100 units/ml. The culture medium was RPMI medium1640 containing 10% FCS and 100 units/ml penicillin/streptomycin. CD8+ T cells were kept in culture without mitogens or cytokines.
Viruses and Productive Infection in Vitro.
The results shown were obtained by performing infections in vitro with HIV-1 BaL (R5) at an multiplicity of infection of 10−2.8. CD4+ T cells (105) were infected in triplicate in 96-well plates with a spinoculation protocol (35). For coculture, 105 CD4+ T cells were mixed with 105 CD8+ T cells (a 1:1 CD8/CD4 ratio) or with 104 CD8+ T cells (a 0.1:1 CD8/CD4 ratio) at the moment of infection. After infection, the cells were washed and cultured for 14 days. HIV-1 replication was monitored every 3–4 days in supernatants by p24 ELISA (Beckman Coulter, Roissy, France). Unless otherwise indicated, infectivity assays were carried out in the presence of 100 units/ml IL-2.
Single-Round Infections.
BaL- and vesicular stomatitis virus glycoprotein-pseudotyped HIV-1 particles were produced by cotransfecting (SuperFect; Qiagen, Courtaboeuf, France) 293T cells with the proviral pNL-Luc-E-R+ (36) or pNL-GFP-E-R+ (37) and BaL-Env or VSV-G expression vectors. CD4+ T cells (105) were infected in triplicate with HIV-1 pseudotypes, as is described above. Luciferase activity was assessed in cell lysates (Luciferase Reporter 1000 Assay System; Promega, Charbonnieres, France) in a Veritas microplate luminometer (Turner BioSystems, Sunnyvale, CA) as described (38).
Peptides.
We used a set of 124 peptides corresponding to known optimal cytotoxic T lymphocyte epitopes (National Institutes of Health HIV Molecular Immunology Database, www.hiv.lanl.gov/content/immunology/index.html). The peptides were synthesized by Neosystem (Strasbourg, France) and were used at a final concentration of 2 μg/ml.
ELISPOT Assay.
IFN-γ secretion by HIV-specific CD8+ T cells was quantified with an ELISPOT assay with the appropriate stimuli (HLA-defined optimal peptides derived from HIV-1 Env, Gag, Pol, and Nef proteins or controls) (20). IFN-γ SFC were counted with a KS-ELISPOT system (Zeiss, Stuttgart, Germany) and expressed as SFC per 106 PBMC after subtracting the background. Wells were considered positive if they contained at least 50 SFC per 106 PBMC and exhibited at least two times the background level.
Antibodies.
The following antibodies were used. CD8-ECD or CD8-PC5 (clone B9.11), CD3-PC5 (UCHT1), CD45RO-ECD (UCHL1), HLA-DR-ECD (Immu-357), CD38-FITC (T16), and CD57-FITC (NC1) were from Beckman Coulter. CD27-FITC (M-T271), perforin-FITC (dG9), and Ki67-FITC (B56) were from BD Biosciences (San Jose, CA).
Tetramer Staining and Phenotyping.
HIV-specific CD8+ T cells were detected with the following PE-conjugated tetramers: HLA-A*0201-SLYNTVATL (HIV gag 77–85), A*0201-ILKEPVHGV (HIV pol 476–484), A*0301-RLRPGGKKK (HIV gag 20–28), A*0301-QVPLRPMTYK (HIV nef 73–82), and B*2705-KRWIILGLNK (HIV gag 263–272), which were from Proimmune (Oxford, United Kingdom), and B*5701-KAFSPEVIPMF (HIV gag 162–172) from Beckman Coulter. PBMC were incubated with tetramers (1 μg/ml) for 30 min and then with relevant antibodies for 15 min. Cells were washed in Cell Wash (BD Biosciences) plus 1% BSA, incubated for 10 min with FACS lysing solution (BD Biosciences), and washed. For intracellular staining, cells were incubated for 10 min with FACS permeabilizing solution (BD Biosciences) before adding antibodies for 30 min. Cells were fixed in 1% paraformaldehyde for flow cytometry with a Beckman Coulter Epics XL cytometer and RXP software (Beckman Coulter).
Proliferation Assay.
The proliferative capacity of HIV-specific CD8+ T cells was evaluated by flow cytometry. PBMC were stained with 0.35 μM carboxyfluorescein diacetate succinimidyl ester (Molecular Probes, Breda, The Netherlands) for 10 min at 37°C and then stimulated for 5 days with 2 μg/ml peptide or medium alone. After labeling with tetramer, anti-CD8, anti-CD3, and/or anti-HLA-DR antibodies, PBMC were fixed in 1% paraformaldehyde.
Statistical Analyses.
All values throughout the text are expressed as means ± SD. P values were calculated with the Mann–Whitney U test. Correlations were identified by simple linear regression analysis and Spearman's rank correlation test.
Supplementary Material
Supporting Figures
Acknowledgments
We thank Laurence Meyer from the SEROCO/HEMOCO cohort, Ioannis Theodorov and Christine Rouzioux for contributing data, the physicians who care for the patients, and the subjects who participated in this study for their cooperation. This work was supported by the French National Agency for Research on AIDS and Viral Hepatitis. A.S.-C. and C.L. were the recipients of postdoctoral fellowships from Sidaction and the French National Agency for Research on AIDS and Viral Hepatitis, respectively.
Abbreviations
HAART
highly active antiretroviral therapy
HIC
HIV controllers
PBMC
peripheral blood mononuclear cell
PHA
phytohaemagglutinin
SFC
spot-forming cells.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Cao Y, Qin L, Zhang L, Safrit J, Ho DD. N Engl J Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 2.de Quiros JC, Shupert WL, McNeil AC, Gea-Banacloche JC, Flanigan M, Savage A, Martino L, Weiskopf EE, Imamichi H, Zhang YM, et al. J Virol. 2000;74:2023–2028. doi: 10.1128/jvi.74.4.2023-2028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrer T, Harrer E, Kalams SA, Elbeik T, Staprans SI, Feinberg MB, Cao Y, Ho DD, Yilma T, Caliendo AM, et al. AIDS Res Hum Retroviruses. 1996;12:585–592. doi: 10.1089/aid.1996.12.585. [DOI] [PubMed] [Google Scholar]
- 4.Kloosterboer N, Groeneveld PH, Jansen CA, van der Vorst TJ, Koning F, Winkel CN, Duits AJ, Miedema F, van Baarle D, van Rij RP, et al. Virology. 2005;339:70–80. doi: 10.1016/j.virol.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Lambotte O, Boufassa F, Madec Y, Nguyen A, Goujard C, Meyer L, Rouzioux C, Venet A, Delfraissy JF. Clin Infect Dis. 2005;41:1053–1056. doi: 10.1086/433188. [DOI] [PubMed] [Google Scholar]
- 6.Wang B, Dyer WB, Zaunders JJ, Mikhail M, Sullivan JS, Williams L, Haddad DN, Harris G, Holt JA, Cooper DA, et al. Virology. 2002;304:246–264. doi: 10.1006/viro.2002.1706. [DOI] [PubMed] [Google Scholar]
- 7.Gea-Banacloche JC, Migueles SA, Martino L, Shupert WL, McNeil AC, Sabbaghian MS, Ehler L, Prussin C, Stevens R, Lambert L, et al. J Immunol. 2000;165:1082–1092. doi: 10.4049/jimmunol.165.2.1082. [DOI] [PubMed] [Google Scholar]
- 8.Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, Van Baarle D, Kostense S, Miedema F, McLaughlin M, et al. Nat Immunol. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 9.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, et al. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores-Villanueva PO, Yunis EJ, Delgado JC, Vittinghoff E, Buchbinder S, Leung JY, Uglialoro AM, Clavijo OP, Rosenberg ES, Kalams SA, et al. Proc Natl Acad Sci USA. 2001;98:5140–5145. doi: 10.1073/pnas.071548198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, et al. Proc Natl Acad Sci USA. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey JR, Williams TM, Siliciano RF, Blankson JN. J Exp Med. 2006;203:1357–1369. doi: 10.1084/jem.20052319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamann D, Kostense S, Wolthers KC, Otto SA, Baars PA, Miedema F, van Lier RA. Int Immunol. 1999;11:1027–1033. doi: 10.1093/intimm/11.7.1027. [DOI] [PubMed] [Google Scholar]
- 14.Speiser DE, Migliaccio M, Pittet MJ, Valmori D, Lienard D, Lejeune F, Reichenbach P, Guillaume P, Luscher I, Cerottini JC, et al. Eur J Immunol. 2001;31:459–466. doi: 10.1002/1521-4141(200102)31:2<459::aid-immu459>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 15.Nethe M, Berkhout B, van der Kuyl AC. Retrovirology. 2005;2:52. doi: 10.1186/1742-4690-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 17.Levy JA. Trends Immunol. 2003;24:628–632. doi: 10.1016/j.it.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Mackewicz CE, Blackbourn DJ, Levy JA. Proc Natl Acad Sci USA. 1995;92:2308–2312. doi: 10.1073/pnas.92.6.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalod M, Dupuis M, Deschemin JC, Sicard D, Salmon D, Delfraissy JF, Venet A, Sinet M, Guillet JG. J Virol. 1999;73:7108–7116. doi: 10.1128/jvi.73.9.7108-7116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lacabaratz-Porret C, Urrutia A, Doisne JM, Goujard C, Deveau C, Dalod M, Meyer L, Rouzioux C, Delfraissy JF, Venet A, et al. J Infect Dis. 2003;187:748–757. doi: 10.1086/368333. [DOI] [PubMed] [Google Scholar]
- 21.Appay V, Dunbar PR, Callan M, Klenerman P, Gillespie GM, Papagno L, Ogg GS, King A, Lechner F, Spina CA, et al. Nat Med. 2002;8:379–385. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 22.Appay V, Nixon DF, Donahoe SM, Gillespie GM, Dong T, King A, Ogg GS, Spiegel HM, Conlon C, Spina CA, et al. J Exp Med. 2000;192:63–75. doi: 10.1084/jem.192.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Champagne P, Ogg GS, King AS, Knabenhans C, Ellefsen K, Nobile M, Appay V, Rizzardi GP, Fleury S, Lipp M, et al. Nature. 2001;410:106–111. doi: 10.1038/35065118. [DOI] [PubMed] [Google Scholar]
- 24.Hess C, Altfeld M, Thomas SY, Addo MM, Rosenberg ES, Allen TM, Draenert R, Eldrige RL, van Lunzen J, Stellbrink HJ, et al. Lancet. 2004;363:863–6. doi: 10.1016/S0140-6736(04)15735-8. [DOI] [PubMed] [Google Scholar]
- 25.van Baarle D, Kostense S, Hovenkamp E, Ogg G, Nanlohy N, Callan MF, Dukers NH, McMichael AJ, van Oers MH, Miedema F. AIDS. 2002;16:2001–2011. doi: 10.1097/00002030-200210180-00004. [DOI] [PubMed] [Google Scholar]
- 26.Doisne JM, Urrutia A, Lacabaratz-Porret C, Goujard C, Meyer L, Chaix ML, Sinet M, Venet A. J Immunol. 2004;173:2410–2418. doi: 10.4049/jimmunol.173.4.2410. [DOI] [PubMed] [Google Scholar]
- 27.Kestens L, Vanham G, Gigase P, Young G, Hannet I, Vanlangendonck F, Hulstaert F, Bach BA. AIDS. 1992;6:793–797. doi: 10.1097/00002030-199208000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Papagno L, Spina CA, Marchant A, Salio M, Rufer N, Little S, Dong T, Chesney G, Waters A, Easterbrook P, et al. PLoS Biol. 2004;2:173–185. doi: 10.1371/journal.pbio.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giorgi JV, Ho HN, Hirji K, Chou CC, Hultin LE, O'Rourke S, Park L, Margolick JB, Ferbas J, Phair JP. J Infect Dis. 1994;170:775–781. doi: 10.1093/infdis/170.4.775. [DOI] [PubMed] [Google Scholar]
- 30.Brenchley JM, Karandikar NJ, Betts MR, Ambrozak DR, Hill BJ, Crotty LE, Casazza JP, Kuruppu J, Migueles SA, Connors M, et al. Blood. 2003;101:2711–2720. doi: 10.1182/blood-2002-07-2103. [DOI] [PubMed] [Google Scholar]
- 31.Walker BD, Chakrabarti S, Moss B, Paradis TJ, Flynn T, Durno AG, Blumberg RS, Kaplan JC, Hirsch MS, Schooley RT. Nature. 1987;328:345–348. doi: 10.1038/328345a0. [DOI] [PubMed] [Google Scholar]
- 32.Walker BD, Plata F. AIDS. 1990;4:177–184. doi: 10.1097/00002030-199003000-00001. [DOI] [PubMed] [Google Scholar]
- 33.Yang OO, Kalams SA, Rosenzweig M, Trocha A, Jones N, Koziel M, Walker BD, Johnson RP. J Virol. 1996;70:5799–5806. doi: 10.1128/jvi.70.9.5799-5806.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lichterfeld M, Kaufmann DE, Yu XG, Mui SK, Addo MM, Johnston MN, Cohen D, Robbins GK, Pae E, Alter G, et al. J Exp Med. 2004;200:701–712. doi: 10.1084/jem.20041270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Doherty U, Swiggard WJ, Malim MH. J Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Connor RI, Chen BK, Choe S, Landau NR. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 37.Amara A, Vidy A, Boulla G, Mollier K, Garcia-Perez J, Alcami J, Blanpain C, Parmentier M, Virelizier JL, Charneau P, et al. J Virol. 2003;77:2550–2558. doi: 10.1128/JVI.77.4.2550-2558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sáez-Cirión A, Nicola MA, Pancino G, Shorte SL. Biotechnol J. 2006;1:682–689. doi: 10.1002/biot.200600045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figures