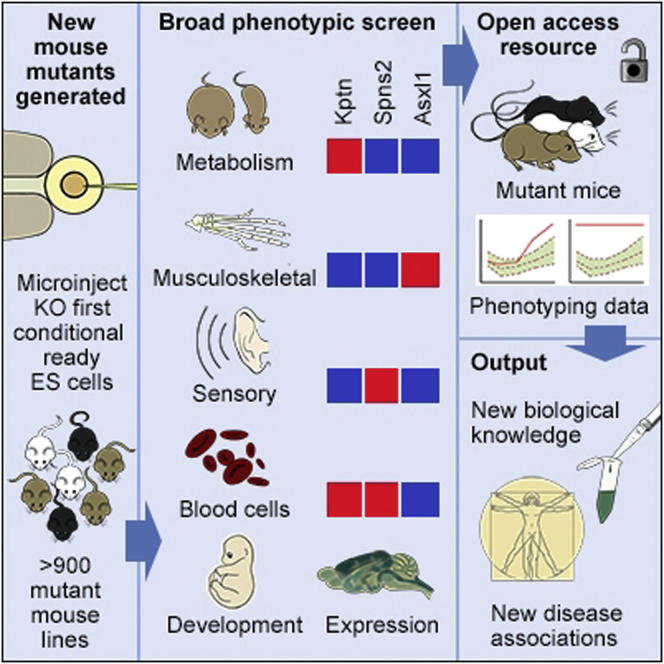
Genome-wide Generation and Systematic Phenotyping of Knockout Mice Reveals New Roles for Many Genes (original) (raw)
Summary
Mutations in whole organisms are powerful ways of interrogating gene function in a realistic context. We describe a program, the Sanger Institute Mouse Genetics Project, that provides a step toward the aim of knocking out all genes and screening each line for a broad range of traits. We found that hitherto unpublished genes were as likely to reveal phenotypes as known genes, suggesting that novel genes represent a rich resource for investigating the molecular basis of disease. We found many unexpected phenotypes detected only because we screened for them, emphasizing the value of screening all mutants for a wide range of traits. Haploinsufficiency and pleiotropy were both surprisingly common. Forty-two percent of genes were essential for viability, and these were less likely to have a paralog and more likely to contribute to a protein complex than other genes. Phenotypic data and more than 900 mutants are openly available for further analysis.
PaperClip
Graphical Abstract
Highlights
- •
Large openly available resource of targeted mouse mutants and phenotypic data - •
Screen for broad range of disease features and traits - •
Many novel phenotypes suggest functions for both studied and unstudied genes - •
Haploinsufficiency and pleiotropy are common
More than 900 new mutant mice lines and a multifaceted phenotypic screening platform reveal unanticipated pleiotropies, widespread effects of haploinsufficiency, potential disease models, and functions for unstudied genes.
Introduction
The availability of well-annotated genome sequences for a variety of organisms has provided a strong foundation on which much biological knowledge has been assembled, including the generation of comprehensive genetic resources. This has been achieved in several model organisms, including E. coli, S. cerevisiae, S. pombe, A. thaliana, C. elegans, and D. melanogaster, greatly facilitating studies focused on single genes and enabling genome-wide genetic screens.
Annotation of the human genome has identified over 20,000 protein-coding genes as well as many noncoding RNAs. Despite the dramatic increase in the knowledge of variation in human genomes, the normal function of many genes is still unknown or predicted from sequence analysis alone, and consequently, the disease significance of rare variants remains obscure. Furthermore, there remains a large bias toward research on a small number of the best-known genes (Edwards et al., 2011). Realizing the full value of the complete human genome sequence requires broadening this focus, and the availability of comprehensive biological resources will facilitate this process.
The mouse is a key model organism for assessing mammalian gene function, providing access to conserved processes such as development, metabolism, and physiology. Genetic studies in mice, mostly via targeted mutagenesis in ES cells, have described a function for 7,229 genes (ftp://ftp.informatics.jax.org/pub/reports/MGI_PhenotypicAllele.rpt, February 2013). The vast majority of these studies have been directed at previously studied (known) genes, driven by previous biological knowledge. Phenotype-driven screens have also identified genes associated with specific phenotypes, although to a smaller extent. Although targeted mutagenesis has been very successful, the global distribution of the effort has resulted in significant heterogeneity in allele design, genetic background of mice used, and their phenotypic analysis. Furthermore, the biological focus of most targeted knockout experiments is constrained by the expertise of the specific research group. As a result, many phenotypes have not been detected, and consequently, the full biological function of many genes studied using knockout mice is significantly underreported.
Some efforts to generate and phenotype sizeable sets of new targeted alleles of genes of interest have been reported previously (e.g., Tang et al., 2010). These studies focused on specific categories of molecules such as secreted and transmembrane proteins or other “drugable” targets. Other research centers have established mouse clinics, with the aim of carrying out a comprehensive analysis of the phenotypes of mutant lines of specific interest (e.g., Fuchs et al., 2012; Wakana et al., 2009; Laughlin et al., 2012).
The genome-wide set of targeted mutations in ES cells established by the KOMP, EUCOMM, and MirKO programs (Skarnes et al., 2011; Prosser et al., 2011; Park et al., 2012) provides an opportunity to conduct systematic, large-scale gene function analysis in a mammalian system without the variables inherent in studies by individual groups. The Sanger Institute’s Mouse Genetics Project (MGP) was one of the first programs to pursue this objective, established in 2006 when the first targeted ES cells became available. The MGP later expanded to contribute to a European phenotyping effort, EUMODIC, and more recently has become a founding member of the International Mouse Phenotyping Consortium (IMPC). Summaries of the developing efforts and aspirations of the IMPC have been reported (e.g., Brown and Moore, 2012; Ayadi et al., 2012). As the first established large-scale project using the KOMP/EUCOMM ES cells, the MGP has provided pilot data to inform the design of the international effort, such as the advantages of a single pipeline design, optimum numbers of mice, and details of variance for specific phenotyping tests. To date, the MGP has generated more than 900 lines of mutants using KOMP/EUCOMM resources (http://www.sanger.ac.uk/mouseportal/), and here, we describe the analysis of 489 of these for viability and fertility and 250 lines that have passed through a systematic screen for adult phenotypes, providing a glimpse into the wealth of biological insight that will emerge from these programs. Publicly available data enable the construction of new hypotheses, and the mouse mutants provide an invaluable resource for follow-up studies.
Results
Genes and Alleles
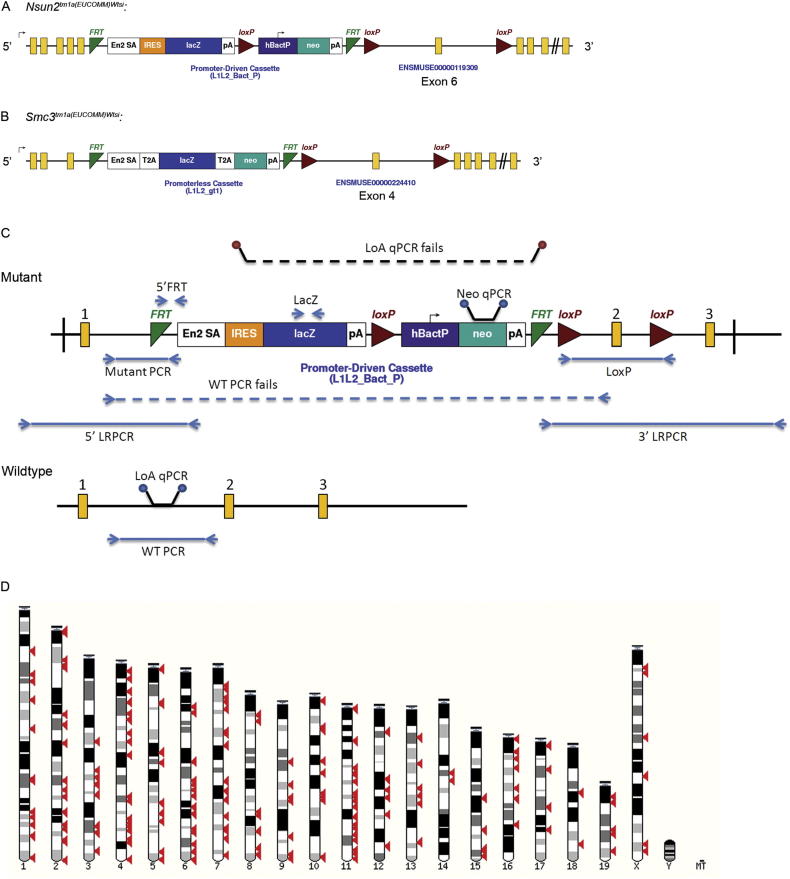
Mice carrying targeted knockout first conditional-ready alleles from the KOMP/EUCOMM ES cell resources (Figures S1A and S1B available online; Skarnes et al., 2011) were established on a C57BL/6 genetic background. The mutants generated are listed in Tables S1 and S2, and all are available through public repositories including EMMA (http://www.emmanet.org/) and KOMP (http://www.komp.org/). Two classes of alleles are represented: those targeted with a promoter-driven selectable marker, and those with promoterless targeting vectors. Most are expected to be null alleles based on previous experience with this design (Mitchell et al., 2001; Testa et al., 2004). Data from 25 alleles showed that most (15) had <0.5% of normal transcript level detected in liver with a minority (4) showing a “leakiness” of ∼20% (column X; Table S2). The structure of each allele was confirmed when established in mice (Figure S1C).
Figure S1.
Allele Design, Genotyping, and Chromosomal Distribution of Genes Selected, Related to Figure 1
(A and B) Examples of the allele designs used. Illustration of the two main alleles used, A, Nsun2 tm1a contains a promotor-driven targeting vector, and B, Smc3 tm1a contains a promotorless targeting vector [gene build Mouse NCBIM37, (Ensembl 66: Feb 2012)]. The promotorless allele design is biased toward genes that are expressed in ES cells. The alleles are expected to be null alleles, but assessment of the degree of knockdown and the extent of off-target effects on nearby genes has not been carried out systematically.
(C) Genotyping and quality control of mice. ES cells: Long-range (LR) PCR, using one primer in the cassette and another outside of the homology arms of the allele design, was used to confirm the targeting on either the 3′ or 5′ side of the vector prior to micro-injection. Mice: To determine the genotype and confirm gene identity, three short-range PCR assays were used: mutant allele-specific, wild-type allele-specific and to detect the lacZ gene. Targeting was confirmed by either LRPCR, loss of the wild-type specific short-range PCR product in homozygotes or a qPCR assay confirming loss of the wild-type allele. Presence of the 3′ LoxP site was detected by either qPCR or short-range PCR assays. Further details of the QC protocols are available from: http://www.knockoutmouse.org/kb/25/. Initially mice were genotyped using a combination of the three short-range PCR assays, but to facilitate high-throughput, we later switched to a qPCR neo cassette counting-based system. Initial genotyping was carried out using ear punches from ∼14 day old mice, so that mice of the desired genotypes for screening could be identified and weaned together. Genotyping was repeated at the far-end of the pipeline after culling, and data were only accepted from mice for which the second genotype was concordant with the 14 day genotype.
(D) Genomic distribution of genes studied. An illustration of the mouse karyotype showing the location of genes targeted (red arrowheads) across all chromosomes except Y.
Viability
Viability was assessed at postnatal day 14 (P14) by genotyping offspring of heterozygous crosses (Figure 1A). Data from 489 targeted alleles are summarized in Figure 2A. Overall, 58% were fully viable, whereas 29% produced no homozygotes at P14 and were classed as lethal, consistent with the proportion of homozygous embryonic/perinatal lethal mutants reported by MGI (2,183 of 7,229 lines of mice [30%]; ftp://ftp.informatics.jax.org/pub/reports/MGI_PhenoGenoMP.rpt, February 2013). A further 13% produced fewer than 13% homozygotes and were considered to be subviable. Genes required for survival included alleles generated with both promoter-driven and promoterless selection cassettes, but the latter were significantly more likely to be lethal (Figure 2B; Table S3) despite a greater level of persistent gene expression (11 of 14 promotor-driven compared with 4 of 11 promotorless alleles with <0.5% expression; column X; Table S2).
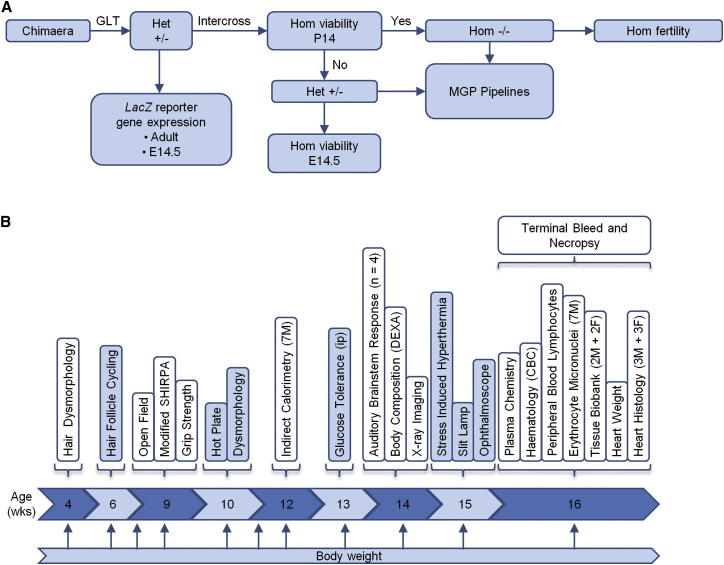
Figure 1.
Illustration of the Phenotyping Pipelines
(A) An overview of the typical workflow from chimera to entry into phenotyping pipelines, encompassing homozygous (Hom) viability, fertility, and target gene expression profiling using the lacZ reporter. Het, heterozygous.
(B) The Sanger Institute MGP clinical phenotyping pipeline showing tests performed during each week. Seven male and seven female mutant mice are processed for each allele screened. In addition, seven male and seven female WT controls per genetic background are processed every week.
See also Figure S1 and Tables S1 and S2.
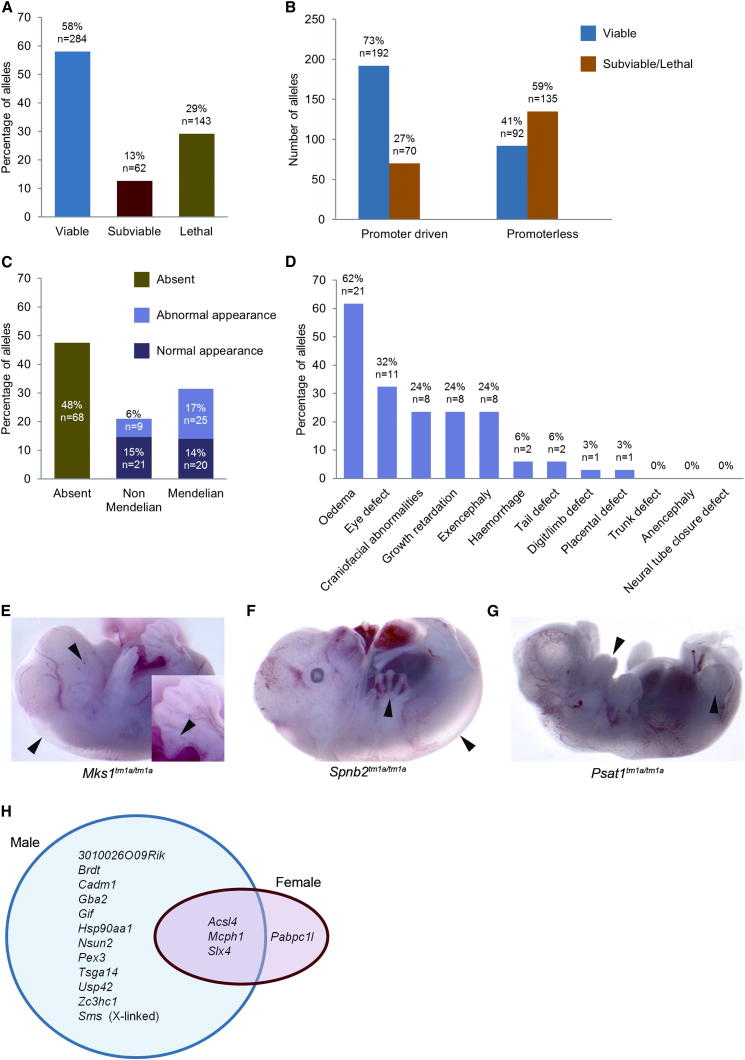
Figure 2.
Homozygous Viability and Fertility Overview
(A) Homozygous viability at P14 was assessed in 489 EUCOMM/KOMP targeted alleles. A minimum of 28 live progeny were required to assign viability status. Lines with 0% homozygotes were classed as lethal, >0% and ≤13% as subviable, and >13% as viable.
(B) Comparison of homozygous viability data from targeted alleles carrying either a promoter-driven or promoterless neomycin selection cassette.
(C) Lines classed as lethal or subviable at P14 were further assessed for viability at E14.5. Of the 205 targeted alleles eligible for this recessive lethality screen, 143 are reported here. A total of 28 embryos were required to assign viability status, and outcomes were categorized by both the number and dysmorphology of homozygous offspring.
(D) A basic dysmorphology screen encompassing 12 parameters was performed on all embryos for the 75 targeted alleles classed as viable or subviable at E14.5. A total of 34 targeted alleles showed one or more abnormality, and the percentage incidence is presented.
(E–G) Examples of E14.5 dysmorphology (arrowheads indicate abnormalities) are presented. Homozygous progeny were detected at a Mendelian frequency in all three examples. Sixty-seven percent (six of nine) Mks1 tm1a/tm1a embryos presented with edema, polydactyly, and eye defects (E). Sixty-two percent (five of eight) Spnb2 tm1a/tm1a embryos presented with edema and hemorrhage (F). Eighty-six percent (six of seven) Psat1 tm1a/tm1a embryos presented with growth retardation, exencephaly, and craniofacial abnormalities (G).
(H) Fertility was assessed in homozygous viable lines (307 mouse lines assessed from a total of 331 eligible lines). At least four independent 6-week-old mice of each sex were mated for a minimum of 6 weeks, and if progeny were born, the line was classed as fertile, regardless of if the progeny survived to weaning. Of note is the strong skew toward male (blue circle) fertility issues (15 of 16 genes) compared to 4 of 15 genes that displayed female (red circle) fertility issues.
See also Table S3.
Alleles classed as lethal or subviable at P14 were further assessed at E14.5 (Figure 2C). From 143 alleles examined, 48% (68 genes) produced no homozygotes, indicating embryonic lethality and complete resorption by E14.5. One-third of alleles (n = 45) produced the expected number of homozygous embryos, whereas 30 (21%) produced fewer than expected homozygotes. Of the 75 mutant lines that produced homozygous embryos, 34 exhibited one or more morphological defect (Figure 2D). Some mutants (n = 23) presented with specific abnormalities including craniofacial defects and polydactyly, whereas 11 lines displayed only generalized indicators of developmental defects, edema, and/or growth retardation (Figure 2D). Examples are illustrated in Figures 2E–2G.
Fertility
Fertility of heterozygotes was assessed from heterozygous intercrosses. Of 489 alleles assessed, all heterozygotes were able to produce offspring. Homozygous mutants for 307 of the viable lines were then assessed. A homozygous infertility rate of 5.2% (n = 16) was observed (Figure 2H), strongly male biased with 15 of 16 genes exhibiting male infertility. A total of 11 genes affected only males, whereas just 1 was female specific (Pabpc1l). Of these 16 genes, 7 have not previously been associated with infertility. Although some were good candidates such as Usp42, expressed during mouse spermatogenesis (Kim et al., 2007), others are novel genes such as 3010026O09Rik and may suggest new pathways or mechanisms influencing fertility.
Adult Phenotypes
We report here the results of our screen of the first 250 lines to complete all primary phenotyping pipelines. In contrast to previous focused screens by Mitchell et al. (2001) and Tang et al. (2010), a broad range of gene products was included. The 250 genes reported span all chromosomes except Y (Figure S1D) and include eight control lines published previously and 87 genes proposed by the research community. For 34 of the 250 genes, no functional information has been published. A comparison of this gene set with all mouse genes indicates minimal GO term enrichment spread over a variety of processes and underrepresentation only in sensory perception of smell, indicating that the gene set can be regarded as a reasonable sample of the genome.
A series of tests was used (Figure 1B), designed to detect robust variations in phenotypes that were key indicators of a broad spectrum of disease categories. Of the 250 reported lines, 104 were either lethal or subviable; most of these were screened as heterozygotes (n = 90), and the remaining lines were screened as homozygotes and/or hemizygotes (n = 160). All mutant lines generated passed through all primary phenotypic screens. For most tests in the pipeline, seven males and seven females were used, tested in small batches so that the data for each genotype were gathered on different days (Figure S2A). Assays culminated in the collection of samples at 16 weeks of age (Table S4). The primary screen included a high-fat diet challenge to exacerbate any latent phenotypes. Separate pipelines included challenges with two infectious agents: Salmonella Typhimurium and Citrobacter rodentium (Table S4).
Figure S2.
Batch Size and Baseline Variation over Time, Related to Experimental Procedures
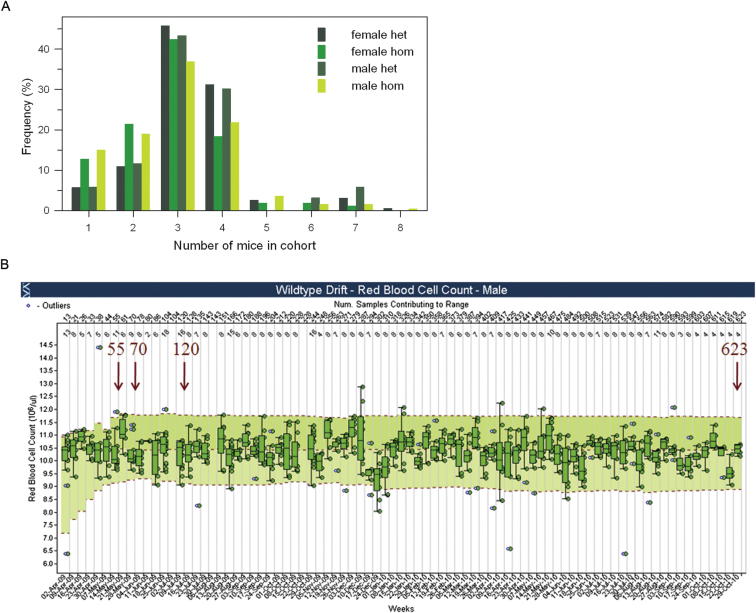
(A) Batch size of mutant mice. Frequency distribution of cohort size of mice of the same genotype issued to the phenotyping pipeline at a time. For each mutant allele, typically 3 mice of a defined sex and zygosity were issued to the Clinical Phenotyping Pipeline at one time. However, the number ranged from 1-8 mice issued in a single batch or cohort.
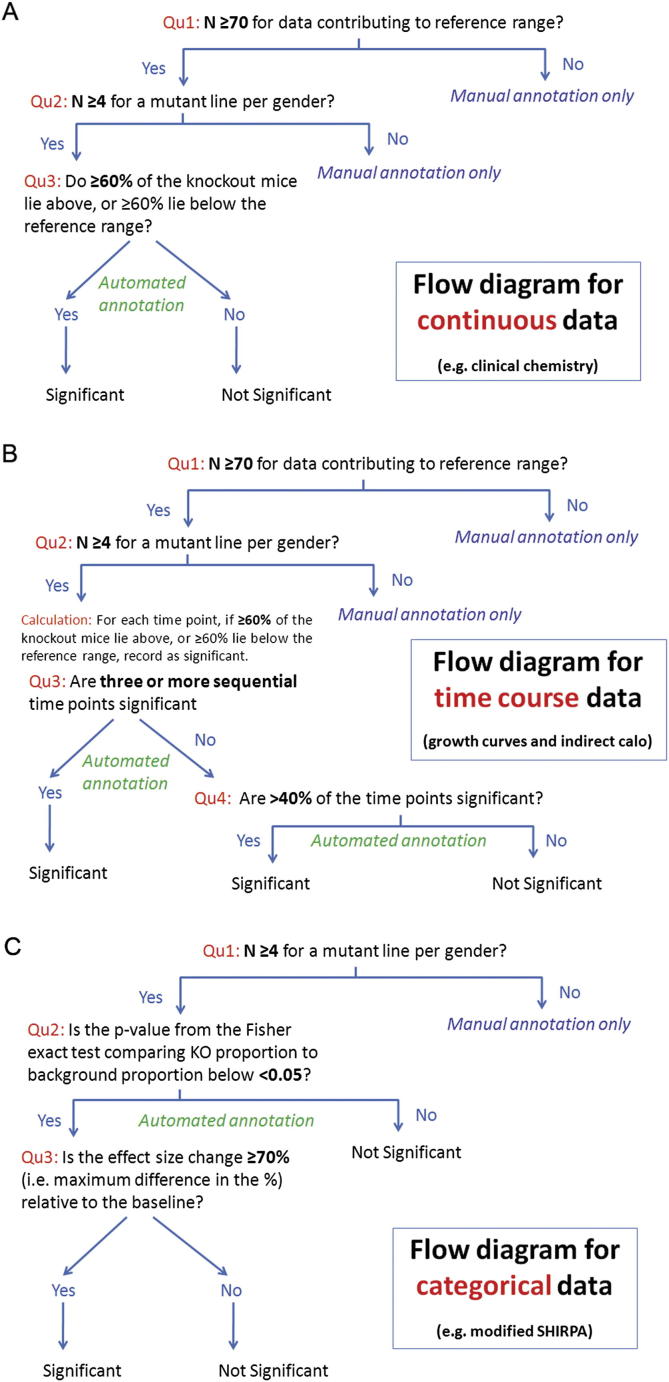
(B) Baseline variation over time. Example of baseline week to week variation seen in the control data. Example shown is red blood cell count presented weekly from 02/04/09 to 29/10/10 for male mice for the strain group B6Brd;B6Dnk;B6N-Tyr c-Brd. Each boxplot represents data collected from control mice in one week. The size of this effect is significant as shown by some of the box plots not overlapping each other, indicating a Cohen’s d > 3. The pale green area indicates the 95% reference range calculated from the 2.5 and 97.5 percentile values as the data accumulate. Red arrows show the cumulative total of animals contributing to the reference range from 55 mice in May 2009 up to 623 mice in October 2010. The reference range becomes stable after about 70 control mice.
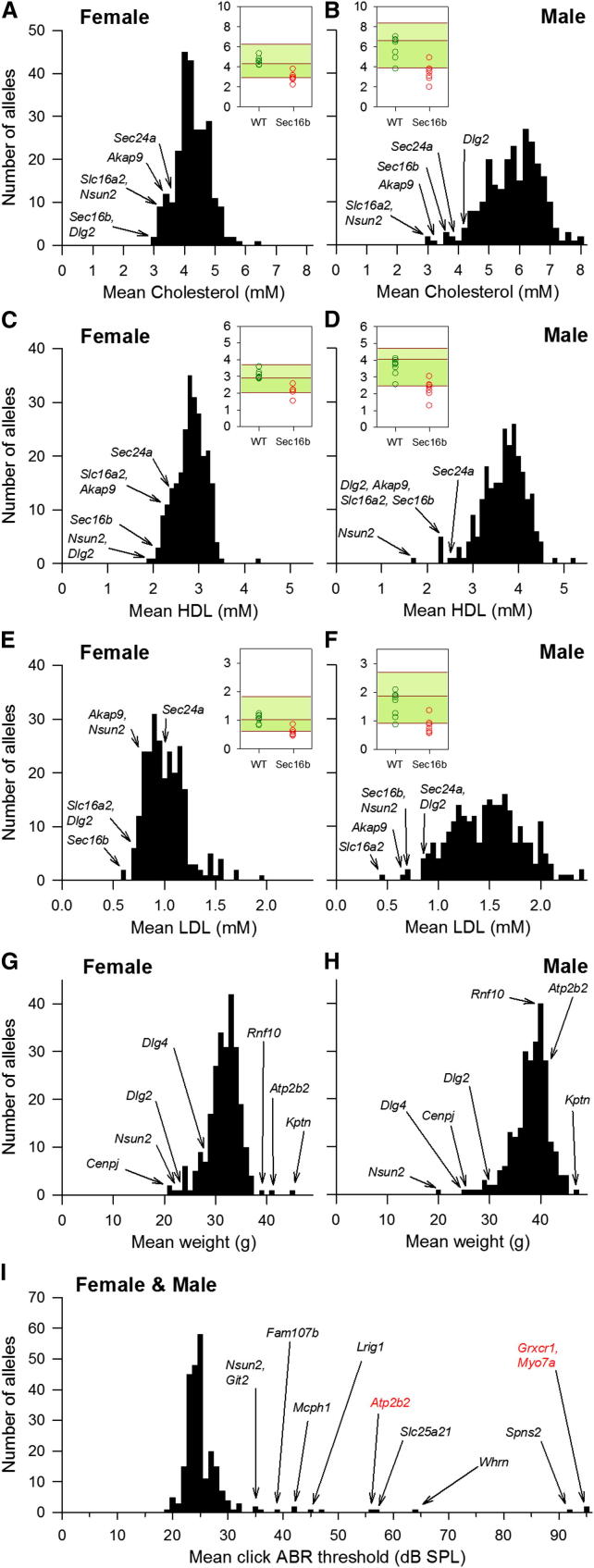
Phenotypic data from the first 250 mutant alleles through the adult pipelines are summarized in Tables S1 and S2, with significant differences from the control baseline (hits) indicated by a red box. To make robust phenotypic calls, a reference range method was implemented that uses accumulated wild-type (WT) data to identify and refine the 95% reference range (Figure S2B). Mutant data were compared to the relevant reference range and variant phenotypes determined using a standardized set of rules (Figure S3). We aimed to highlight phenotypes with large effect sizes. This approach results in conservative calls and minimizes false positives. There was very little missing data (2.14% of all calls; Table S2). The maximum number of parameters collected per line was 263. Of these, 147 were categorical variables, for example normal or abnormal teeth, whereas 116, such as plasma magnesium levels, exhibited a continuous distribution from which outliers were identified. Examples of parameters with continuous variables (cholesterol, high-density lipoprotein [HDL], low-density lipoprotein [LDL], mean weights, and auditory brainstem responses [ABR]) are illustrated in Figure 3.
Figure S3.
Decision-Making Process for Calling Hits, Related to Experimental Procedures
The figures show the process we used to call significant hits for three different types of data: (A) continuous, (B) time course and (C) categorical.
Figure 3.
Data Distributions for Selected Parameters
(A–F) Distribution of mean total cholesterol (A and B), mean HDL cholesterol (C and D), and mean LDL cholesterol (E and F) at 16 weeks of age in both sexes for 250 unique alleles. Outliers are identified by gene name. The insets in (A)–(F) present the data for one outlier, Sec16b tm1a/tm1a (red circles represent individual mice), compared to the WT controls processed during the same week (green circles), and a cumulative baseline of all WT mice of that age, sex, and genetic background (>260 WT mice) is presented as the median and 95% confidence interval.
(G and H) Distribution of mean body weight at 16 weeks in (G) female and (H) male mutant lines of mice. Outliers are identified by gene name.
(I) Distribution of mean click ABR threshold at 14 weeks (typically n = 4, independent of sex). Outliers are identified by gene name including positive controls highlighted in red.
Gene expression was examined by whole-mount lacZ reporter gene expression in 41 tissues and organs of adults, typically using heterozygotes (≥6 weeks old; n = 243 lines; Table S1). Ubiquitous expression was recorded for eight lines (3.3%) and complete absence of expression in nine lines (3.7%). Of the remaining lines, 168 (69.1%) showed expression in <20 of the tissues, suggesting a relatively specific expression pattern, whereas 58 (23.9%) were more broadly expressed (≥20 tissues with detectable lacZ expression).
The data and images can be viewed on the Sanger Institute’s mouse portal, accompanied by step-by-step examples of how to access the data (http://www.sanger.ac.uk/mouseportal/). Much of the raw data can be downloaded from the MGP Phenotyping Biomart (http://www.sanger.ac.uk/htgt/biomart/martview/) for further analysis. Summaries can be found by searching for each gene of interest in Wikipedia (http://en.wikipedia.org/wiki/Category:Genes_mutated_in_mice) and Mouse Genome Informatics (http://www.informatics.jax.org/).
Many Unexpected Phenotypes Discovered
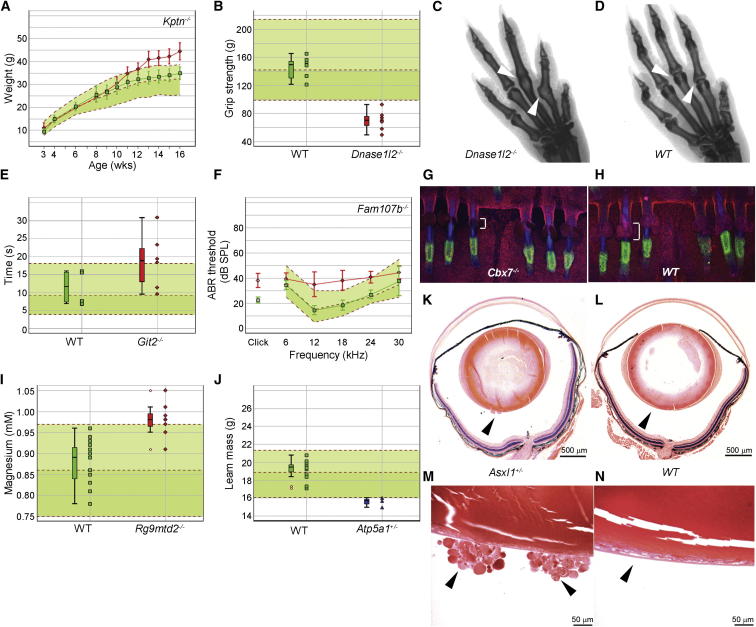
A few examples of the wide range of phenotypes we discovered are illustrated in Figure 4. Body weight and fat/lean composition were among the most common anomalies, with both overweight (n = 2) and underweight (n = 21) mutants discovered. The Kptn mutant is an example of an unexpected phenotype. Kptn is a putative actin binding protein proposed as a candidate for deafness because it is expressed in sensory hair cells (Bearer et al., 2000). Instead, the homozygous Kptn mutant has increased body weight on a high-fat diet (Figure 4A) and increased bacterial counts following Salmonella Typhimurium challenge but normal hearing (Table S2). Additional new phenotypes were detected in genes that had been published previously, such as reduced grip strength and ankylosis of the metacarpophalangeal joints in Dnase1l2 mutants (Fischer et al., 2011), delayed response in the hot plate test in Git2 mutants (Schmalzigaug et al., 2009), and small sebaceous glands in Cbx7 mutants (Forzati et al., 2012) (Figures 4B–4E, 4G, and 4H). Phenotypes were also detected in genes that had not been published previously, such as impaired hearing in Fam107b mutants and elevated plasma magnesium concentration in Rg9mtd2 mutants (Figures 4F and 4I, respectively). These examples demonstrate that many phenotypes will be missed unless they are specifically looked for and illustrate the value of carrying out a broad range of screens with all mutants going through all screens. They also reveal our collective inability to predict phenotypes based on sequence or expression pattern alone.
Figure 4.
Examples of Novel Phenotypes from a Wide Range of Assays with Particular Focus on Novel Genes
(A) Elevated body weight gain of Kptn tm1a/tm1a females (n = 7) fed a high-fat diet from 4 weeks of age. Mean ± SD body weight is plotted against age for Kptn tm1a/tm1a females (red line) and local WT controls run during the same weeks (n = 16; green line). The median and 95% reference range (2.5% and 97.5%; dotted lines) for all WT mice of the same genetic background and sex (n = 956 females) are displayed on the pale green background.
(B) Reduced grip strength in Dnase1l2 tm1a/tm1a males (n = 7) (red symbols) compared with controls (n = 8) (green symbols) and the reference range (n = 289). Each mouse is represented as a single symbol on the graph. Median, 25th and 75th percentile (box), and the lowest and highest data point still within 1.5× the interquartile range (IQR) (whiskers) are shown.
(C and D) Ankylosis of the metacarpophalangeal joints (arrowheads) shown by X-ray in Dnase1l2 tm1a/tm1a mice (C) (six of seven males; five of seven females) compared with WT controls (D) correlates with reduced grip strength (B).
(E) Increased latency to respond to heat stimulus in Git2 Gt(XG510)Byg/ Gt(XG510)Byg females (n = 6) (red symbols) compared with controls (n = 4) (green symbols) and the reference range (n = 115), with box and whisker plots on the left (see Figure 4B legend).
(F) Mild hearing impairment at the middle range of frequencies in Fam107b tm1a/tm1a mutants (n = 8) (red line shows mean ± SD) compared with controls (n = 10) and the reference range (n = 440).
(G) Smaller sebaceous glands (indicated by bracket) in Cbx7 tm1a/tm1a mutant tail skin hairs compared with WT (H).
(I) Increased plasma magnesium levels in Rg9mtd2 tm1a/tm1a males (n = 8) (red symbols) compared with local controls (n = 15) (green symbols) and the reference range (n = 241), with box and whisker plots on the left (see Figure 4B legend).
(J) Decreased lean mass in Atp5a1 tm1a/+ females (n = 3) (blue symbols) compared with local controls (n = 15) (green symbols) and the reference range (n = 757), with box and whisker plots on the left (see Figure 4B legend).
(K and L) Histopathology showed opacities in the vitreous of eyes from Asx11 tm1a/+ mice (K) (arrowheads; scale bar, 500 μm) compared with empty vitreous in WT (L).
(M and N) Higher magnification revealed round opacities extending from the lens into the vitreous (arrowheads; scale bar, 50 μm) in Asx11 tm1a/+ mice (M) compared with a normal lens contained within the lens capsule in WT mice (N).
See also Table S5.
Haploinsufficient and Nonessential Genes
Haploinsufficient phenotypes were detected in 38 of 90 (42%) of these lines. Thus, haploinsufficiency is relatively common, suggesting that screening heterozygotes of knockout lines can yield valuable insight into gene function and provide models for dominantly inherited human disorders. All 90 genes screened as heterozygotes had at least 1 hit (usually viability) and together gave a total of 181 hits (ranging from 1 to 14 per line), an average of 2.0 per line, or 1.0 per line if we consider that abnormal viability is a feature of the homozygote. The distributions of phenotypic hits are shown in Figures 5A and 5B. Two examples of haploinsufficiency are illustrated in Figures 4J–4N.
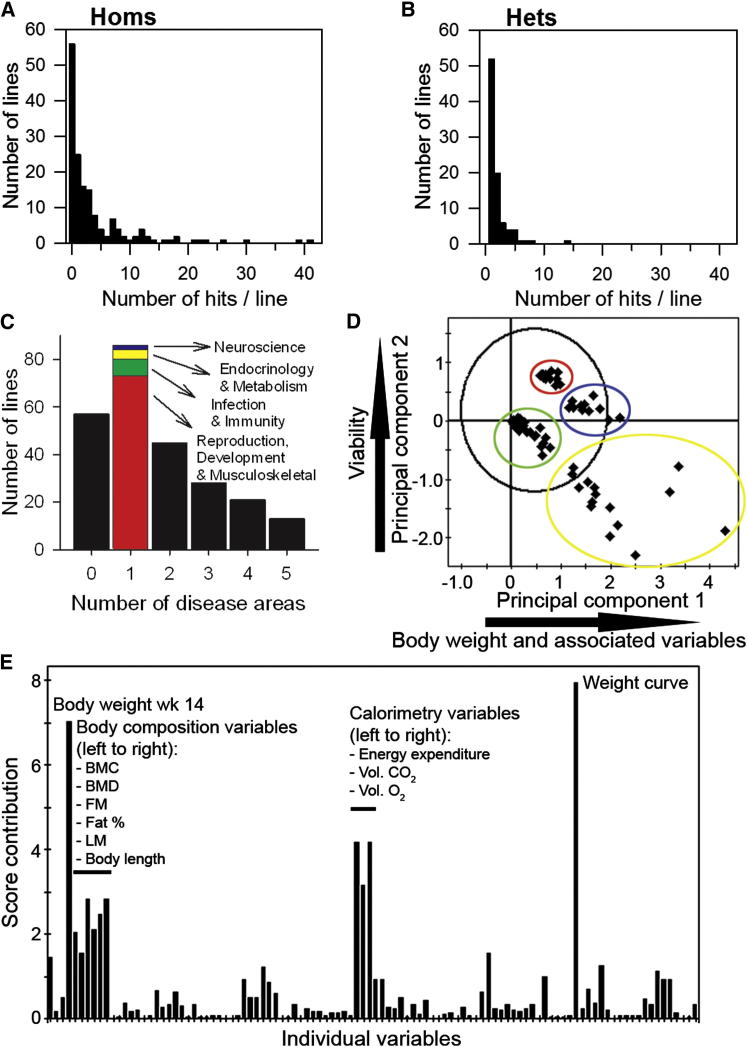
Figure 5.
Characteristics of Phenotypic Hits Detected
(A) Distribution of the number of phenotypic hits in each line screened as homozygotes showing the peak at no hits but a long tail of lines with multiple hits up to 41.
(B) Distribution of hits in lines screened as heterozygotes; all lines had at least one hit (for viability) with a spread up to 14 hits.
(C) Distribution of lines with hits in different disease areas showing a peak of lines with just one area affected (colors indicate which areas) but some lines with multiple disease areas involved, indicating a high degree of pleiotropy.
(D) Principal component analysis score scatterplot showing the deviation of each gene from the first two principal components to visualize the clustering in genes within the multidimensional space. The black ovoid represents the Hotelling’s T2 95% confidence limits. Colored ovoids mark four different clusters of mutant lines. The two main principal components (or latent variables) in the model are significant in explaining 19.2% and 11.7% of the variation, respectively, and are predictive.
(E) Principal component analysis contribution plot indicating the contribution of the variables to the separation between the red and green clusters compared to the blue and yellow clusters in (D). Major phenotypic contributions are labeled.
Key to variables is presented in Table S7.
A total of 837 phenotypic variants were detected in the 250 mutant lines, 1.27% of the total calls (Tables S1 and S2). Of the lines screened as homozygotes or hemizygotes, 35% (56 of 160) appeared completely normal in our screen. There are several possible explanations for the lack of a detected phenotype, such as incomplete inactivation of the gene, a subtle change in phenotype not detected by our screen, or the gene may be nonessential. So far, there is no overlap between the 56 mouse lines with no detected phenotype and genes homozygously inactivated in humans, but both data sets are limited in coverage to date (MacArthur et al., 2012). The remaining 104 homozygous/hemizygous lines gave a total of 656 hits (range 0–41 per line), an average of 6.3 hits per line.
Sensitivity of the MGP Screen
To assess the sensitivity of our screen, the phenotypes were compared with published data on alternative alleles where available. A total of 91 of 250 genes had published data reported in MGI (Table S5), and for 61 of these, our observations detected features of the published phenotypes. Importantly, for 56 genes, a new phenotype was detected by our screen (column K, Table S5). For 31 genes, features of the published phenotype were assessed but not detected by our pipeline. For example, Asxl1 tm1Bc/tm1Bc mice are published as being viable (Fisher et al., 2010), but we found that Asxl1 tm1a(EUCOMM)Wtsi homozygotes were lethal, with none detected among 276 progeny from heterozygous intercrosses (χ2 = 95.13, df = 2; p < 2.2 × 10−16). These discrepant cases may reflect differences in the allele and/or genetic background. In other cases (77 genes), the reported characteristics required a specialized test not included in our screen, such as the calcium signaling defect in cardiomyocytes of Anxa6 tm1Moss/tm1Moss mutants (Hawkins et al., 1999).
New Mouse Models for Human Disease
The data set reported here includes 59 orthologs of known human disease genes. We compared our data with human disease features described in OMIM (Table S6). Approximately half (27) of these mutants exhibited phenotypes that were broadly consistent with the human phenotype. However, many additional phenotypes were detected in the mouse mutants suggesting additional features that might also occur in patients that have hitherto not been reported. Interestingly, a large proportion of genes underlying recessive disorders in humans are homozygous lethal in mice (17 of 37 genes), possibly because the human mutations are not as disruptive as the mouse alleles. Of the 59 genes, 26 represent the first mouse mutant with publicly available data. Three examples (Sms, Ap4e1, and Smc3) representing the first targeted mouse mutant for each gene are illustrated in Figure 6, and all show similar phenotypic features to their human counterparts.
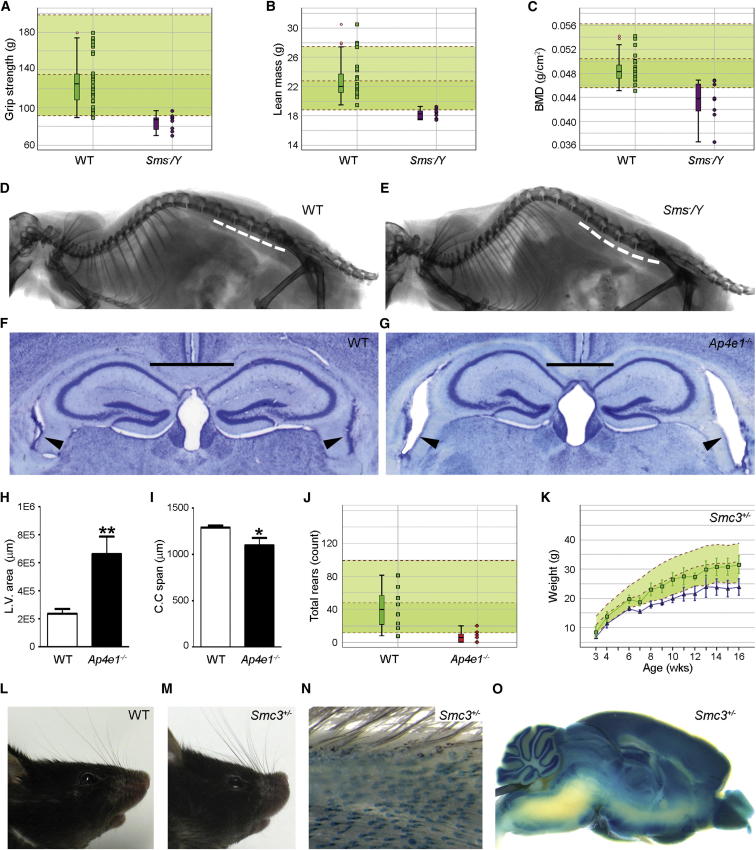
Figure 6.
Correlated Disease Characteristics in Knockouts of Three Known Human Disease Genes
(A–E) Male hemizygotes for the Sms mutation showed similar features to X-linked Snyder-Robinson syndrome.
(A) Reduced grip strength in Sms/Y mice (n = 8) (purple symbols) compared with WT controls (n = 30) (green symbols) and the reference range (n = 793). Each mouse is represented as a single symbol on the graph, with box and whisker plots on the left (see Figure 4B legend).
(B and C) Decreased lean mass (B) and bone mineral density (C) in Sms/Y mice (n = 8) (purple symbols) compared with controls (n = 27) (green symbols) and the reference range (n = 753), with box and whisker plots on the left (see Figure 4B legend).
(D and E) Lumbar lordosis shown by X-ray (seven of eight males) in Sms/Y (E) compared with WT (D).
(F–J) Ap4e1 tm1a/tm1a mice displayed similarities to spastic quadriplegic cerebral palsy 4.
(F–I) Increased lateral ventricle area (arrowheads in F and G) and decreased corpus callosum span (solid lines in F and G) in Ap4e1 tm1a/tm1a mice (G) compared with WT mice (F) with measurements plotted (mean ± SD) in (H) and (I), respectively (∗p < 0.05, ∗∗ p < 0.01; n = 3 mutant males and 34 WT males). Error bars in (H) and (I) are SD.
(J) Decreased rearing in Ap4e1 tm1a/tm1a females (n = 7) (red symbols) compared with WT controls (n = 8) (green symbols) and the reference range (n = 180), with box and whisker plots on the left (see Figure 4B legend).
(K–O) Surviving Smc3 tm1a/+ mice showed similar features to Cornelia de Lange syndrome 3.
(K) Decreased body weight in Smc3 tm1a/+ females (n = 7) fed on high-fat diet. Mean ± SD body weight is plotted against age for Smc3 tm1a/+ females (blue line), WT mice (n = 24; green line), and the reference range (n = 948).
(L and M) Distinct craniofacial abnormalities in Smc3 tm1a/+ mice including upturned snout (M) (three of seven males, one of seven females), which was not observed in WTs (L) (n = 850 male and 859 female).
(N and O) The lacZ reporter gene revealed a distinct Smc3 expression pattern including (N) hair follicles and (O) key brain substructures, noteworthy because of the hirsutism and neurodevelopmental delay aspects of Cornelia de Lange syndrome 3.
See also Table S6.
SMS mutations in humans cause X-linked Snyder-Robinson syndrome involving hypotonia, unsteady gait, diminished muscle mass, kyphoscoliosis, osteoporosis, facial asymmetry, and intellectual disability (Cason et al., 2003). Hemizygous Sms mutant male mice showed reduced muscle strength, lean mass and bone mineral density, lumbar lordosis (Figures 6A–6E), and growth retardation, recapitulating features of the human disease. In addition, male infertility was detected, suggesting a feature that may not have been recognized in humans with SMS mutations.
Spastic paraplegia 51, autosomal recessive, is caused by mutations in AP4E1 and leads to spastic tetraplegia with hyperreflexia and generalized hypertonia, microcephaly, intellectual disability and dilated ventricles, cerebellar atrophy, and/or abnormal white matter (Abou Jamra et al., 2011; Moreno-De-Luca et al., 2011). Homozygous Ap4e1 mutant mice displayed several similarities such as increased lateral ventricle area, decreased corpus callosum span, and decreased rearing (Figures 6F–6J). In addition, hematological changes suggestive of anemia were detected in female Ap4e1 mutants, which have not been reported in humans.
The third example is SMC3, associated with dominantly inherited Cornelia de Lange syndrome 3, featuring facial dysmorphism, hirsutism, growth retardation, neurodevelopmental delay, and upper-limb anomalies (Deardorff et al., 2007). Smc3 mutant mice displayed homozygous lethality (prior to E14.5) and reduced heterozygote viability at P14 (45% instead of the expected 67%; p′ = 0.0064). Surviving Smc3 heterozygotes showed reduced body weight, and a subset showed a distinct craniofacial morphology (Figures 6K–6M). Distinct Smc3 expression in hair follicles and key brain substructures was revealed using lacZ (Figures 6N and 6O), noteworthy because of the hirsutism and neurodevelopmental delay aspects of Cornelia de Lange syndrome 3. In addition, an increase in the number of helper and cytotoxic T cells was observed in the mutant mice, again indicating an aspect that might contribute to the phenotype of patients but that has not yet been reported.
Pleiotropic Effects of Mutations
The phenotypes detected in this study vary from discrete specific defects (e.g., decreased platelet cell number in Crlf3 tm1a/tm1a mutants) to complex phenotypes in which many organ systems are involved (e.g., Spns2 tm1a(KOMP)Wtsi homozygotes show eye, hearing, and immune defects; Nijnik et al., 2012). The distribution of phenotypic hits is shown in Figures 5A and 5B for homozygous and heterozygous mutants, respectively. The peak for homozygotes was the category with no detected abnormalities, whereas the second biggest group consists of mutants with just one phenotypic call. The lines examined as heterozygotes all have at least one hit (viability), but 20 lines have in addition one other abnormal phenotype, and a handful have several. Classifying parameters into five disease categories, we analyzed the distribution of disease areas represented across all 250 mouse lines. The most common phenotypic call was in the category reproduction, development, and musculoskeletal (Figure 5C).
Some abnormal phenotypes are clearly not primary effects; for example, reduced weight may be a secondary consequence of a number of different primary defects. Given that certain phenotypic features would be expected to co-occur frequently, reflecting physiological or developmental associations, a principal component analysis was conducted to look for correlated patterns in the data. Plotting principal component 1 against 2 revealed four main clusters of mouse lines (colored ovoids in Figure 5D). The separation along principal component 2 arises from viability. The remaining separation of clusters marked by red and green from clusters marked blue and yellow (Figure 5D) arises from body weight and associated variables, including DEXA measurements and energy use (Figure 5E). Body weight is a common covariable in disease (Reed et al., 2008), so it is not surprising that it dominates the principal component analysis.
Features of Essential Genes
Genes are generally defined as essential if they are required for survival or fertility. Studies in yeast and worms suggest that genes with paralogs are much less likely to be essential, presumably because the paralog can compensate for the function of the inactivated gene (Gu et al., 2003; Conant and Wagner, 2004). Previous analyses of published data on mouse knockouts did not find a significant difference in essential genes between singleton and duplicated genes (Liang and Li, 2007; Liao and Zhang, 2007). However, the published gene set is biased toward genes involved in development (Makino et al., 2009). In contrast, we found that genes in our set without a paralog were more than twice as likely to be essential, a significant effect (Table S3; Figure 7A).
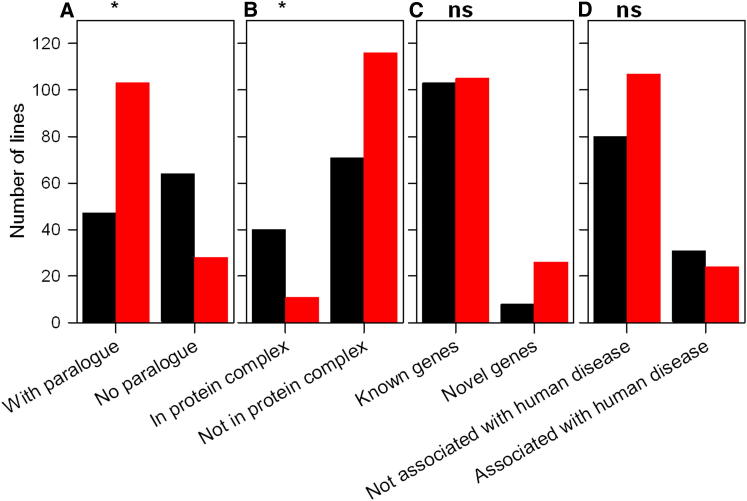
Figure 7.
Features Associated with Essential Genes
Essential genes (black bars) are compared with genes that are not essential for viability (red bars). The asterisk (∗) indicates significant difference. ns, no significant difference in proportion of essential genes between the two categories. Statistics are presented in Table S3.
(A) Genes with no paralog show a significantly larger proportion of essential lines than genes with at least one paralog.
(B) Genes predicted to contribute to protein complexes showed a significantly larger proportion of essential lines than genes not predicted to contribute to a complex.
(C) Novel genes showed no significant difference in proportion of essential genes or number of hits than known genes.
(D) Genes known to underlie human disease were no more likely to be essential than genes not yet associated with human disease.
We next asked if the essential genes in our gene set are more likely to be involved in a protein complex, using an experimentally validated data set of human protein complexes from the CORUM database (Ruepp et al., 2010). We found that genes with a human ortholog that is part of a complex were significantly more likely to be essential (Table S3; Figure 7B).
Finally, we asked if there were certain types of gene products that were more likely to be important for viability/fertility than others. In humans, transcription factor mutations appear enhanced in prenatal disease, and enzymes are overrepresented in diseases with onset in the first year after birth (Jimenez-Sanchez et al., 2001). We investigated four classes of protein identified by GO terms: transcription factors (n = 7), transmembrane proteins (n = 50), enzymes (n = 131), and chromatin-associated proteins (n = 24). Numbers of each were limited, but there was no significant enrichment for essential genes among any of the four groups (Tables S2 and S3).
In summary, we found that essential genes were less likely to have a paralog and more likely to be part of a protein complex, but no specific class of protein appeared more likely to be predictive of essentiality.
Annotating the Function of Novel Genes
There is a large bias in the literature toward analysis of known genes (Edwards et al., 2011), but are genes that have yet to be examined experimentally less likely to underlie disease? Genes in our set that had no associated publications (other than high-throughput genome-wide reports) were compared with genes where some aspect of their function had been described. The proportion of essential genes among the novel set was not significantly different from the known genes (Figure 7C). Furthermore, there was no significant difference in the number of hits observed per line between known and novel genes (Tables S2 and S3). As a second test, we asked if genes with orthologs involved in human disease (having an OMIM disease ID) were enriched in essential genes or the number of phenotypic hits compared with genes not (yet) ascribed to human disease, but there was no significant difference (Tables S2 and S3; Figure 7D). Finally, we compared genes that had been proposed for inclusion by the community (n = 87) with those with no specific request to ask if genes of interest to the community were more likely to be essential or to have detected phenotypes. There was no significant difference between the two groups (Table S3). Thus, known genes are no more likely to be involved in disease than novel genes, emphasizing that much new biology will be uncovered from the analysis of mutations in novel genes.
Discussion
Genetic studies in mice via targeted mutagenesis of ES cells have been successful at illuminating selected aspects of the function of more than 7,000 mammalian genes. However, until recently, these studies have been conducted by individual laboratories and largely directed at previously studied genes. The focused collection of phenotypic information from these mutants has been very information rich, but many aspects remain undetected because they are outside the area of interest of the laboratory generating the mutant. Individual endeavors have led to wide variation in allele design and genetic backgrounds used, and all too often, the mutant is not available to other groups for further analysis. In contrast, the mutant mice described here have the advantage of a common genetic background and a standard allele design with the option of generating conditional mutations, and all are available from public repositories.
The phenotyping described here was not intended to provide an exhaustive characterization of the phenotype of the mutant lines but, rather, to place mutant alleles into broad categories by using screens, generating a pool of genetic resources from which individual mutants can be selected based on their phenotype for secondary follow-up studies. Several of the mutants have been analyzed further following an initial phenotypic observation in the screen, and these add to the depth of our knowledge of biological mechanisms of disease (e.g., Nijnik et al., 2012; Crossan et al., 2011). As the assembled data expands, it will become possible to discern patterns between phenotypes and come to more holistic conclusions about categories of genes. Genes linked by common phenotypes can be grouped together to test for regulatory or other functional interactions and ultimately placed into pathways that in turn will implicate other genes in the disease process. For example, of the four genes associated with abnormal fasting glucose levels in our data set, Slc16a2 can be linked to Ldha via regulation of L-triiodothyronine (Friesema et al., 2006; Miller et al., 2001), but the other two genes, Nsun2 and Cyb561, have no reported regulatory links apart from in vitro protein-protein interactions, so these represent candidates to investigate further. Already some broad conclusions can be drawn from the data set, such as the value of analyzing novel genes, the increased incidence of essentiality in genes with no paralog, and the increased number of genes required for male compared to female fertility. Many completely unexpected associations between genes and phenotypes have been discovered, illustrating the value of a broad-based screen.
Another aspect of our study was the examination of heterozygous mutants, a genotype that often is not studied by individual laboratories. Although this was restricted to mutants that displayed lethality or subviability of homozygotes, it revealed a number of genes with haploinsufficiency, a feature commonly associated with mutations in the human genome but rarely described in mouse knockouts.
The tests used in screening varied considerably in their complexity, cost, and suitability in a high-throughput scenario. The performance of these tests across 250 alleles provided insight into those that should be included or excluded in the efforts to examine 5,000 alleles through the activities of the IMPC. Key considerations are variance in the control group, specificity, sensitivity, effect size, and redundancy.
The major contribution of null alleles will be an improved understanding of biological processes and molecular mechanisms of disease. The null allele will give insight into the temporal and spatial requirements for the gene and will contribute to the establishment of gene networks involved in mammalian disease processes. Furthermore, our data set demonstrates that many features of human Mendelian diseases can be found in the corresponding mouse mutant. The mouse alleles studied here are expected to be null alleles or strong hypomorphs, which may not always reflect the consequence of the human mutation. However, null alleles should reveal haploinsufficiency and recessive effects due to deleterious mutations such as frameshift and nonsense mutations. Null alleles in the mouse are likely to make the largest impact upon understanding human diseases caused by rare variants of large effect size. Complex multifactorial diseases, which may depend on human-specific variants with small effect size or more specific molecular effects such as gain-of-function mutations, will require more customized approaches such as knockin of specific human mutations. Alternative approaches using the mouse for discovering loci underlying complex disease include the Hybrid Mouse Diversity panel and the Collaborative Cross (reviewed by Flint and Eskin, 2012). These allow interrogation of many different loci simultaneously and study of epistatic interactions and can lead to identification of single gene variants causing disease (e.g., Orozco et al., 2012; Andreux et al., 2012), when variants affecting the trait of interest are present in the founders. ENU mutagenesis is another powerful technique that can be used to produce allelic series of mutations with differing effects upon function of single genes (e.g., Andrews et al., 2012). However, the null alleles that we describe here are a complement to these alternative approaches and will be invaluable for defining mechanisms of gene function on a standard genetic background.
The study described in this report builds on the large KOMP/EUCOMM resource of targeted mutations in mouse ES cells (Skarnes et al., 2011) and illustrates the breadth of phenotypic information that can be garnered from an organized effort. The Clinical Phenotyping Pipeline optimized here has been adopted by several other programs within the IMPC; multiple groups are now working together to extend what is described in this report for 250 genes to 5,000 genes over the next 4 years with the vision that this will eventually cover all protein-coding genes. The primary phenotypes and genetic resources emerging from these programs will make a significant contribution to our understanding of mammalian gene function.
Experimental Procedures
Animals
Mice carrying knockout first conditional-ready alleles (Figures S1A and S1B) were generated from the KOMP/EUCOMM targeted ES cell resource using standard techniques. Eight in-house lines were included as known mutant controls. Details of the 250 lines can be found in Table S2. All lines are available from http://www.knockoutmouse.org/; or mouseinterest@sanger.ac.uk. Mice were maintained in a specific pathogen-free unit under a 12 hr light, 12 hr dark cycle with ad libitum access to water and food. The care and use of mice were in accordance with the UK Home Office regulations, UK Animals (Scientific Procedures) Act of 1986.
Genotyping and Allele Quality Control
Short-range, long-range and quantitative PCR strategies (http://www.knockoutmouse.org/kb/25/) were used to evaluate the quality of each allele (Figure S1C). A subset of these assays was used to genotype offspring. The degree of knockdown in homozygotes was assessed by qRT-PCR of adult liver in a subset known to show expression in liver. Details are given in Extended Experimental Procedures.
Extended Experimental Procedures.
Online Database
Results can be accessed at http://www.sanger.ac.uk/mouseportal/, accompanied by step-by-step examples of how to navigate the data. Alternatively, much of the raw data can be downloaded from the MGP Phenotyping Biomart at http://www.sanger.ac.uk/htgt/biomart/martview/. Advice on navigating this Biomart is provided at ftp://ftp.sanger.ac.uk/pub/mgp/extracting_mouse_genetic_program_raw_phenotyping_data.docx. Results have also been summarized in Wikipedia (http://en.wikipedia.org/wiki/Category:Genes_mutated_in_mice) and Mouse Genome Informatics (http://www.informatics.jax.org/).
Animals
The mutant lines screened are listed in Table S2. We generated most of the mutant lines (242/250) reported in detail here using the EUCOMM/KOMP knockout first conditional-ready targeted ES cell resource on a C57BL/6N background (Skarnes et al., 2011) (Figures S1A and S1B). We maintained the mice on a consistent inbred C57BL/6N background (n = 47 lines), or for early lines on mixed C57BL/6 backgrounds (e.g., 190 lines were maintained on a C57BL/6N;C57BL/6Brd-Tyr c-Brd background), to minimize variation in screening results due to strain variation and to facilitate comparison across mutant lines. Eight mutant lines with known phenotypes from other sources were included in early screening as positive controls. All lines are available from http://www.knockoutmouse.org/; or mouseinterest@sanger.ac.uk. Mice were maintained ad libitum on Mouse Breeders Diet (LabDiets 5021-3, IPS, Richmond, USA) unless otherwise stated.
Genotyping and Allele Quality Control
ES cell QC was performed as described (Skarnes et al., 2011). Furthermore, extensive quality control of each allele was performed in mice using a panel of short-range PCR assays (specific for the mutant or wild-type allele, the lacZ reporter gene, 5′ FRT site or 3′ loxP site), quantitative (q) PCR assays (neo cassette and loss of wild-type allele counting systems), and long-range (LR) PCR assays (5′ and 3′ using one primer in the cassette and another outside of the homology arms of the allele design) as summarized in Figure S1C. Further details of the QC protocols are given at http://www.knockoutmouse.org/kb/25/.
Typically, mice were genotyped at postnatal day (P)14 using a combination of the three short-range PCR assays or the qPCR neo cassette allele-counting assay. Upon completion of phenotyping, genotyping was repeated and data were only accepted from mice for which the second genotype was concordant with the P14 genotype.
A subset of 25 lines, selected because of previously-reported gene expression in liver, was used to assess the degree of knockdown resulting from the targeting event. A TaqMan assay was devised to detect wild-type splicing between the exons on either side of the mutagenic cassette. Samples showing wild-type expression by qRTPCR were confirmed by end-point RTPCR and sequencing. ∼350ng of total RNA extracted from liver was used in each reaction, performed in triplicate as duplex reactions using the RNA-to-Ct one step kit (Applied Biosystems) with Gapdh or B2m as endogenous controls and analyzed using a 7900HT qPCR machine with RQ manager software v1.2 (Applied Biosystems).
Phenotyping Pipeline and Tests
Viability was assessed at P14 and, for those lines classed as lethal or sub-viable at P14, again at E14.5 (Figure 1A). A minimum of 28 genotype-confirmed, live progeny from heterozygous intercrosses were required to assess viability. Based on exact binomial probability calculations, zero homozygotes from 28 progeny gave 95% confidence that the probability of homozygote survival was ≤ 40%. Lines were classed as homozygous fertile if offspring were born from homozygous parents, regardless of whether the offspring survived to weaning.
At 4 weeks of age, mice undergoing the Clinical Phenotyping Pipeline (Figure 1B) were transferred from Mouse Breeders Diet to a high fat (21.4% fat by crude content; 42% calories provided by fat) dietary challenge (Special Diet Services Western RD 829100, SDS, Witham, UK) for the remainder of the pipeline. This pipeline included the established phenotyping tests described in Table S4. For most tests in this pipeline 7 male and 7 female mutants were used, with the exceptions of the Auditory Brainstem Response (n = 4, independent of gender) and erythrocyte micronuclei (7 males), both deemed to be sufficient with the reduced numbers, and indirect calorimetry (7 males) and biobanking of 41 tissues and organs in paraffin blocks (2 males, 2 females), both limited by operational constraints. If homozygotes were lethal, difficult to produce due to sub-viability or were unsuitable for screening due to welfare concerns, we used heterozygotes for screening. For each genetic background, control cohorts (7M + 7F) were run each week.
A second primary pipeline included challenges with two infectious agents, Citrobacter rodentium (8 females) and Salmonella Typhimurium (8 males), with matched controls run simultaneously (Table S4). In both challenges we looked at colonization of target tissues at 14 and 28 days post infection. Tissue was biobanked in paraffin and serum taken from the Salmonella challenged animals to measure antigen specific IgG (and subclass) antibodies.
Statistical and Bioinformatic Analysis
For GO term enrichment using TermFinder (Boyle et al., 2004) only high quality experimental evidence codes were included (EXP, IDA, IMP, IPI, IGI, IEP and IC). Positive control lines were excluded from the gene set for this analysis. For GO term enrichment using FuncAssociate v2.0 (Berriz et al., 2009) (http://llama.mshri.on.ca/funcassociate/), the evidence code IEA (Inferred from Electronic Annotation) was excluded. This software used a gene association file downloaded from ftp.geneontology.org on 26th September 2011. Of the 14139 GO terms queried, 83 (0.59%) were classed as being over-represented and 6 (0.04%) under-represented in our gene set. Revigo was used to reduce the GO term redundancy to give a representative subset of terms (Supek et al., 2011). The gene set was found to be under-represented in only one area: “Sensory perception of smell.” The gene set was found to be over-represented in 20 spread over a variety of processes with no one area dominating.
Principal components analysis, performed in SIMCA-P (http://www.umetrics.com), was used to assess gene clustering based on variation across the phenotypic hits. The methodology was as default, except that data rescaling was turned off as it was not required for this binary data set. Model predictivity was confirmed by a cross validation procedure (Wold, 1978). The differences between the clusters were investigated using a contribution plot, which showed the differences in scaled units between the highlighted groups for all variables in the model.
Using G∗Power (Version 3.1.3) sensitivity approach (Erdfelder et al., 1996) we calculated the power of our gene set to ask if specific types of protein were more likely to be important for viability and fertility. For a target power of 80%, with the numbers of genes we had in each category, the transcription factor data set needed an effect size (ES) of 45%, the transmembrane proteins needed an ES of 19.3%, the enzyme data set needed an ES of 19% and the chromatin-associated proteins needed an ES of 47% to be detected at the 0.05 threshold.
For protein characterization, amino acid sequences were obtained from Ensembl, release 59, and the longest protein product was chosen as a representative for each gene. Paralog assignments were also obtained from Ensembl (Flicek et al., 2011). Transcription factor annotations were obtained from the DBD database (Wilson et al., 2008) and transmembrane regions were predicted with Phobius (Käll et al., 2004). Involvement in a protein complex was inferred from CORUM, an experimentally-validated data set of human protein complexes (Ruepp et al., 2010).
Phenotyping Pipeline and Tests
The typical workflow from chimera to primary phenotyping pipelines and an outline of the clinical phenotyping pipeline are presented in Figure 1. Details of batch size are given in Figure S2A. These pipelines include established tests used to characterize systematically every line of mice as described in Table S4.
Histochemical Analysis of the lacZ Reporter
Adult whole-mount lacZ reporter gene expression was carried out essentially as described by Valenzuela et al. (2003).
Statistical and Bioinformatic Analysis
For continuous data, including time course, a reference range approach was used to identify phenotypic variants as detailed in Figures S3A and S3B. Fisher’s exact test was used to assess categorical data (Figure S3C). These automated calls were complemented by a manual assessment made by biological experts. An example of the establishment of the reference range is given in Figure S2B.
Downstream data analysis was performed using SPSS (version 17.0.2), R, and SIMCA-P (V-12.0, Umetrics). The data structure and biological question determined the statistical test used; details are in Table S3. Principal components analysis was performed in SIMCA-P (http://www.umetrics.com).
Further details of analyses and gene annotations are given in Extended Experimental Procedures.
Acknowledgments
We thank Alex Bateman, Lars Barquist, Steve O’Rahilly, Keith Burling, Seth Grant, Pentao Liu, and Lorraine Everett for advice and the EUMODIC consortium for discussions. This work was supported by the Wellcome Trust (grant Nos. 098051 to Wellcome Trust Sanger Institute and RG45277 PCAG/116 to F.M.W.), Medical Research Council (to K.P.S. and F.M.W.), European Commission (EUMODIC contract No. LSHG-CT-2006-037188), NIH (EY08213 to S.H.T. and 5K08EY020530-02 to V.B.M.), Research to Prevent Blindness (to S.H.T. and V.B.M.), Australian Research Council (DP1092723 to I.S.), and Cancer Research UK (to D.J.A.). J.K.W. and K.P.S. conceived and devised the single phenotyping pipeline and principles of analysis and presentation of the data; R.R.S., J.K.W., R.H., J.R.B., and E.R. managed mouse production and genotyping; J.N.B. and J.S. managed mouse breeding; A.K.G., C.P., J.E., D.S., N.I., and J.K.W. managed mouse phenotyping and analyzed data; the Sanger Institute Mouse Genetics Project team contributed to all aspects of the work; S.C. and G.D. led the infectious challenge screen; V.B.M. and S.H.T. led the eye histopathology screen; I.S. and F.M.W. led the skin screen; J.F. led the brain histopathology screen; D.J.A. led the micronucleus screen; J.N.B. and J.R.B. managed mouse distribution; J.K.W., A.K.G., M.B., and K.P.S. compiled Tables S1, S2, S5, and S6; N.A.K., D.M., D.S., and M.B. carried out annotation and statistical analysis; N.C.A., D.G.M., and W.C.S. led development of informatics support; D.W.L. wrote Wiki pages for the mutants; J.K.W., D.J.A., R.R.S., and K.P.S. led the project; and K.P.S., A.B., and J.K.W. wrote the paper with contributions from all authors.
Published: July 18, 2013
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information includes Extended Experimental Procedures, three figures, seven tables, and a complete list of contributors from the Sanger Institute Mouse Genetics Project and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.06.022.
Contributor Information
Karen P. Steel, Email: karen.steel@kcl.ac.uk.
The Sanger Institute Mouse Genetics Project:
Lauren Baker, Caroline Barnes, Ryan Beveridge, Emma Cambridge, Damian Carragher, Prabhjoat Chana, Kay Clarke, Yvette Hooks, Natalia Igosheva, Ozama Ismail, Hannah Jackson, Leanne Kane, Rosalind Lacey, David Tino Lafont, Mark Lucas, Simon Maguire, Katherine McGill, Rebecca E. McIntyre, Sophie Messager, Lynda Mottram, Lee Mulderrig, Selina Pearson, Hayley J. Protheroe, Laura-Anne Roberson, Grace Salsbury, Mark Sanderson, Daniel Sanger, Carl Shannon, Paul C. Thompson, Elizabeth Tuck, Valerie E. Vancollie, Lisa Brackenbury, Wendy Bushell, Ross Cook, Priya Dalvi, Diane Gleeson, Bishoy Habib, Matt Hardy, Kifayathullah Liakath-Ali, Evelina Miklejewska, Stacey Price, Debarati Sethi, Elizabeth Trenchard, Dominique von Schiller, Sapna Vyas, Anthony P. West, John Woodward, Elizabeth Wynn, Arthur Evans, David Gannon, Mark Griffiths, Simon Holroyd, Vivek Iyer, Christian Kipp, Morag Lewis, Wei Li, Darren Oakley, David Richardson, Damian Smedley, Chukwuma Agu, Jackie Bryant, Liz Delaney, Nadia I. Gueorguieva, Helen Tharagonnet, Anne J. Townsend, Daniel Biggs, Ellen Brown, Adam Collinson, Charles-Etienne Dumeau, Evelyn Grau, Sarah Harrison, James Harrison, Catherine E. Ingle, Helen Kundi, Alla Madich, Danielle Mayhew, Tom Metcalf, Stuart Newman, Johanna Pass, Laila Pearson, Helen Reynolds, Caroline Sinclair, Hannah Wardle-Jones, Michael Woods, Liam Alexander, Terry Brown, Francesca Flack, Carole Frost, Nicola Griggs, Silvia Hrnciarova, Andrea Kirton, Jordan McDermott, Claire Rogerson, Gemma White, Pawel Zielezinski, Tia DiTommaso, Andrew Edwards, Emma Heath, Mary Ann Mahajan, and Binnaz Yalcin
Supplemental Information
Table S1. Heatmap Showing Phenotypic Hits, Related to Figure 1
Heatmap of 250 genes, extracted from the public heatmap on Sanger Institute mouse portal. Each row represents the phenotype data from the assessed genotype, with red boxes indicating a hit, pale blue boxes indicating data falling within the reference range, and white boxes indicating that phenotyping was not completed. The key is at the top right of the table. Where a ≥ symbol occurs, clicking on the box will lead to further details of the data. Each data column represents a set of variables assessed using the indicated screening test, and a red box indicates that at least one variable in that test was classed as significant in either male or female groups or both. A complete, up-to-date heatmap with all lines screened can be downloaded from http://www.sanger.ac.uk/mouseportal/.
Table S2. Expanded Heatmap to Show All Variables Assessed and Gene Annotations, Related to Figure 1
In this heatmap, we expanded the columns to show one column for each variable scored in the phenotypic screening. Significant differences from the control baseline (hits) are shown by 1 and are shaded pink, unfilled boxes marked 0 indicate the results were not significantly different to controls, and a blank box shows the test was not done. A box is shaded pink if either the male, the female, or both groups show a significant difference. Missing data amounted to only 2.14% of the total. We assessed a total of 263 parameters, including 147 categorical variables and 116 quantitative variables. Further annotation of each mouse line and gene is included. Most are self-explanatory. (Column I) A line is classed as known if either there is a published mutant mouse allele on MGI, or as searched through Pubmed there is a paper published including the gene function or gene cloned in any species. A gene is classed as novel if the only publication is a high throughput or mapping paper or no publication. (Column U) For protein complex annotations, we took Entrez IDs of human homologs from the HMD_Human5.rpt table (from the MGI db) and checked whether these were in the CORUM database of known protein complexes. (Column Z) A line is classed as haploinsufficient (HI) if heterozygotes were screened and showed some phenotypic hits in addition to being lethal or sub-viable; A line is classed as essential (E) if homozygotes were either lethal or sub-viable; A line is classed as essential for fertility (EF) if the tested genotype showed fertility defects; A line is classed as non-essential (NE) if homozygotes show no phenotypic hits.
Table S3. Summary of Statistical Test Results, Related to Figure 2
Table S4. Brief Descriptions of Phenotypic Screening Tests, Related to Experimental Procedures
Table S5. Comparison of Our Phenotypic Data with Published Data, Related to Figure 4
We compared our data with existing published data on the same or other mutations of the same gene. Comparisons are listed according to whether they agreed with published data, we would not have seen the feature because we did not screen for it, we did not see the feature even though we screened for it, and phenotypic observations not noted in previous publications.
Table S6. Comparison of Our Phenotypic Data with Human Disease Features, Related to Figure 6
We compared our data with human disease features where an orthologous gene was involved in a human disease. We found 27/59 (45%) showed something consistent with the human disease, 21/59 (36%) where the mouse phenotype did not match the report of the human disease at all, and 11/59 (19%) where there appears to be a legitimate reason why we might not have seen something, for example, cases where the disease is recessive in humans but the mouse homozygotes are lethal and so we screened heterozygotes. In a number of cases (Column H) we detected phenotypes in the mouse that have not been reported in humans but may be worth looking for. For 26 genes, our findings represent the first mouse mutant reported, and these may serve as models for the respective human diseases.
Table S7. Key to Individual Variables in Figure 5E
Listed left to right.
Document S1. Complete Author List
Document S2. Article plus Supplemental Information
References
- Abou Jamra R., Philippe O., Raas-Rothschild A., Eck S.H., Graf E., Buchert R., Borck G., Ekici A., Brockschmidt F.F., Nöthen M.M. Adaptor protein complex 4 deficiency causes severe autosomal-recessive intellectual disability, progressive spastic paraplegia, shy character, and short stature. Am. J. Hum. Genet. 2011;88:788–795. doi: 10.1016/j.ajhg.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreux P.A., Williams E.G., Koutnikova H., Houtkooper R.H., Champy M.F., Henry H., Schoonjans K., Williams R.W., Auwerx J. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. 2012;150:1287–1299. doi: 10.1016/j.cell.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews T.D., Whittle B., Field M.A., Balakishnan B., Zhang Y., Shao Y., Cho V., Kirk M., Singh M., Xia Y. Massively parallel sequencing of the mouse exome to accurately identify rare, induced mutations: an immediate source for thousands of new mouse models. Open Biol. 2012;2:120061. doi: 10.1098/rsob.120061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayadi A., Birling M.C., Bottomley J., Bussell J., Fuchs H., Fray M., Gailus-Durner V., Greenaway S., Houghton R., Karp N. Mouse large-scale phenotyping initiatives: overview of the European Mouse Disease Clinic (EUMODIC) and of the Wellcome Trust Sanger Institute Mouse Genetics Project. Mamm. Genome. 2012;23:600–610. doi: 10.1007/s00335-012-9418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer E.L., Chen A.F., Chen A.H., Li Z., Mark H.F., Smith R.J.H., Jackson C.L. 2E4/Kaptin (KPTN)— a candidate gene for the hearing loss locus, DFNA4. Ann. Hum. Genet. 2000;64:189–196. doi: 10.1046/j.1469-1809.2000.6430189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S.D., Moore M.W. Towards an encyclopaedia of mammalian gene function: the International Mouse Phenotyping Consortium. Dis. Model. Mech. 2012;5:289–292. doi: 10.1242/dmm.009878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cason A.L., Ikeguchi Y., Skinner C., Wood T.C., Holden K.R., Lubs H.A., Martinez F., Simensen R.J., Stevenson R.E., Pegg A.E., Schwartz C.E. X-linked spermine synthase gene (SMS) defect: the first polyamine deficiency syndrome. Eur. J. Hum. Genet. 2003;11:937–944. doi: 10.1038/sj.ejhg.5201072. [DOI] [PubMed] [Google Scholar]
- Conant G.C., Wagner A. Duplicate genes and robustness to transient gene knock-downs in Caenorhabditis elegans. Proc. Biol. Sci. 2004;271:89–96. doi: 10.1098/rspb.2003.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossan G.P., van der Weyden L., Rosado I.V., Langevin F., Gaillard P.H., McIntyre R.E., Gallagher F., Kettunen M.I., Lewis D.Y., Brindle K., Sanger Mouse Genetics Project Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat. Genet. 2011;43:147–152. doi: 10.1038/ng.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff M.A., Kaur M., Yaeger D., Rampuria A., Korolev S., Pie J., Gil-Rodríguez C., Arnedo M., Loeys B., Kline A.D. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am. J. Hum. Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A.M., Isserlin R., Bader G.D., Frye S.V., Willson T.M., Yu F.H. Too many roads not taken. Nature. 2011;470:163–165. doi: 10.1038/470163a. [DOI] [PubMed] [Google Scholar]
- Fischer H., Szabo S., Scherz J., Jaeger K., Rossiter H., Buchberger M., Ghannadan M., Hermann M., Theussl H.C., Tobin D.J. Essential role of the keratinocyte-specific endonuclease DNase1L2 in the removal of nuclear DNA from hair and nails. J. Invest. Dermatol. 2011;131:1208–1215. doi: 10.1038/jid.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher C.L., Pineault N., Brookes C., Helgason C.D., Ohta H., Bodner C., Hess J.L., Humphries R.K., Brock H.W. Loss-of-function Additional sex combs like 1 mutations disrupt hematopoiesis but do not cause severe myelodysplasia or leukemia. Blood. 2010;115:38–46. doi: 10.1182/blood-2009-07-230698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., Eskin E. Genome-wide association studies in mice. Nat. Rev. Genet. 2012;13:807–817. doi: 10.1038/nrg3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forzati F., Federico A., Pallante P., Abbate A., Esposito F., Malapelle U., Sepe R., Palma G., Troncone G., Scarfò M. CBX7 is a tumor suppressor in mice and humans. J. Clin. Invest. 2012;122:612–623. doi: 10.1172/JCI58620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesema E.C., Kuiper G.G., Jansen J., Visser T.J., Kester M.H. Thyroid hormone transport by the human monocarboxylate transporter 8 and its rate-limiting role in intracellular metabolism. Mol. Endocrinol. 2006;20:2761–2772. doi: 10.1210/me.2005-0256. [DOI] [PubMed] [Google Scholar]
- Fuchs H., Gailus-Durner V., Neschen S., Adler T., Afonso L.C., Aguilar-Pimentel J.A., Becker L., Bohla A., Calzada-Wack J., Cohrs C. Innovations in phenotyping of mouse models in the German Mouse Clinic. Mamm. Genome. 2012;23:611–622. doi: 10.1007/s00335-012-9415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Steinmetz L.M., Gu X., Scharfe C., Davis R.W., Li W.H. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- Hawkins T.E., Roes J., Rees D., Monkhouse J., Moss S.E. Immunological development and cardiovascular function are normal in annexin VI null mutant mice. Mol. Cell. Biol. 1999;19:8028–8032. doi: 10.1128/mcb.19.12.8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Sanchez G., Childs B., Valle D. Human disease genes. Nature. 2001;409:853–855. doi: 10.1038/35057050. [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Kim Y.S., Yoo K.J., Lee H.J., Lee D.R., Yeo C.Y., Baek K.H. The expression of Usp42 during embryogenesis and spermatogenesis in mouse. Gene Expr. Patterns. 2007;7:143–148. doi: 10.1016/j.modgep.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Laughlin M.R., Lloyd K.C., Cline G.W., Wasserman D.H., Mouse Metabolic Phenotyping Centers Consortium NIH Mouse Metabolic Phenotyping Centers: the power of centralized phenotyping. Mamm. Genome. 2012;23:623–631. doi: 10.1007/s00335-012-9425-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Li W.H. Gene essentiality, gene duplicability and protein connectivity in human and mouse. Trends Genet. 2007;23:375–378. doi: 10.1016/j.tig.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Liao B.Y., Zhang J. Mouse duplicate genes are as essential as singletons. Trends Genet. 2007;23:378–381. doi: 10.1016/j.tig.2007.05.006. [DOI] [PubMed] [Google Scholar]
- MacArthur D.G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., Jostins L., Habegger L., Pickrell J.K., Montgomery S.B., 1000 Genomes Project Consortium A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino T., Hokamp K., McLysaght A. The complex relationship of gene duplication and essentiality. Trends Genet. 2009;25:152–155. doi: 10.1016/j.tig.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Miller L.D., Park K.S., Guo Q.M., Alkharouf N.W., Malek R.L., Lee N.H., Liu E.T., Cheng S.Y. Silencing of Wnt signaling and activation of multiple metabolic pathways in response to thyroid hormone-stimulated cell proliferation. Mol. Cell. Biol. 2001;21:6626–6639. doi: 10.1128/MCB.21.19.6626-6639.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell K.J., Pinson K.I., Kelly O.G., Brennan J., Zupicich J., Scherz P., Leighton P.A., Goodrich L.V., Lu X., Avery B.J. Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat. Genet. 2001;28:241–249. doi: 10.1038/90074. [DOI] [PubMed] [Google Scholar]
- Moreno-De-Luca A., Helmers S.L., Mao H., Burns T.G., Melton A.M., Schmidt K.R., Fernhoff P.M., Ledbetter D.H., Martin C.L. Adaptor protein complex-4 (AP-4) deficiency causes a novel autosomal recessive cerebral palsy syndrome with microcephaly and intellectual disability. J. Med. Genet. 2011;48:141–144. doi: 10.1136/jmg.2010.082263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijnik A., Clare S., Hale C., Chen J., Raisen C., Mottram L., Lucas M., Estabel J., Ryder E., Adissu H., Sanger Mouse Genetics Project The role of sphingosine-1-phosphate transporter Spns2 in immune system function. J. Immunol. 2012;189:102–111. doi: 10.4049/jimmunol.1200282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco L.D., Bennett B.J., Farber C.R., Ghazalpour A., Pan C., Che N., Wen P., Qi H.X., Mutukulu A., Siemers N. Unraveling inflammatory responses using systems genetics and gene-environment interactions in macrophages. Cell. 2012;151:658–670. doi: 10.1016/j.cell.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.Y., Jeker L.T., Carver-Moore K., Oh A., Liu H.J., Cameron R., Richards H., Li Z., Adler D., Yoshinaga Y. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 2012;1:385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser H.M., Koike-Yusa H., Cooper J.D., Law F.C., Bradley A. A resource of vectors and ES cells for targeted deletion of microRNAs in mice. Nat. Biotechnol. 2011;29:840–845. doi: 10.1038/nbt.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed D.R., Lawler M.P., Tordoff M.G. Reduced body weight is a common effect of gene knockout in mice. BMC Genet. 2008;9:4. doi: 10.1186/1471-2156-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A., Waegele B., Lechner M., Brauner B., Dunger-Kaltenbach I., Fobo G., Frishman G., Montrone C., Mewes H.W. CORUM: the comprehensive resource of mammalian protein complexes—2009. Nucleic Acids Res. 2010;38(Database issue):D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalzigaug R., Rodriguiz R.M., Phillips L.E., Davidson C.E., Wetsel W.C., Premont R.T. Anxiety-like behaviors in mice lacking GIT2. Neurosci. Lett. 2009;451:156–161. doi: 10.1016/j.neulet.2008.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T., Li L., Tang J., Li Y., Lin W.Y., Martin F., Grant D., Solloway M., Parker L., Ye W. A mouse knockout library for secreted and transmembrane proteins. Nat. Biotechnol. 2010;28:749–755. doi: 10.1038/nbt.1644. [DOI] [PubMed] [Google Scholar]
- Testa G., Schaft J., van der Hoeven F., Glaser S., Anastassiadis K., Zhang Y., Hermann T., Stremmel W., Stewart A.F. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 2004;38:151–158. doi: 10.1002/gene.20012. [DOI] [PubMed] [Google Scholar]
- Valenzuela D.M., Murphy A.J., Frendewey D., Gale N.W., Economides A.N., Auerbach W., Poueymirou W.T., Adams N.C., Rojas J., Yasenchak J. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat. Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- Wakana S., Suzuki T., Furuse T., Kobayashi K., Miura I., Kaneda H., Yamada I., Motegi H., Toki H., Inoue M. Introduction to the Japan Mouse Clinic at the RIKEN BioResource Center. Exp. Anim. 2009;58:443–450. doi: 10.1538/expanim.58.443. [DOI] [PubMed] [Google Scholar]
Supplemental References
- Berriz G.F., Beaver J.E., Cenik C., Tasan M., Roth F.P. Next generation software for functional trend analysis. Bioinformatics. 2009;25:3043–3044. doi: 10.1093/bioinformatics/btp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdfelder E., Faul F., Buchner A. GPOWER: a general power analysis program. Behav. Res. Methods Instrum. Comput. 1996;28:1–11. [Google Scholar]
- Flicek P., Amode M.R., Barrell D., Beal K., Brent S., Chen Y., Clapham P., Coates G., Fairley S., Fitzgerald S. Ensembl 2011. Nucleic Acids Res. 2011;39(Database issue):D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L., Krogh A., Sonnhammer E.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D., Charoensawan V., Kummerfeld S.K., Teichmann S.A. DBD—taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 2008;36(Database issue):D88–D92. doi: 10.1093/nar/gkm964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S. Cross-validatory estimation of the number of components in factor and principal components models. Technometrics. 1978;20:397–405. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Heatmap Showing Phenotypic Hits, Related to Figure 1
Heatmap of 250 genes, extracted from the public heatmap on Sanger Institute mouse portal. Each row represents the phenotype data from the assessed genotype, with red boxes indicating a hit, pale blue boxes indicating data falling within the reference range, and white boxes indicating that phenotyping was not completed. The key is at the top right of the table. Where a ≥ symbol occurs, clicking on the box will lead to further details of the data. Each data column represents a set of variables assessed using the indicated screening test, and a red box indicates that at least one variable in that test was classed as significant in either male or female groups or both. A complete, up-to-date heatmap with all lines screened can be downloaded from http://www.sanger.ac.uk/mouseportal/.
Table S2. Expanded Heatmap to Show All Variables Assessed and Gene Annotations, Related to Figure 1
In this heatmap, we expanded the columns to show one column for each variable scored in the phenotypic screening. Significant differences from the control baseline (hits) are shown by 1 and are shaded pink, unfilled boxes marked 0 indicate the results were not significantly different to controls, and a blank box shows the test was not done. A box is shaded pink if either the male, the female, or both groups show a significant difference. Missing data amounted to only 2.14% of the total. We assessed a total of 263 parameters, including 147 categorical variables and 116 quantitative variables. Further annotation of each mouse line and gene is included. Most are self-explanatory. (Column I) A line is classed as known if either there is a published mutant mouse allele on MGI, or as searched through Pubmed there is a paper published including the gene function or gene cloned in any species. A gene is classed as novel if the only publication is a high throughput or mapping paper or no publication. (Column U) For protein complex annotations, we took Entrez IDs of human homologs from the HMD_Human5.rpt table (from the MGI db) and checked whether these were in the CORUM database of known protein complexes. (Column Z) A line is classed as haploinsufficient (HI) if heterozygotes were screened and showed some phenotypic hits in addition to being lethal or sub-viable; A line is classed as essential (E) if homozygotes were either lethal or sub-viable; A line is classed as essential for fertility (EF) if the tested genotype showed fertility defects; A line is classed as non-essential (NE) if homozygotes show no phenotypic hits.
Table S3. Summary of Statistical Test Results, Related to Figure 2
Table S4. Brief Descriptions of Phenotypic Screening Tests, Related to Experimental Procedures
Table S5. Comparison of Our Phenotypic Data with Published Data, Related to Figure 4
We compared our data with existing published data on the same or other mutations of the same gene. Comparisons are listed according to whether they agreed with published data, we would not have seen the feature because we did not screen for it, we did not see the feature even though we screened for it, and phenotypic observations not noted in previous publications.
Table S6. Comparison of Our Phenotypic Data with Human Disease Features, Related to Figure 6
We compared our data with human disease features where an orthologous gene was involved in a human disease. We found 27/59 (45%) showed something consistent with the human disease, 21/59 (36%) where the mouse phenotype did not match the report of the human disease at all, and 11/59 (19%) where there appears to be a legitimate reason why we might not have seen something, for example, cases where the disease is recessive in humans but the mouse homozygotes are lethal and so we screened heterozygotes. In a number of cases (Column H) we detected phenotypes in the mouse that have not been reported in humans but may be worth looking for. For 26 genes, our findings represent the first mouse mutant reported, and these may serve as models for the respective human diseases.
Table S7. Key to Individual Variables in Figure 5E
Listed left to right.
Document S1. Complete Author List
Document S2. Article plus Supplemental Information