Stabilization of Actin Bundles by a Dynamin 1/Cortactin Ring Complex Is Necessary for Growth Cone Filopodia (original) (raw)
Abstract
Dynamin GTPase, a key molecule in endocytosis, mechanically severs the invaginated membrane upon GTP hydrolysis. Dynamin functions also in regulating actin cytoskeleton, but the mechanisms are yet to be defined. Here we show that dynamin 1, a neuronal isoform of dynamin, and cortactin form ring complexes, which twine around F-actin bundles and stabilize them. By negative-staining EM, dynamin 1–cortactin complexes appeared as “open” or “closed” rings depending on guanine nucleotide conditions. By pyrene actin assembly assay, dynamin 1 stimulated actin assembly in mouse brain cytosol. In vitro incubation of F-actin with both dynamin 1 and cortactin led to the formation of long and thick actin bundles, on which dynamin 1 and cortactin were periodically colocalized in puncta. A depolymerization assay revealed that dynamin 1 and cortactin increased the stability of actin bundles, most prominently in the presence of GTP. In rat cortical neurons and human neuroblastoma cell line, SH-SY5Y, both dynamin 1 and cortactin localized on actin filaments and the bundles at growth cone filopodia as revealed by immunoelectron microscopy. In SH-SY5Y cell, acute inhibition of dynamin 1 by application of dynamin inhibitor led to growth cone collapse. Cortactin knockdown also reduced growth cone filopodia. Together, our results strongly suggest that dynamin 1 and cortactin ring complex mechanically stabilizes F-actin bundles in growth cone filopodia. Thus, the GTPase-dependent mechanochemical enzyme property of dynamin is commonly used both in endocytosis and regulation of F-actin bundles by a dynamin 1–cortactin complex.
Introduction
Dynamin 1, a neuronal isoform of dynamin, has primarily been studied as a key molecule involved in clathrin-mediated endocytosis of synaptic vesicles. Dynamin 1 assembles into a helical ring at the neck of endocytic pits (Takei et al., 1995), and the conformational changes of these polymerized dynamin upon GTP hydrolysis provides the mechanical force to sever the endocytic pit (Sweitzer and Hinshaw, 1998; Takei et al., 1998; Marks et al., 2001; Roux et al., 2006; Ramachandran and Schmid, 2008).
Dynamin's function as a mechanochemical enzyme in membrane fission is strongly supported by the recently revealed crystal structure of dynamin 1 (dynamin ΔPRD) comprised of the GTPase domain, the bundle signaling element (BSE), and the stalk and the pleckstrin homology (PH) domain (Faelber et al., 2011; Ford et al., 2011). The stalk is responsible for dimerization of dynamin in a criss-cross fashion, and for interaction with the PH domain and BSE of the neighboring dynamin molecule in a polymerized helix. In addition, dynamin 1 has a proline/arginine-rich domain (PRD) at the C terminus that interacts with various SH3 domain-containing synaptic endocytic proteins, including amphiphysin 1 (David et al., 1996; Takei et al., 1999; Yoshida et al., 2004), endophilin (Farsad et al., 2001), sorting nexin 9 (Ramachandran and Schmid, 2008), syndapin (Kessels and Qualmann, 2004), and intersectin (Yamabhai et al., 1998).
Cortactin, an F-actin-binding protein, binds to the ubiquitously expressed dynamin 2 (McNiven et al., 2000). Cortactin has an N-terminal acidic domain that binds to Arp2/3 complexes, F-actin-binding “cortactin repeats,” and a C-terminal SH3 domain. Thus, dynamin 2 is thought to be involved in the regulation of the actin cytoskeleton (Schafer et al., 2002; Cao et al., 2003; Zhu et al., 2005; Mooren et al., 2009; Yamada et al., 2009a). Several studies implicate dynamin's GTPase activity in the regulation of actin dynamics, including remodeling of actin filaments (Mooren et al., 2009), actin comet formation (Lee and De Camilli, 2002; Orth et al., 2002) or podosomes (Ochoa et al., 2000; Bruzzaniti et al., 2005), and maintenance of cell shape (Damke et al., 1994). It is currently unknown whether dynamin functions as a GTP-driven mechanochemical enzyme in actin regulation.
Actin is enriched in growth cones that are implicated in axon guidance. Growth cone are stabilized when they encounter attractive guidance molecules, and they retract upon contact with repellent guidance molecules (Dent and Gertler, 2003; Geraldo and Gordon-Weeks, 2009; Lowery and Van Vactor, 2009). Growth cone is spread by radially arranged filopodia, which are supported by actin bundles (Lewis and Bridgman, 1992), and the stability of the actin bundles is crucial for the growth cone morphology. Although F-actin cross-linking proteins, including α-actinin or Fascin, are thought to stabilize the F-actin bundles (Courson and Rock, 2010), precise mechanisms are hardly unknown.
In this study, we demonstrate that dynamin 1 and cortactin form ring-shaped complexes, and the rings grab the F-actin bundles using dynamin's mechanochemical properties and stabilize the bundles. This mechanism is implicated in the formation of growth cone filopodia, and essential for the stabilization of growth cones.
Materials and Methods
Antibodies and reagents.
The rabbit polyclonal antibody against dynamin 1 (CK(633)EKASETEENGSDSF(647); PA1-660), mouse monoclonal antibody against clathrin heavy chain (MA1-065), and rabbit polyclonal anti-dynamin 3 antibody (PA1-662) were purchased from Thermo Scientific. The rabbit polyclonal anti-myc antibody (C3956), mouse monoclonal anti-β-actin antibody (A5441), and dynasore were purchased from Sigma-Aldrich. The goat polyclonal anti-dynamin 2 antibody (sc-6400), and mouse monoclonal antibodies against myc (sc-40) and GFP (sc-9996) were purchased from Santa Cruz Biotechnology. The mouse monoclonal antibody against cortactin (05-180) was purchased from Millipore. The rabbit polyclonal dynamin 1 antibody (EP801) was purchased from Epitomics Inc. Alexa Fluor 488-conjugated anti-rabbit IgG, Rhodamine-conjugated anti-mouse IgG, and Rhodamine- and Alexa Fluor 488-labeled phalloidin were all purchased from Life Technologies. Goat anti-mouse IgG- and goat anti-rabbit IgG-conjugated gold particles were purchased from British BioCell International. Rabbit α-actinin was purchased from Cytoskeleton Inc.
Cell culture.
Cortical neurons from postnatal day 0 (P0)–P2 mouse brains of either sex were cultured for 4–6 d, as described previously (Hayashi et al., 2008). SH-SY5Y (CRL-2266, ATCC) and NG108-15 (HB-12317, ATCC) cells were cultured in DMEM with 10% FBS at 37°C under 5% CO2 and were differentiated with 10 μm retinoic acid or 1 mm dibutyryl cAMP, respectively, 2–3 d before experiments. Poly-d-lysine was used as a coating substrate for cortical neurons and NG108-15 cells, while fibronectin was used for SH-SY5Y cells.
Purification of recombinant proteins.
A point mutation for dynamin 1 K44A was introduced into WT human dynamin 1 using the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies). His-tagged dynamin 1 or dynamin 1 K44A was expressed using the Bac-to-Bac baculovirus expression system (Life Technologies) and purified as described previously (Warnock et al., 1997). The purified dynamin solutions were concentrated using Centriplus YM50 (Millipore). By reversed phase HPLC analysis, it was confirmed that >98% of the purified dynamin 1 was guanine nucleotide free form. Rat dynamin 2 cDNA in baculovirus was a kind gift provided by Prof. Handa (Tokyo Institute of Technology, Tokyo, Japan), and the recombinant protein was purified as described for dynamin 1 above.
The cDNA encoding rat full-length cortactin (Cort WT) and the cortactin deletion mutants [1-80 aa, 1-450 aa (Cort ΔSH3), 284-450 aa and 451-509 aa (Cort SH3)] were prepared by PCR (McNiven et al., 2000) and subcloned into the plasmid pGEX-6P vector following digestion with BamH1-EcoR1. GST-tagged cortactin W525K (Cort W525K) was generated by mutation of pGEX-6p-cortactin using the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies) (Schafer et al., 2002). For expression in cells, Cort WT or Cort ΔSH3 was subcloned into the pEF1 myc-His vector (Life Technologies) as EcoR1-Xba1 fragments. Cort WT or Cort W525K was subcloned into pIRES2-AcGFP1 expression vector (Clontech) as EcoR1-BamH1 fragments. The resulting plasmid was transformed into bacterial BL21(DE3) pLysS strain for protein expression. The expression of GST-fusion proteins was induced by 0.1 mm isopropyl-1-thio-d-galactopyranoside at 37°C for 3–6 h in LB media supplemented with 100 μg/ml ampicillin at _A_600 = 0.8. GST-fusion proteins were then purified as described previously (Slepnev et al., 2000). The GST was cleaved using a PreScission protease (GE Healthcare) according to the manufacturer's instructions, and purified on Mono Q columns equilibrated in 20 mm Tris-HCl, pH 7.7, and 0.2 m NaCl. The nucleotide sequences of the constructs used in this study were verified with DNA sequence analysis. All of the purified protein solutions (2–5 mg/ml) were stored at −80°C and thawed at 37°C before use.
Preparation of brain cytosol.
Brain cytosol was prepared as described previously with slight modifications (Yamada et al., 2009b). Briefly, 20 male mouse brains were homogenized in 5 ml of buffer (XB: 10 mm HEPES, pH 7.4, 100 mm KCl, 2 mm MgCl2, 0.1 mm CaCl2, 5 mm EGTA, 50 mm sucrose, 1 mm DTT, leupeptin at 1 μg/ml, pepstatin at 5 μg/ml, and PMSF at 0.4 mg/ml). The homogenate was centrifuged at 3000 × g for 20 min and then 10,000 × g for 20 min. The resultant supernatant was diluted with XB fourfold and centrifuged at 400,000 × g for 1 h. The clear supernatant was carefully collected and concentrated to one-fourth of the volume using Centriprep-10 concentrators (Millipore). The prepared cytosol (40–50 mg/ml) was supplemented with an ATP generating system (1 mm ATP, 8 mm creatine phosphate, 8 U/ml phosphocreatine kinase, 1 mm MgCl2, 200 mm sucrose) and stored at −80°C.
In vitro actin assembly and disassembly assays.
Actin assembly was quantified using pyrene-actin (Yamada et al., 2009b). Briefly, 12 mg/ml brain cytosol and 9.4 μm pyrene-actin (Cytoskeleton Inc.) in Assay buffer (20 mm HEPES, pH 7.4, 100 mm KCl, 1 mm MgCl2, 0.1 mm EDTA, 1 mm DTT, and 0.1 mm EGTA) was preincubated with an ATP regenerating system in a quartz cuvette for 10 min at room temperature. Then, 120 μm small unilamellar liposomes consisting of 50% phosphatidylserine (PS) and 50% phosphatidylcholine were added. Pyrene fluorescence at 407 nm (10 nm slit width) with excitation at 365 nm was then measured using an F-2500 fluorescence spectrophotometer (Hitachi Ltd.).
Actin disassembly was quantitatively analyzed as described previously (Okamoto et al., 2007). F-actin was formed from 4.7 μm pyrene-actin at room temperature for 30 min in F-buffer (0.2 mm CaCl2, 50 mm KCl, 2 mm MgCl2, and 1 mm ATP in 5 mm Tris-HCl, pH 7.5). F-actin was then depolymerized by diluting at 1:10 with F-buffer in the absence or presence of 5 μm each of dynamin 1 and cortactin under the indicated guanine nucleotide conditions. To assess the effect of an actin depolymerization toxin on F-actin disassembly, 2 μm Mycalolide B diluted with G-buffer (0.2 mm CaCl2 in 5 mm Tris-HCl, pH 8.0) was added to the preformed F-actin. Pyrene fluorescence was detected with a fluorescent microplate reader (MTP-600F, Corona Electric Co. Ltd., Japan). All steps were done at room temperature.
siRNA-mediated dynamin 1 or cortactin knockdown.
Pre-annealed siRNA designed using sequences for human dynamin 1 or cortactin were synthesized and purified by Life Technologies. The target sequence for dynamin 1 was 5′-GCACUGCAAGGGAAAGAAAtt-3′.
The target sequences for cortactin were 5′-CCGAAUGGAUAAGUCAGUCtt-3′ for oligo 1; 5′-GGUUUCGGCGGCAAAUACGtt-3′ for oligo2; 5′-CGAAUAUCAGUCGAAACUUtt-3′ for oligo 3.
Scrambled RNA that has no significant sequence homology to mouse, rat, or human gene sequences was also purchased to use as a negative control. One day before transfection, cells were plated in six-well plates (5.0 × 104 cells/well). Duplex siRNA (100 pmol) was transfected into the cells using 4 μl of Lipofectamine RNAiMax (Life Technologies). After 72 h, the cells were used for experiments. Three different siRNAs for cortactin were effective and yielded the same results. For rescue experiments, SH-SY5Y cells were first treated with Oligo 3 for cortactin knockdown, and cultured for 24 h. Cells (1 × 105/coverslip) were then transfected with 0.25 μg of cDNA of rat Cort WT or Cort W525K cloned into pIRES2-AcGFP1 expression vector (Clontech), and then 10 μm retinoic acid was added for differentiation. After 48 h, the cells were fixed.
Quantification of actin bundles by a low-speed sedimentation assay.
Non-muscle actin (Cytoskeleton Inc.) was polymerized in F-buffer containing 10 mm Tris-HCl, 0.2 mm DTT, 0.2 mm CaCl2, 2 mm MgCl2, 50 mm KCl, and 0.5 mm ATP, pH 7.5, for 1 h. Dynamin 1 and cortactin at indicated concentrations were then incubated with 3.3 μm F-actin in the presence or absence of 0.1 mm GTP, GDP, or GTPγS for 1 h. Actin bundles were sedimented by low-speed centrifugation, at 5000 × g, for 10 min (Dixon et al., 2008). The pellet and supernatant were separated by SDS-PAGE, stained with SYPRO Orange (Life Technologies), and quantitated by densitometry using ImageJ. All steps were performed at room temperature.
Pull-down assay and immunoprecipitation.
GST pull-down assays were performed as described previously (Yamada et al., 2009b). GST-fusion proteins (100 μg) bound to glutathione-Sepharose beads (GE Healthcare) were incubated with 1 mg of mouse brain extract in 1% TX-100, 100 mm KCl, 0.5 mm EDTA, 10 mm NaF, 20 mm HEPES/KOH, pH 7.4, and a Protease Inhibitor Cocktail tablet (Roche Diagnostics). Dynamin 1 that bound to the beads was then analyzed by Western blotting. For detection of the direct interaction between dynamin 1 PRD and Cort WT, 5 μg of GST-Cort WT or Cort W525K bound to glutathione beads was incubated with an equal amount of recombinant dynamin 1 PRD at 4°C for 2 h. Dynamin 1 PRD bound to the beads was then analyzed by SDS-PAGE and stained by Coomassie Brilliant Blue.
For the immunoprecipitation assay, differentiated NG108-15 cells were cotransfected with GFP-tagged dynamin 1 and either myc-tagged cortactin or cortactin ΔSH3. The cell lysates were extracted with 1% NP-40, 100 mm KCl, 0.5 mm EDTA, 10 mm NaF, 20 mm HEPES/KOH, pH 7.4, and a Protease Inhibitor Cocktail tablet (Roche Diagnostics). The protein complexes were immunoprecipitated from 1 mg of cell extract using either 5 μg of polyclonal anti-myc antibodies or preimmune IgG, and were then visualized by Western blotting with monoclonal anti-GFP or anti-myc antibody.
Fluorescent microscopy.
Transfection of cultured cells was performed using a Fugene 6 Transfection Reagent (Roche Diagnostics) or Lipofectamine LTX (Life Technologies). Cells were fixed with 4% paraformaldehyde and stained by immunofluorescence as described previously (Hayashi et al., 2008).
For in vitro F-actin bundle formation, 3.3 μm F-actin was incubated for 1 h with 5 μm dynamin 1 and/or cortactin, then for an additional 30 min with 3 μm Alexa Fluor 488-phalloidin. For immunolocalization of dynamin 1 and cortactin to F-actin bundles, samples were incubated for an additional 30 min with 3 μm phalloidin to stabilize the filaments and were then centrifuged at 5000 × g for 10 min. The pellet was resuspended with 50 μl of F-buffer and then immunostained in suspension for 30 min with 1 μl of primary antibody. The mixture was centrifuged at 5000 × g and the pellet was then washed with F-buffer. The samples were incubated with secondary antibodies and washed as was done for the primary antibody. All steps were performed at room temperature. The samples were dispersed on glass slides and mounted. Samples were examined using a spinning disc confocal microscope system (CSU10, Yokogawa Electric Co.) combined with an inverted microscope (IX-71, Olympus Optical Co., Ltd.) and a CoolSNAP-HQ camera (Roper Industries). The confocal system was controlled by MetaMorph Software (Molecular Devices). When necessary, images were processed using Adobe Photoshop CS3 or Illustrator CS3 software.
Electron microscopy.
For negative staining, actin bundles were formed in vitro as above and diluted 1:20 with F-buffer. Dynamin 1–cortactin complexes were formed by incubating 1 μm each of the proteins in cytosolic buffer (25 mm HEPES–KOH, 25 mm KCl, 2.5 mm magnesium acetate, 100 mm potassium glutamate, pH 7.2) at 37°C for 15 min. The samples were absorbed to a formvar- and carbon-coated copper grid and then stained with 3% uranyl acetate in ddH2O for 2 min. For morphometric analysis of ring complex formation, negative-stained samples with similar density were photographed at 30,000–40,000×. For each condition, 5 areas corresponding to 0.5 μm2 (total 2.5 μm2) were randomly selected, and the number of rings was counted. Close rings and open rings that have more than three quarters were defined as “rings.”
Preembedding immunoelectron microscopy was performed as described previously (Nakagawa et al., 2001). Briefly, SH-SY5Y cells were fixed with cytoskeleton buffer (10 mm MES, 150 mm NaCl, 5 mm EGTA, 5 mm MgCl2 and 5 mm glucose, pH 6.0) containing 10 μg/ml phalloidin, 0.1% Triton X-100, and 3% formaldehyde for 1 min, and then for an additional 30 min without Triton X-100, before being washed with 5 μg/ml phalloidin in PBS. After incubation in blocking solution (10 μg/ml phalloidin, 2 mg/ml BSA, and 100 mm glycine in PBS), samples were incubated with primary antibody diluted in blocking solution, washed with 5 μg/ml phalloidin in PBS, incubated with 5 or 10 nm gold conjugated with secondary antibodies, and then fixed with 2.5% glutaraldehyde and 5 μg/ml phalloidin in PBS. The samples were postfixed with 1% OsO4 in 0.1 m sodium cacodylate buffer for 1 h, dehydrated, and embedded in Epon 812 for ultrathin sectioning.
For whole-mount immunoelectron microscopy, SH-SY5Y cells cultured on fibronectin- and formvar-coated gold grids were fixed and immunostained by the preembedding method described above, and then negatively stained with 3% uranyl acetate. Electron microscopic observation was performed using Hitachi H-7100 transmission electron microscope.
Morphometric analysis.
For measurement of filopodium length, SH-SY5Y cells were fixed and stained with rhodamine- or Alexa Fluor 488-conjugated phalloidin. Actin-rich structures that possessed lamellipodia and filopodia at tips of neurites were defined as “growth cones” and digital images were acquired at 100× magnification. Up to three filopodia for each growth cone were randomly selected, and their length was measured with ImageJ software (v. 1.40 g).
To assess colocalization of dynamin 1 with cortactin or clathrin, immunostained cells were photographed, and the immunoreactivities along randomly selected filopodia were measured using MetaMorph software. To quantify immunocytochemical expression of dynamin 1, immunostained cells were photographed at fixed exposure of 1000 ms for dynamin 1, and 1500 ms for phalloidin. Images of randomly selected 35 filopodia were used to measure “average fluorescence intensities (F.I.) in filopodia” using MetaMorph software after background subtraction
Dynamin GTPase assay.
GTP hydrolysis was measured at 1 mm GTP using a colorimetric assay to detect Pi release as previously described (Soulet et al., 2005). Dynamin 1 was incubated with the equal amount of either cortactin or Cort W525K mutant in 150 mm NaCl, 2 mm MgCl2, 20 mm Tris-Cl, pH 7.5, containing 0.25 μm proteins each in 160 μl of total volume at 37°C for 60 min.
In-gel digestion and MS analysis.
Proteins binding with dynamin 1 were separated by SDS-PAGE and stained by Coomassie Brilliant Blue. Gel pieces of interest were cut, and incubated with 10 mm DTT for 15 min and 100 mm Iodoacetamide for 15 min. After washing with methanol/water/acetic acid (50:40:10) 4 times, the gel pieces were soaked with 50 mm ammonium bicarbonate for 10 min and acetonitrile for 10 min. Gel pieces were then completely dried in a Speed Vac evaporator (Thermo Savant). The dried gel pieces were swollen in 2 ml of 50 mm ammonium bicarbonate containing 0.02 mg of trypsin (Promega). Another 30 ml of 50 mm ammonium bicarbonate was then added and the mixture was incubated in 37°C for overnight. After the in-gel digestion, dried samples were redissolved in 5 μl of acetonitrile/water/TFA (trifluoroacetic acid; 5:95:0.1), and a 0.5 μl aliquot was used for matrix-assisted laser desorption-ionization mass spectrometry (MALDI-MS) analysis. The matrix solution was prepared by saturating 2,5-dihydroxybenzoic acid in acetonitrile/water/TFA (33:67:0.1) and was diluted fourfold before being mixed with the peptide solution. Cocrystallization was performed by depositing 0.5 μl of peptide solution and 0.5 μl of the diluted matrix solution together to the same spot of a MALDI plate, followed by drying at room temperature. MALDI-MS spectra were obtained using a Voyager Linear DE (Applied Biosystems). The Voyager was operated in delayed-extraction mode and the spectra were calibrated using the internal standards, angiotensin III [average molecular weight (MW) 931.1 Da] and oxidized insulin B chain (average MW 3495.9). MASCOT (Matrix Sciences) was used as the search engine for peptide mass fingerprinting.
Statistical analysis.
Data were analyzed using KaleidaGraph (v 4.1) for Macintosh (Synergy Software) to perform statistical analysis. ANOVA and Tukey's HSD post hoc test were applied for several different groups, and Student's t test for two different groups. Based on these analyses, *p < 0.05, **p < 0.01 is considered significant.
Results
Dynamin 1 is implicated in actin dynamics in brain cytosol
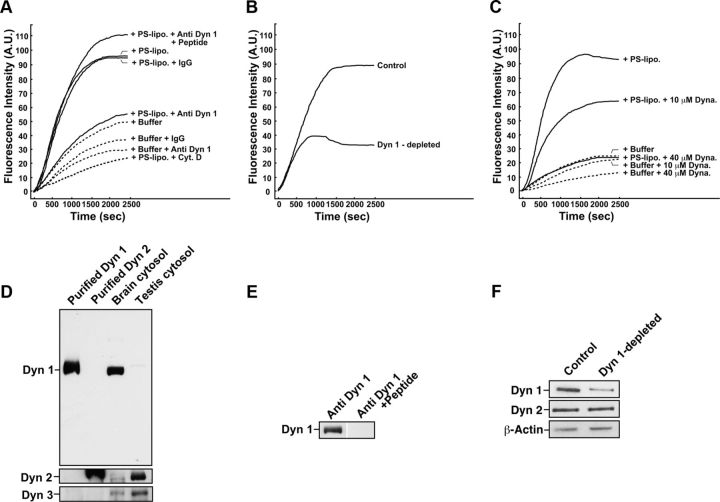
To examine whether dynamin 1 is involved in actin dynamics in mouse brain cytosol, we performed an actin assembly assay, in which actin polymerization can be quantitatively monitored using pyrene-conjugated actin added to the cytosol. Rapid actin assembly was observed in the brain cytosol in the presence of PS-liposomes (Fig. 1A), confirming previously reported acidic phospholipid-stimulated actin polymerization (Yamada et al., 2009b). Addition of anti-dynamin 1 antibody, but not preimmune IgG to the cytosol, strongly inhibited the PS-dependent actin assembly. This inhibitory effect was blocked by the preincubation of the antibody with the corresponding antigenic peptides (Fig. 1A), suggesting that the dynamin 1 antibody acted in a specific manner. Incubation of the dynamin 1 antibodies with rat brain cytosol resulted in an ∼60% immune-depletion of dynamin 1 (Fig. 1F). Actin assembly was significantly reduced in the dynamin 1-depleted brain cytosol (Fig. 1B). Specificity of the dynamin 1 antibody used for the immune-depletion was suggested by the fact that the antibody did not recognize purified dynamin 2 or either dynamin 2 or dynamin 3 in testis cytosol (Fig. 1D; Kusumi et al., 2007). Furthermore, the dynamin 1 antibody was inactivated by preincubation with the corresponding antigenic peptides (Fig. 1E). Dynasore, a potent inhibitor for dynamins (Macia et al., 2006), also inhibited the actin assembly in a dose-dependent manner (Fig. 1C). These results strongly indicate that dynamin 1 participates in the formation and/or stabilization of F-actin in the brain cytosol.
Figure 1.
Inhibition of dynamin 1 reduces actin dynamics in brain cytosol. A, Anti-dynamin 1 antibodies strongly inhibit PS-induced actin assembly. Mouse brain cytosol was either treated with 120 μm PS-containing liposomes (+PS-lipo.) (solid lines) or incubated alone (+Buffer) (dotted lines). The time 0 indicates the point of liposome addition. In same samples, the cytosol was preincubated for 10 min with 7.5 μg of anti-dynamin 1 antibodies (+Anti Dyn 1) or nonspecific rabbit IgG (+IgG). To inactivate anti-dynamin 1 antibodies, the antibodies were pretreated with 7.5 μg of corresponding antigenic peptides for 30 min at room temperature (+Peptide). Very few actin polymerization was observed in the presence of 10 μm Cytocharasin D (+ Cyt. D). B, Actin assembly was reduced in dynamin 1-depleted cytosol. For the immunodepletion, rat brain cytosol was incubated with 10 μg of anti-dynamin 1 antibodies for 2 h at 4°C and then for an additional 1 h with protein G-Sepharose beads. The dynamin 1-antibody complexes bound to the protein G-Sepharose beads were removed by centrifugation. C, Inhibition of PS-induced actin assembly by dynasore. Brain cytosol was pretreated with 10 or 40 μm dynasore for 10 min. For negative control for dynasore, 1% DMSO is used. Dotted lines represent samples in which PS-liposomes were omitted. D, Anti-dynamin 1 antibodies did not recognize dynamin 2 or dynamin 3. Mouse brain cytosol or mouse testis cytosol (5 μg), which is enriched with dynamin 3, was analyzed by Western blotting. For controls, 0.25 μg of recombinant human dynamin 1 or rat dynamin 2 were used. E, Inactivation of anti-dynamin 1 antibody by the antigenic peptides. Five micrograms of the antibody was incubated with 500 μg of the corresponding antigenic peptides for 2 h. Mouse brain cytosol (5 μg) was analyzed by Western blotting with either untreated anti-dynamin 1 antibody (left), or the inactivated antibody (right). F, Immunodepletion of dynamin 1 from rat brain cytosol. Rat brain cytosol was incubated with 10 μg of anti-dynamin 1 antibody for 2 h at 4°C, and then for an additional 1 h with protein G-Sepharose beads. The dynamin 1–antibody complexes bound to the protein G-Sepharose beads were removed by centrifugation. The resultant cytosol (10 μg) was examined by Western blotting.
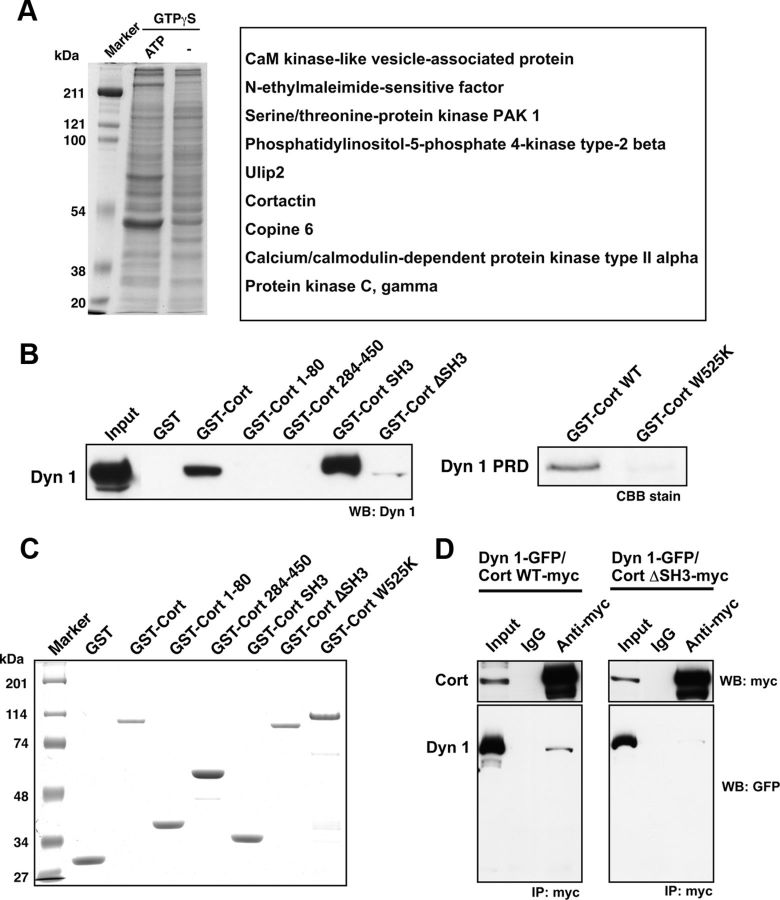
In lipid-induced actin assembly, actin polymerization is initiated at the liposome surface (Ma et al., 1998), to which dynamin is also recruited (Otsuka et al., 2009). To search for actin-regulating proteins that cooperatively function with dynamin 1, actin assembly in mouse brain cytosol was stimulated with liposomes, and proteins that cosedimented with the liposomes were analyzed by MALDI-MS (Fig. 2A). Using this method, we identified cortactin, an F-actin associated protein, as a putative binding partner for dynamin 1. A specific interaction between dynamin 1 and cortactin was then found by pull-down assay and immunoprecipitation. The dynamin1-cortactin interaction was mediated by dynamin 1 PRD and cortactin SH3 as demonstrated using Cort ΔSH3 or Cort W525K, a mutant deficient in dynamin-binding (Schafer et al., 2002; Fig. 2B,C). Furthermore, we found that exogenously expressed myc-tagged Cort WT, but not Cort ΔSH3, coprecipitated with GFP-tagged dynamin 1 in NG108-15 cells (mouse neuroblastoma × rat glioma hybrid cells) (Fig. 2D). Thus, we found a specific interaction between cortactin SH3 domain and dynamin 1 PRD.
Figure 2.
Dynamin 1 directly binds to cortactin in vitro and in vivo. A, Identification of proteins interacting with dynamin 1 during lipid-induced actin polymerization by mass spectrometry. Mouse brain cytosol (50 μg) was incubated at 37°C for 15 min in the presence of 0.1 mm GTPγS, 40 μg of liposomes consisting of 74% Folch fraction, 6% phosphatidylinositol (4,5)-bisphosphate and 20% cholesterol, and with or without an ATP generating system. The liposomes collected by sedimentation were separated by SDS-PAGE, and all proteins in the gel were treated with trypsin. The digested peptides were then analyzed by MALDI-MS. Nine potential proteins were identified. B, GST pull-down assay demonstrating that both GST-Cortactin and GST-Cortactin SH3 bound to dynamin 1. The binding was not detected with cortactin mutants lacking SH3 (GST-Cort 1–80, GST-Cort 284–450, GST-Cort ΔSH3). Brain extract (20 μg) was also loaded (Input) (left). Dynamin 1 PRD directly bound to Cort WT but not to its SH3 point mutant, W525K (right). C, SDS-PAGE of GST fusion proteins used in (B). Five micrograms of purified GST fused cortactin mutants indicated were analyzed. D, Immunoprecipitation demonstrating an in vivo interaction between dynamin 1 and cortactin. Differentiated NG108-15 cells were cotransfected with GFP-tagged dynamin 1 and either myc-tagged cortactin (Cort WT-myc, left) or cortactin ΔSH3 (Cort ΔSH3-myc, right). The protein complexes were immunoprecipitated using polyclonal anti-myc antibodies or preimmune IgG, and then visualized by Western blotting with monoclonal anti-GFP or anti-myc antibodies. Total cell lysates (5 μg) were also loaded (Input).
Dynamin 1 and cortactin colocalize in puncta arranged along growth cone filopodia, and accumulates at the F-actin bundles
As demonstrated above, neuronal dynamin 1 is involved in the regulation of actin assembly (Fig. 1). In addition, cortactin is implicated in the regulation of actin dynamics (Ammer and Weed, 2008). Therefore, we next examined the localization of these proteins in actin-rich neuronal growth cones. In primary cultured cortical neurons, dynamin 1 and cortactin were visible in puncta aligned along the growth cone filopodia, and they were often colocalized (Fig. 3A, bottom). While clathrin was enriched in the “palm” region of growth cones, only trace amounts were present in the filopodia (Fig. 3B). This suggested that participation in clathrin-mediated endocytosis is not a major function of dynamin 1 or cortactin in the filopodia.
Figure 3.
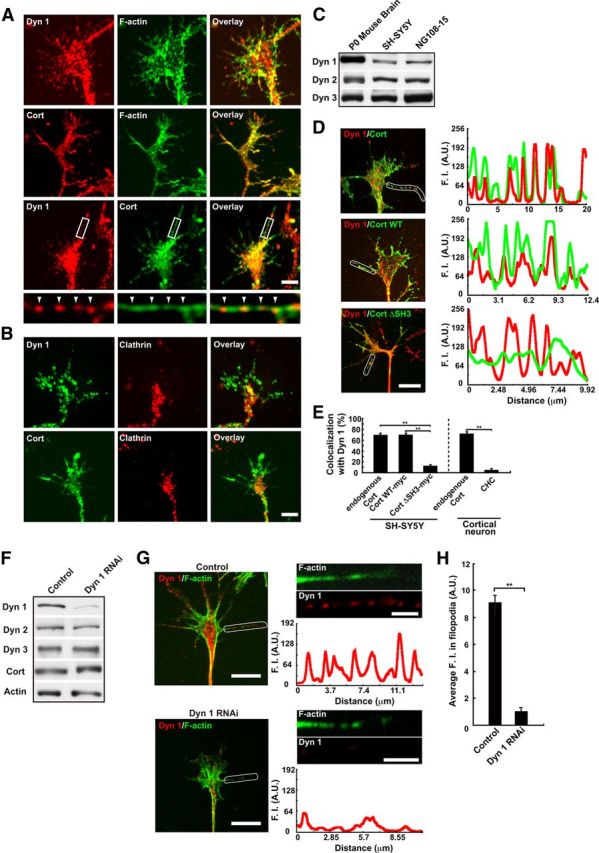
Dynamin 1 colocalizes with cortactin in puncta aligned along filopodia of growth cones. A, Immunofluorescence demonstrating the presence of dynamin 1 (top) and cortactin (middle) in the growth cones of primary cultured cortical neuron. Colocalization of dynamin 1 and cortactin was visualized by double-immunofluorescence. Framed areas are enlarged below (bottom). Note that dynamin 1- and cortactin-positive puncta were present periodically along the filopodia (arrowheads). Scale bar: top, middle, and bottom panels, 5 μm; insets, 1.6 μm. B, Clathrin is not enriched in the growth cone filopodia. Double-immunofluorescence demonstrating that the localization of clathrin was distinct from that of dynamin 1 and cortactin in the growth cones of cortical neurons. Note that virtually no immunoreactivity for clathrin was observed in the filopodia. Scale bar, 5 μm. C, Expression of dynamin isoforms in newly born mouse P0 brain or, neuroblastoma cell lines analyzed by Western blotting. P0 mouse brain homogenate (10 μg), SH-SY5Y cells (20 μg), and NG108-15 cells (20 μg) were loaded per lane. D, Periodic colocalization of dynamin 1 and cortactin along growth cone filopodia of SH-SY5Y cells (top left). Localization of Cort WT-myc (middle left) or Cort ΔSH3-myc (bottom left) with dynamin 1 was examined in SH-SY5Y cells. Scale bar, 10 μm. Line scanning of fluorescence intensity for dynamin 1 (red) and cortactin (green) in the area indicated with a white line (right panels) is shown. E, Colocalization measurement of endogenous or exogenous cortactin and dynamin 1 in growth cone filopodia of SH-SY5Y cells or rat cortical neurons. The data shown are the mean ± SEM. In each sample, 60–100 filopodia of growth cones from three experiments were analyzed. **p < 0.01. F–H, Disappearance of periodic presence of dynamin 1 along growth cone filopodia in dynamin 1-depleted SH-SY5Y cells. Western blotting showing suppression of dynamin 1 by RNAi in SH-SY5Y cells (F). β-Actin was used as the control. Cell lysate (3 μg) was loaded in each lane (F). Periodic localization of dynamin 1 in growth cone filopodia (upper left in G) was impaired in dynamin 1-depleted SH-SY5Y cells (lower left in G). Line scanning of fluorescence intensity (F.I.) for dynamin 1 (red) and phalloidin (green) in the area indicated with a white line is shown in the right panels in G. Filopodia were determined by phalloidin staining. Scale bar: left panels, 10 μm; right panels, 3 μm. H, Average F.I. for dynamin 1 in growth cone filopodia was also decreased by dynamin 1 RNAi (n = 35 each, **p < 0.01).
To further elucidate function of dynamin 1 and cortactin in growth cones, we used SH-SY5Y cells (a human neuroblastoma cell line), widely used as model cells to study growth cones (Meyerson et al., 1992; Hynds et al., 2003; Johnsson and Karlsson, 2012). As confirmed by Western blotting, all the dynamin isoforms were expressed both in SH-SY5Y cells and NG108-15 cells as was the case in P0 mouse brain. (Fig. 3C). As in primary cultured neurons, dynamin 1 and cortactin in SH-SY5Y cells also localized as periodically aligned dots along the growth cone filopodia (Fig. 3D, top), and ∼70% of dynamin-positive puncta were positive for cortactin (Fig. 3E, left column). Using antibody against C terminus of dynamin 1 (EP801), similar immunolocalization was observed, and the dynamin-positive immunoreactivities well colocalized with cortactin (73.2 ± 2.3%, n = 50). Exogenously expressed Cort WT, but not Cort ΔSH3, colocalized with endogenous dynamin 1 in the filopodia of SH-SY5Y cells (Fig. 3D, middle and bottom, E), indicating that SH3-mediated interaction is essential for the periodic localization of dynamin 1 in filopodia.
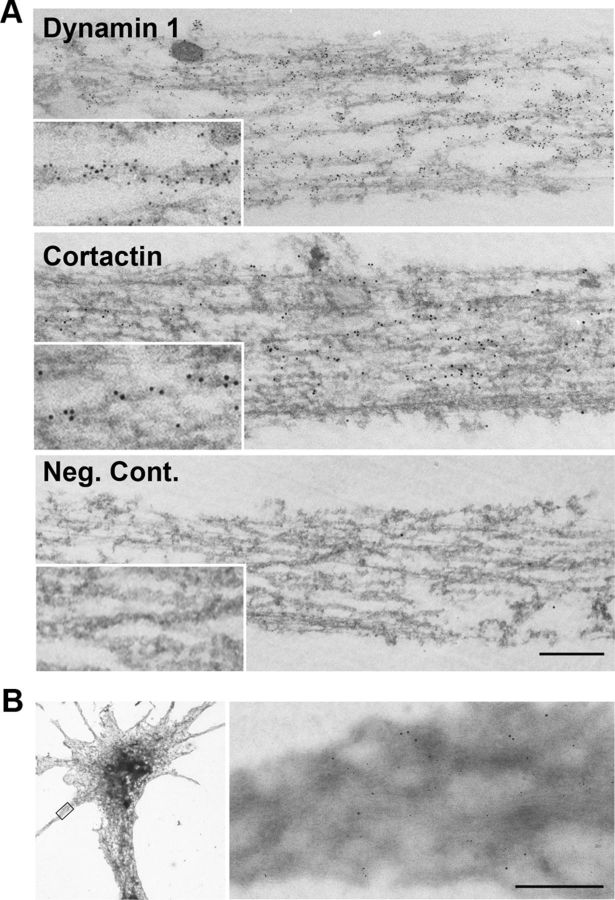
We further confirmed, by RNAi depletion, that periodic localization of dynamin 1 along filopodia is specific to the protein. By RNAi expression of dynamin 1 was selectively knocked down in SH-SY5Y cells without disturbing the expression of other dynamin isoforms and cortactin (Fig. 3F). Morphologically, the RNAi reduced the formation of growth cone filopodia (Fig. 3G, bottom left), and the fluorescence intensity for dynamin 1 was reduced to ∼10% (Fig. 3H). Characteristic periodic staining for dynamin 1 was disappeared in growth cone filopodia formed in this condition (Fig. 3G, bottom right). Thus, specificity of the periodic localization of dynamin 1 along filopodia was confirmed. Interestingly, immunoelectron microscopy revealed that both dynamin 1 and cortactin were associated with actin filaments in SH-SY5Y cells (Fig. 4A). Using whole-mount immunogold staining, we often observed dynamin 1 and cortactin on F-actin bundles in growth cone filopodia (Fig. 4B).
Figure 4.
Dynamin 1 and cortactin associate with actin filaments. A, Preembedding immunoelectron microscopy for dynamin 1 (top) and cortactin (middle) in SH-SY5Y cells. An anti-dynamin 1 antibody (PA1-660) or an anti-cortactin antibody was detected using 5 nm colloidal gold-conjugated anti-rabbit IgG, or 10 nm colloidal gold-conjugated anti-mouse IgG, respectively. The primary antibodies were omitted in the negative control (bottom). Scale bar: A, 300 nm; insets, 150 nm. B, Double staining for dynamin 1 and cortactin in growth cone filopodia of whole-mount SH-SY5Y cells. The primary antibodies and immunogold-conjugated secondary antibodies were used as in A. A filopodium indicated with a rectangle in the low-magnification EM (left), is enlarged on the right. Note that dynamin (5 nm gold) and cortactin (10 nm gold) closely localize on a actin bundle (right). Scale bar: left, 8 μm; right, 300 nm.
Dysfunction of dynamin 1 and cortactin destabilizes filopodia of growth cones
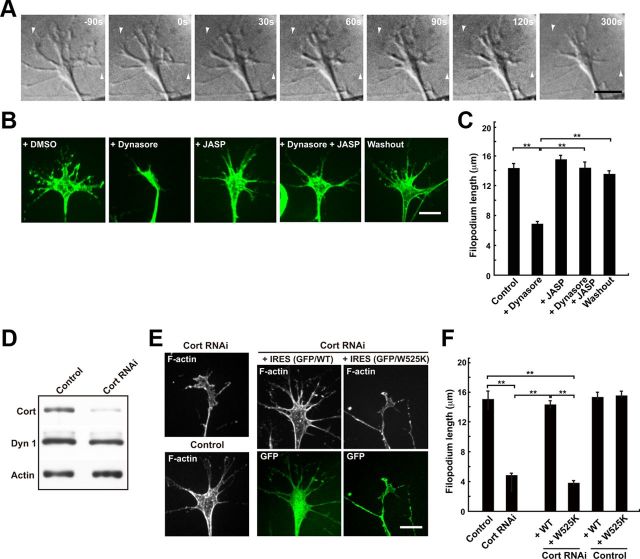
The striking colocalization of dynamin 1 and cortactin in growth cone filopodia, and their presence on actin filaments prompted us to investigate whether dynamin 1 and cortactin function in the filopodia. Time-lapsed observation of growth cones of SH-SY5Y cells revealed that the application of dynasore, a dynamin inhibitor, resulted in rapid retraction of the growth cone filopodia. Within 300 s of dynasore application, the mean filopodium length was decreased by ∼50% (Fig. 5A–C). Furthermore, the retraction of filopodia was recovered by washing out dynasore (Fig. 5B,C), indicating that dynasore-induced retraction was reversible. The dynasore-induced filopodia retraction was abolished by the presence of Jasplakinolide, an actin depolymerization inhibitor (Bubb et al., 2000; Fig. 5B,C), indicating that inhibition of dynamin by dynasore leads to destabilization of F-actin.
Figure 5.
Inhibition of dynamin 1 or knockdown of cortactin destabilizes growth cone filopodia. A, Sequential DIC images of growth cones of SH-SY5Y cells are shown. Dynasore (80 μm) was applied to the samples 90 s after the beginning of imaging. Note the rapid filopodium retraction that occurs within 120 s of dynasore addition. Arrowheads indicate the positions of filopodia tip at time 0. Scale bar, 10 μm. B, Jasplakinolide prevented dynasore-induced filopodium retraction. SH-SY5Y cells were preincubated with or without 50 nm Jasplakinolide (+JASP) for 30 min, and then treated with 80 μm dynasore (+Dynasore) for 5 min. As a negative control, cells were treated with 1% DMSO (+DMSO). For wash out experiment, dynasore-treated cells were incubated in medium without dynasore for 60 min (Washout). Growth cones were visualized by staining with Alexa Fluor 488-phalloidin. Scale bar, 10 μm. C, Morphometric analysis of the length of growth cone filopodium in B. All results are represented as the mean ± SEM. In each sample, 90–120 growth cones filopodia were analyzed from three experiments. (**p < 0.01). D, Western blotting showing suppression of cortactin expression by RNAi in SH-SY5Y cells. β-Actin was used as the control. Cell lysate (3 μg) was loaded in each lane. E, Growth cones of SH-SY5Y cells visualized by Alexa Fluor 488-phalloidin staining. In cortactin-depleted cells, the mean length of filopodia is shorter and the area of growth cones is smaller than that of the control cells (left panels). Cortactin depleted SH-SY5Y cells were transfected with cDNA of rat cortactin (middle panels) or Cort W525K (right panels) cloned into pIRES2-AcGFP1 expression vector. Transfected cells were determined by GFP expression. Scale bar, 10 μm. F, The mean length of growth cone filopodia in SH-SY5Y cells in (E). All results represent the mean ± SEM. In each sample, 82–100 growth cones filopodia were analyzed from three experiments. **p < 0.01.
Furthermore, cortactin RNAi resulted in smaller growth cones and shorter filopodia in SH-SY5Y cells (Fig. 5E,F). These suppressive phenotypes were cancelled by expression of Cort WT, but not of Cort W525K mutant (Fig. 5E,F). These results strongly suggested that both dynamin 1 and cortactin are crucial for the stabilization of growth cone filopodia. Consistently, overexpression of cortactin in hippocampal neurons results in the increase of size and spreading of growth cones (Kurklinsky et al., 2011).
Dynamin 1–cortactin complexes bundle F-actin
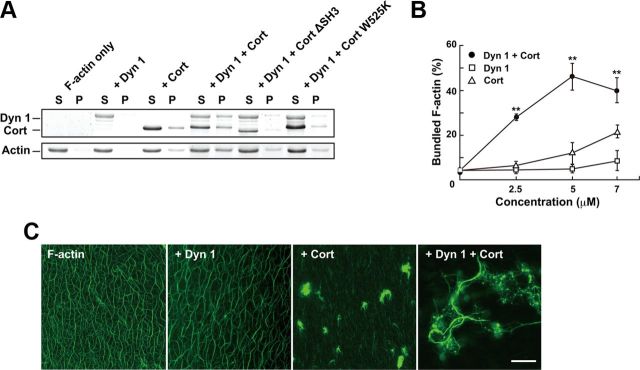
Given the presence of dynamin 1 and cortactin on actin filaments, and the strong functional link to filopodia, which are supported by F-actin bundles, we next examined the effects of dynamin 1 and cortactin on actin filaments in vitro. F-actin bundling by dynamin 1 and cortactin was quantitatively assessed using a low-speed sedimentation assay (Dixon et al., 2008), in which F-actin bundles are recovered in the pellet. Only trace amount of F-actin bundles were observed when F-actin was incubated with either dynamin 1 or cortactin alone. On the other hand, formation of F-actin bundles was drastically increased (approximately fourfold) when 5 μm both dynamin 1 and cortactin were added (Fig. 6A,B). In addition, considerable amounts of dynamin 1 and cortactin were coprecipitated in the pellet (Fig. 6A,B). F-actin bundle formation was not increased when cortactin was replaced either with Cort ΔSH3, or Cort W525K, mutants that are unable to bind to dynamin (Fig. 6A).
Figure 6.
Direct interaction between dynamin 1 and cortactin is essential for actin filament bundling. A, F-actin bundle formation by dynamin 1 and either WT or mutant cortactin in the presence of GTP detected by a low-speed sedimentation assay. Dynamin 1 and cortactin were used at 5 μm. Note that significant amount of F-actin bundles were formed in the presence of both dynamin 1 and Cort WT. B, Quantitation of F-actin bundle formation with various concentrations of dynamin 1 and cortactin. The F-actin bundles recovered in the low-speed pellet were measured. All results represent the mean ± SEM (n = 3). **p < 0.01. C, Long F-actin bundles are formed in the presence of both dynamin 1 and cortactin. Preformed actin filaments were incubated with or without dynamin 1 and cortactin in the presence of GTP. F-actin was visualized by adding Alexa Fluor 488-phalloidin. Actin filaments alone appear as uniformly dispersed thin threads (F-actin). Dynamin 1 alone induced no visible change in the filament (+Dyn 1). Cortactin induced clustering of F-actin (+Cort). Long and thick F-actin bundles formed in the presence of both dynamin 1 and cortactin (+Dyn 1+Cort). Scale bar, 30 μm.
To morphologically visualize the formation of F-actin bundles by dynamin 1 and cortactin, preformed F-actin was incubated with or without cortactin and dynamin 1 in the presence of GTP, and then observed by fluorescent microscopy. F-actin alone appeared as uniform threads, and addition of dynamin 1 to F-actin did not produce any visible changes (Fig. 6C). F-actin incubated only with cortactin often formed small clusters, consistent with a previous report (Fig. 6C, Huang et al., 1997). Surprisingly, the presence of both dynamin 1 and cortactin resulted in the formation of long and thick F-actin bundles, 135.1 ± 8.3 μm in length (n = 15) (Fig. 6C, right).
Next, we examined the localization of dynamin 1 and cortactin on the thick F-actin bundles using immunofluorescence. As shown in Figure 7A, both dynamin 1 and cortactin were observed as bright puncta periodically aligned along the F-actin bundles, where the two proteins were often colocalized. The immunoreactive puncta were arranged along F-actin bundles with an average interval of 1.33 ± 0.03 μm (n = 159).
Figure 7.
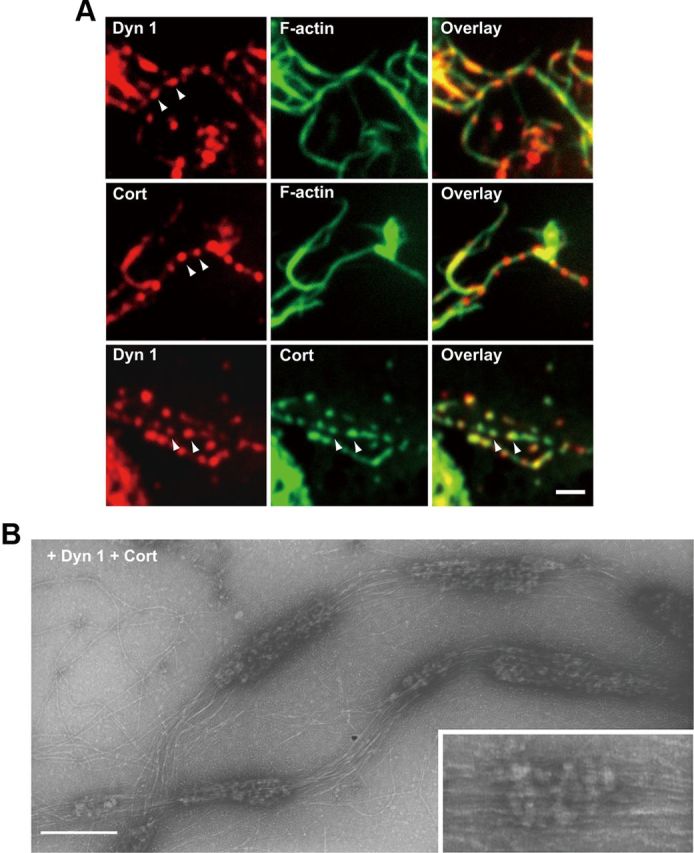
Dynamin 1 colocalizes with cortactin in puncta along F-actin bundles. A, Immunofluorescence demonstrating periodical presence of dynamin 1 and cortactin along F-actin bundles (top and middle). Colocalization of dynamin 1 and cortactin on F-actin bundles was revealed by double-immunofluorescence (bottom). Arrowheads indicate the periodic localization. Scale bar, 2 μm. B, Electron micrographs of negatively stained F-actin bundles formed by dynamin 1 and cortactin. The incubations were performed as in A. Note the periodical presence of protein clusters along the F-actin bundles. Note that dynamin 1-cortactin complexes twine around several actin filaments (inset). Scale bar: B, 250 nm; inset, 120 nm.
Electron microscopic observation of the same samples revealed that F-actin bundles generated by dynamin 1 and cortactin were periodically decorated with protein clusters (Fig. 7B). The interval between the protein clusters was in good agreement with that observed by immunofluorescence (1.03 ± 0.05 μm, n = 54) (Fig. 7B). Occasionally, rows of protein complexes twined around approximately three to four actin filaments, which were suggestive of a spiral, were observed (Fig. 7B, inset). It is very likely that the protein clusters observed on the F-actin bundles in this electron micrograph represent dynamin 1–cortactin complexes.
Dynamin 1 and cortactin copolymerize into ring-shaped structures that are sensitive to dynamin inhibition
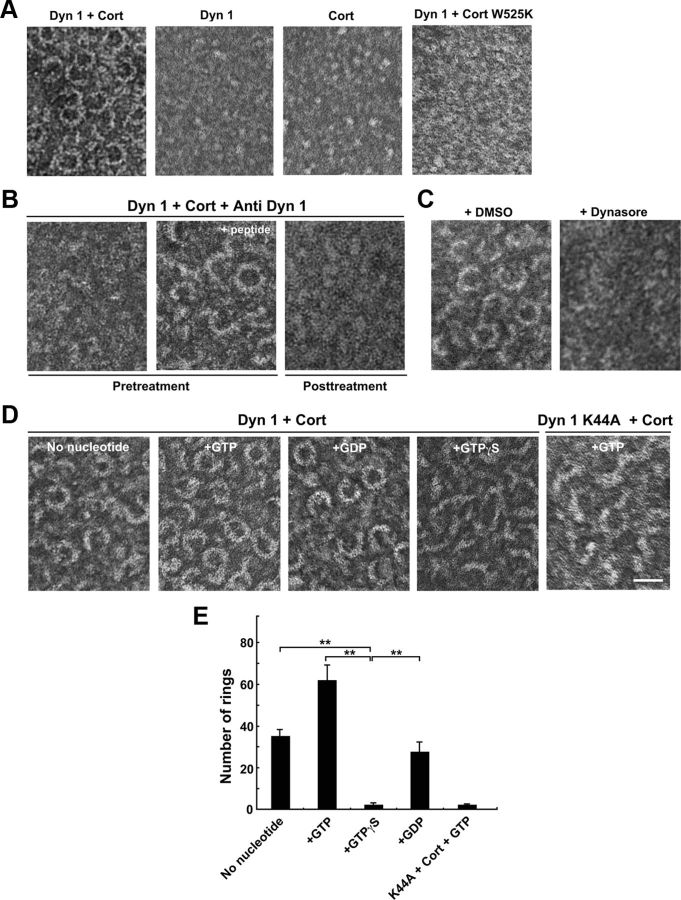
Dynamin 1 assembles into rings and spirals by itself under a low ionic condition (Hinshaw and Schmid, 1995), or in the presence of liposomes (Takei et al., 1998; Stowell et al., 1999). Dynamin 1 also coassembles into rings with amphiphysin or endophilin (Takei et al., 1999; Farsad et al., 2001). Likewise, dynamin 1 and cortactin formed ring-shaped complexes in the presence of GTP as revealed by negative-stain electron microscopy (Fig. 8A). Both open and closed ring complexes were observed, and the size of the closed rings was 25 ± 1.0 nm along the inner diameter (n = 20), and 42.3 ± 1.0 nm along the outer diameter (n = 24). Neither dynamin 1 nor cortactin alone formed rings using the same buffer conditions. Cort W525K failed to assemble into ring complexes with dynamin 1 (Fig. 8A).
Figure 8.
Dynamin 1 and cortactin copolymerize into ring-shaped complex, which undergoes open-and-close conformational change upon GTP hydrolysis. A, Electron micrographs of negatively stained dynamin 1–cortactin complexes formed in the presence of GTP (left). The complexes were not formed with dynamin 1 alone, cortactin alone or dynamin 1 and Cort W525K (right three panels). B, Preincubation of dynamin 1 with 7.5 μg of anti-dynamin 1 antibody (PA1-660) at room temperature for 30 min prevented the ring-shaped complex formation (left). This inhibitory effect was reversed by the inactivation of the antibody with corresponding 7.5 μg of antigenic peptides (middle). Addition of the antibodies to preformed dynamin 1–cortactin ring complexes (post-treatment) also resulted in complex disruption (right). C, Dynasore (80 μm) inhibited the formation of dynamin 1–cortactin ring-shaped complexes (right). As a control, 1% DMSO was used (left). D, Dynamin 1–cortactin complexes formed under indicated guanine nucleotides conditions. Ring-shaped complex were formed without nucleotide, or with GTP or GDP. The rings are widely open or almost linear in the presence of GTPγS. Dynamin 1 K44A plus cortactin in the presence of GTP also failed to form rings. Scale bar: A–D, 50 nm. E, Morphometric analysis of the ring-shaped complex formation in D. Closed rings and open rings that have more than three quarters were considered as rings as described in Materials and Methods. (**p < 0.01).
The formation of these ring complexes was sensitive to functional inhibition of dynamin 1. Anti-dynamin 1 antibody that suppressed actin assembly in brain cytosol (Fig. 1) prevented the formation of dynamin 1–cortactin ring complexes (Fig. 8B, left). This inhibitory effect of the antibody was prevented by preabsorption of the antibody with the corresponding peptides (Fig. 8B, middle). The dynamin 1 antibody also disrupted preformed dynamin 1–cortactin ring complexes (Fig. 8B, right). Furthermore, dynasore suppressed formation of the ring complexes (Fig. 8C).
Since dynamin functions in membrane fission as a mechanochemical enzyme driven by GTP hydrolysis (Sweitzer and Hinshaw, 1998; Takei et al., 1998; Marks et al., 2001; Roux et al., 2006; Ramachandran and Schmid, 2008; Faelber et al., 2011; Ford et al., 2011), we next examined whether guanine nucleotide conditions affect the conformation of dynamin 1–cortactin ring complex. The ring formation was observed even without guanine nucleotides, or with GDP, but the rings were the most prominent in the presence of GTP (Fig. 8D). Interestingly, in the presence of GTPγS, an unhydrolyzable GTP analog, or substitution of dynamin 1 WT with dynamin 1 K44A, a GTPase defective mutant (Damke et al., 1994), most of dynamin 1-cortactin rings were widely open or almost linear-shaped (Fig. 8D,E). These observations clearly indicate that the conformation of the dynamin 1–cortactin complex is altered upon GTP hydrolysis.
Dynamin 1 and cortactin ring complexes stabilize actin filaments
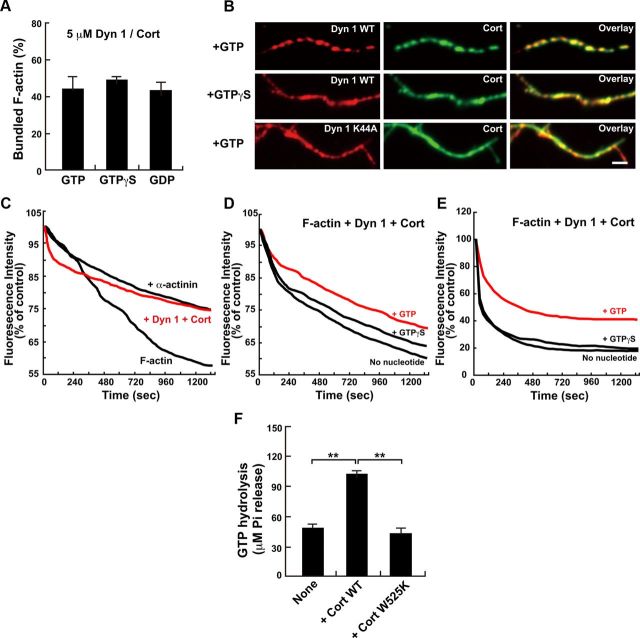
As conformational change of dynamin-cortactin ring was induced by guanine nucleotide conditions (Fig. 8D,E), we further examined whether dynamin 1-cortactin-dependent F-actin bundle formation is affected by an alteration of guanine nucleotide conditions. As revealed by a low-speed sedimentation assay, F-actin bundle formation was unchanged by alteration of guanine nucleotide conditions (Fig. 9A). Consistently, F-actin bundles were morphologically apparent in the presence of GTPγS, an unhydrolyzable analog. Moreover, substitution of WT dynamin 1 is with dynamin 1 K44A, a GTPase defective mutant (Damke et al., 1994), also resulted in F-actin bundle formation (Fig. 9B, bottom).
Figure 9.
Dynamin 1–cortactin ring complexes use GTP hydrolysis to stabilize actin filaments. A, Actin bundle formation induced by dynamin 1 and cortactin at different guanine nucleotide conditions as quantified by a low-speed sedimentation assay. Dynamin 1 and cortactin were used at 5 μm. All results represent the mean ± SEM (n = 3). B, Immunofluorescence showing differential distributions of dynamin 1 and cortactin along F-actin bundles formed under distinct guanine nucleotide conditions. The periodic localization of dynamin 1 and cortactin is most prominent in the presence of GTP (top), but it is obscure in the presence of GTPγS (middle) or when dynamin 1 is replaced with dynamin 1 K44A in the presence of GTP (bottom). Scale bar, 2 μm. C, Kinetics of F-actin disassembly induced by tenfold dilution. Note the prominent suppression of disassembly in the presence of dynamin 1 and cortactin with GTP (red line) as well as in the presence of 5 μm α-actinin. D, E, Kinetics of F-actin disassembly with dynamin 1 and cortactin and distinct guanine nucleotide conditions. Actin bundles were disassembled by dilution (D) or by Mycalolide B (E). In both sets of experiments, the rate of disassembly was slowest in the presence of GTP (red lines). F, Dynamin 1 GTPase activity was stimulated by Cort WT but not by Cort W525K. All results represent the mean ± SEM (n = 3). **p < 0.01.
Interestingly, however, the localization of dynamin 1 and cortactin on F-actin bundles was dependent on guanine nucleotide conditions. While both proteins were periodically present along the F-actin bundles in the presence of GTP (Fig. 9B, top, see also Fig. 7A), their localization became diffuse in the presence of GTPγS or when using dynamin1 K44A (Fig. 9B, middle and bottom).
We next wondered whether the altered localization of dynamin 1 and cortactin caused any changes in the stability of F-actin bundles. To test this hypothesis, we analyzed the depolymerization rate of F-actin to G-actin under various conditions. To this end, the kinetics of preformed pyrene-labeled F-actin depolymerization was examined either by tenfold dilution (Okamoto et al., 2007) or by the addition of Mycalolide B (Saito et al., 1994). In the presence of dynamin 1 and cortactin, the rate of depolymerization by dilution was clearly decreased as potent as by α-actinin, an actin-crosslinking protein, indicating that dynamin 1 and cortactin stabilize F-actin bundles.(Fig. 9C). We next tested whether guanine nucleotide conditions affect the stability of the F-actin bundles decorated with dynamin 1 and cortactin. As shown in Figure 9, D and E, the F-actin bundles were most resistant to the depolymerization treatments in the presence of GTP compared with GTPγS or no nucleotide. We also found that cortactin, but not the Cort W525K mutant, stimulated dynamin GTPase activity ∼2-fold (Fig. 9F).Together, these results suggest that F-actin bundles are stabilized by GTP hydrolysis that is mediated by dynamin 1–cortactin complexes.
Discussion
This study demonstrated that dynamin 1–cortactin complexes stabilize F-actin bundles, which is necessary for the proper formation of growth cone filopodia. Thus, neuronal dynamin 1 plays an essential role in actin regulation in addition to its well studied function in synaptic vesicle endocytosis.
Dynamin 1 is involved in actin dynamics in vivo and in vitro
Growth cone filopodia require bundled F-actin, and the loss of the bundles leads to growth cone collapse (Zhou and Cohan, 2001). In this study, acute inhibition of dynamin 1 in neurons by dynasore application caused rapid collapse of the growth cone filopodia. This effect was prevented by application of Jasplakinolide, an actin depolymerization inhibitor (Fig. 5B,C). In addition, actin assembly in vitro was drastically decreased by the inhibition of dynamin 1 (Fig. 1). These results clearly indicated that dynamin 1 was essential for the stabilization of F-actin. Supporting this notion, immunoelectron microscopy revealed that dynamin 1 was present on actin filaments and F-actin bundles in the growth cone filopodia (Fig. 4). Consistently, it is recently reported that dynamin directly binds to F-actin and it can promote the elongation of individual actin filaments by displacement of actin capping proteins from actin filaments (Gu et al., 2010).
A dynamin 1–cortactin complex is required for bundling of the actin filaments
Dynamin 1 and cortactin copolymerized into ring-shaped complexes (Fig. 8), and in the presence of F-actin, the complexes were often observed twined around F-actin bundles (Fig. 7B, inset). Replacing Cort WT with Cort W525K mutant inhibited the ring formation, indicating that the interaction between dynamin 1 PRD and cortactin SH3 domain was essential for the complex formation. Furthermore, the formation of the ring complexes was disturbed by dynasore application and an antibody against amino acid residues 633–647 of dynamin 1 (Fig. 8B,C). Since the anti-dynamin 1 antibody recognizes a region adjacent to the dynamin stalk domain (Faelber et al., 2011; Ford et al., 2011), in all likelihood, the presence of this antibody would interfere with intramolecular interactions via stalk domain of dynamin 1 that is required for dynamin dimerization and polymerization. It is noteworthy that acute inhibition of dynamin 1 by both dynasore application and the dynamin 1 antibody drastically decreased actin assembly in vitro (Fig. 1) and caused rapid retraction of growth cone filopodia (Fig. 5). Furthermore, depletion of cortactin by RNAi also reduced filopodium formation, which is recovered by expression of cortactin WT but not by a dynamin binding-deficient mutant (Fig. 5). These observations strongly suggest that the dynamin 1–cortactin complex functions to form F-actin bundles in vivo and inhibition of the complex formation yields destabilized F-actin bundles in growth cone filopodia. In support of this notion, similar periodic localizations of dynamin 1 and cortactin in puncta were observed along both F-actin bundles formed in vitro (Fig. 7) and growth cone filopodia (Fig. 3).
Stabilization of actin filaments by a dynamin 1–cortactin complex
In the presence of dynamin 1 and cortactin, F-actin became resistant to depolymerization induced by either dilution or a depolymerizing toxin (Fig. 9C–E). Furthermore, this effect was more prominent in the presence of GTP compared with unhydrolyzable GTPγS or in the absence of GTP (Fig. 9D,E). These observations indicate that dynamin 1–cortactin complex is implicated in stabilizing F-actin bundles in GTP hydrolysis-dependent manner.
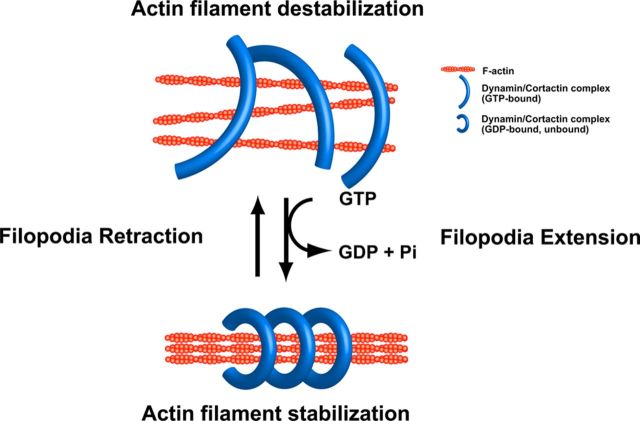
How might GTP hydrolysis by the dynamin 1–cortactin complex increase the stability of F-actin bundles? In synaptic vesicle endocytosis, dynamin 1 polymerizes into a ring, and the conformational change of dynamin upon GTP hydrolysis provides the mechanical force necessary for membrane fission (Sweitzer and Hinshaw, 1998; Takei et al., 1998; Marks et al., 2001; Roux et al., 2006; Ramachandran and Schmid, 2008). It is conceivable that dynamin 1–cortactin complexes use this mechanochemical enzyme property of dynamin 1. The ring-shaped dynamin 1–cortactin complex would hold several actin filaments and then mechanically grab the F-actin bundle more tightly upon GTP hydrolysis (Fig. 10). In support of this notion, dynamin 1–cortactin ring complexes were observed both at open and closed conformations, and, in the presence of GTPγS, the complexes were widely open or almost linear-shaped (Fig. 8), indicating that the conformation of the dynamin 1–cortactin ring complex is altered upon GTP hydrolysis. The ring complexes were twined around several actin filaments that were interconnected each other to form thicker F-actin bundles (Fig. 7B). Such conformational changes in dynamin 1–cortactin complexes may have led to different localization patterns of dynamin 1 and cortactin on F-actin bundles in the presence of GTP or GTPγS (Fig. 9B).
Figure 10.
Model of mechanical bundling of actin filaments by dynamin 1–cortactin ring complex. Dynamin 1–cortactin polymerizes into ring-shaped complex, which undergoes open and closed conformational change. The rings are open at GTP bound state, and closed at GDP bound state. Actin filaments are cross-linked by the complex, and upon GTP hydrolysis, the filaments would be tightened by a closing motion of the ring complex. Increased dynamin GTPase activity by cortactin would facilitate this process.
The presence of dynamin 1–cortactin complexes around F-actin bundles would physically obstructs treadmilling of F-actin. Alternatively, the interaction between dynamin 1–cortactin complexes and actin filaments might interfere with the affinity of F-actin for actin depolymerizing molecules. It has been reported that presence of dynamin and cortactin alter the affinity of F-actin to cofilin (Mooren et al., 2009) or a capping protein gelsolin (Gu et al., 2010). In either case, however, the mechanisms are currently unknown.
Our results indicate that ring-shaped dynamin 1–cortactin complexes stabilize F-actin bundles both in vitro and in vivo. It is noteworthy that the complex uses the GTP-dependent mechanochemical enzyme property of dynamin 1 (Fig. 10). Now that the mode of action of dynamin has been largely clarified by crystallography studies (Faelber et al., 2011; Ford et al., 2011), future studies will be needed to clarify how the dynamin 1–cortactin ring complexes may be involved in neuronal development.
Footnotes
This work was supported in part by grants from the Ministry of Education, Science, Sports, and Culture of Japan to H.Y. (22616004) and K. Takei (23370089) and the Astellas Foundation for Research on Metabolic Disorders (H.Y.). We thank Central Research Laboratory Okayama University Medical School. We acknowledge Dr. Pietro DeCamilli (Yale University) for critical reading of the manuscript.
The authors declare no competing financial interests.
References
- Ammer AG, Weed SA. Cortactin branches out: roles in regulating protrusive actin dynamics. Cell Motil Cytoskeleton. 2008;65:687–707. doi: 10.1002/cm.20296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzaniti A, Neff L, Sanjay A, Horne WC, De Camilli P, Baron R. Dynamin forms a Src kinase-sensitive complex with Cbl and regulates podosomes and osteoclast activity. Mol Biol Cell. 2005;16:3301–3313. doi: 10.1091/mbc.E04-12-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb MR, Spector I, Beyer BB, Fosen KM. Effects of jasplakinolide on the kinetics of actin polymerization. An explanation for certain in vivo observations. J Biol Chem. 2000;275:5163–5170. doi: 10.1074/jbc.275.7.5163. [DOI] [PubMed] [Google Scholar]
- Cao H, Orth JD, Chen J, Weller SG, Heuser JE, McNiven MA. Cortactin is a component of clathrin-coated pits and participates in receptor-mediated endocytosis. Mol Cell Biol. 2003;23:2162–2170. doi: 10.1128/MCB.23.6.2162-2170.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courson DS, Rock RS. Actin cross-link assembly and disassembly mechanics for alpha-Actinin and fascin. J Biol Chem. 2010;285:26350–26357. doi: 10.1074/jbc.M110.123117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C, McPherson PS, Mundigl O, de Camilli P. A role of amphiphysin in synaptic vesicle endocytosis suggested by its binding to dynamin in nerve terminals. Proc Natl Acad Sci U S A. 1996;93:331–335. doi: 10.1073/pnas.93.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Gertler FB. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 2003;40:209–227. doi: 10.1016/s0896-6273(03)00633-0. [DOI] [PubMed] [Google Scholar]
- Dixon RDS, Arneman DK, Rachlin AS, Sundaresan NR, Costello MJ, Campbell SL, Otey CA. Palladin is an actin cross-linking protein that uses immunoglobulin-like domains to bind filamentous actin. J Biol Chem. 2008;283:6222–6231. doi: 10.1074/jbc.M707694200. [DOI] [PubMed] [Google Scholar]
- Faelber K, Posor Y, Gao S, Held M, Roske Y, Schulze D, Haucke V, Noé F, Daumke O. Crystal structure of nucleotide-free dynamin. Nature. 2011;477:556–560. doi: 10.1038/nature10369. [DOI] [PubMed] [Google Scholar]
- Farsad K, Ringstad N, Takei K, Floyd SR, Rose K, De Camilli P. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MG, Jenni S, Nunnari J. The crystal structure of dynamin. Nature. 2011;477:561–566. doi: 10.1038/nature10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo S, Gordon-Weeks PR. Cytoskeletal dynamics in growth-cone steering. J Cell Sci. 2009;122:3595–3604. doi: 10.1242/jcs.042309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Yaddanapudi S, Weins A, Osborn T, Reiser J, Pollak M, Hartwig J, Sever S. Direct dynamin-actin interactions regulate the actin cytoskeleton. EMBO J. 2010;29:3593–3606. doi: 10.1038/emboj.2010.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Raimondi A, O'Toole E, Paradise S, Collesi C, Cremona O, Ferguson SM, De Camilli P. Cell- and stimulus-dependent heterogeneity of synaptic vesicle endocytic recycling mechanisms revealed by studies of dynamin 1-null neurons. Proc Natl Acad Sci U S A. 2008;105:2175–2180. doi: 10.1073/pnas.0712171105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Huang C, Ni Y, Wang T, Gao Y, Haudenschild CC, Zhan X. Down-regulation of the filamentous actin cross-linking activity of cortactin by Src-mediated tyrosine phosphorylation. J Biol Chem. 1997;272:13911–13915. doi: 10.1074/jbc.272.21.13911. [DOI] [PubMed] [Google Scholar]
- Hynds DL, Spencer ML, Andres DA, Snow DM. Rit promotes MEK-independent neurite branching in human neuroblastoma cells. J Cell Sci. 2003;116:1925–1935. doi: 10.1242/jcs.00401. [DOI] [PubMed] [Google Scholar]
- Johnsson AK, Karlsson R. Synaptotagmin 1 causes phosphatidyl inositol lipid-dependent actin remodeling in cultured non-neuronal and neuronal cells. Exp Cell Res. 2012;318:114–126. doi: 10.1016/j.yexcr.2011.10.009. [DOI] [PubMed] [Google Scholar]
- Kessels MM, Qualmann B. The syndapin protein family: linking membrane trafficking with the cytoskeleton. J Cell Sci. 2004;117:3077–3086. doi: 10.1242/jcs.01290. [DOI] [PubMed] [Google Scholar]
- Kurklinsky S, Chen J, McNiven MA. Growth cone morphology and spreading are regulated by a dynamin–cortactin complex at point contacts in hippocampal neurons. J Neurochem. 2011;117:48–60. doi: 10.1111/j.1471-4159.2011.07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi N, Watanabe M, Yamada H, Li SA, Kashiwakura Y, Matsukawa T, Nagai A, Nasu Y, Kumon H, Takei K. Implication of amphiphysin 1 and dynamin 2 in tubulobulbar complex formation and spermatid release. Cell Struct Funct. 2007;32:101–113. doi: 10.1247/csf.07024. [DOI] [PubMed] [Google Scholar]
- Lee E, De Camilli P. Dynamin at actin tails. Proc Natl Acad Sci U S A. 2002;99:161–166. doi: 10.1073/pnas.012607799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AK, Bridgman PC. Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J Cell Biol. 1992;119:1219–1243. doi: 10.1083/jcb.119.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, Van Vactor D. The trip of the tip: understanding the growth cone machinery. Nat Rev Mol Cell Biol. 2009;10:332–343. doi: 10.1038/nrm2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Cantley LC, Janmey PA, Kirschner MW. Corequirement of specific phosphoinositides and small GTP-binding protein Cdc42 in inducing actin assembly in Xenopus egg extracts. J Cell Biol. 1998;140:1125–1136. doi: 10.1083/jcb.140.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Marks B, Stowell MHB, Vallis Y, Mills IG, Gibson A, Hopkins CR, McMahon HT. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature. 2001;410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- McNiven MA, Kim L, Krueger EW, Orth JD, Cao H, Wong TW. Regulated interactions between dynamin and the actin-binding protein cortactin modulate cell shape. J Cell Biol. 2000;151:187–198. doi: 10.1083/jcb.151.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson G, Pfenninger KH, Påhlman S. A complex consisting of pp60c-src/pp60c-srcN and a 38 kDa protein is highly enriched in growth cones from differentiated SH-SY5Y neuroblastoma cells. J Cell Sci. 1992;103:233–243. doi: 10.1242/jcs.103.1.233. [DOI] [PubMed] [Google Scholar]
- Mooren OL, Kotova TI, Moore AJ, Schafer DA. Dynamin 2 GTPase and cortactin remodel actin filaments. J Biol Chem. 2009;284:23995–24005. doi: 10.1074/jbc.M109.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Miki H, Ito M, Ohashi K, Takenawa T, Miyamoto S. N-WASP, WAVE and Mena play different roles in the organization of actin cytoskeleton in lamellipodia. J Cell Sci. 2001;114:1555–1565. doi: 10.1242/jcs.114.8.1555. [DOI] [PubMed] [Google Scholar]
- Ochoa GC, Slepnev VI, Neff L, Ringstad N, Takei K, Daniell L, Kim W, Cao H, McNiven M, Baron R, De Camilli P. A functional link between dynamin and the actin cytoskeleton at podosomes. J Cell Biol. 2000;150:377–389. doi: 10.1083/jcb.150.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Narayanan R, Lee SH, Murata K, Hayashi Y. The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proc Natl Acad Sci U S A. 2007;104:6418–6423. doi: 10.1073/pnas.0701656104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Krueger EW, Cao H, McNiven MA. The large GTPase dynamin regulates actin comet formation and movement in living cells. Proc Natl Acad Sci U S A. 2002;99:167–172. doi: 10.1073/pnas.012607899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka A, Abe T, Watanabe M, Yagisawa H, Takei K, Yamada H. Dynamin 2 is required for actin assembly in phagocytosis in Sertoli cells. Biochem Biophys Res Commun. 2009;378:478–482. doi: 10.1016/j.bbrc.2008.11.066. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Schmid SL. Real-time detection reveals that effectors couple dynamin's GTP-dependent conformational changes to the membrane. EMBO J. 2008;27:27–37. doi: 10.1038/sj.emboj.7601961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A, Uyhazi K, Frost A, De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- Saito S, Watabe S, Ozaki H, Fusetani N, Karaki H. Mycalolide B, a novel actin depolymerizing agent. J Biol Chem. 1994;269:29710–29714. [PubMed] [Google Scholar]
- Schafer DA, Weed SA, Binns D, Karginov AV, Parsons JT, Cooper JA. Dynamin 2 and cortactin regulate actin assembly and filament organization. Curr Biol. 2002;12:1852–1857. doi: 10.1016/s0960-9822(02)01228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepnev VI, Ochoa GC, Butler MH, De Camilli P. Tandem arrangement of the clathrin and AP-2 binding domains in amphiphysin 1 and disruption of clathrin coat function by amphiphysin fragments comprising these sites. J Biol Chem. 2000;275:17583–17589. doi: 10.1074/jbc.M910430199. [DOI] [PubMed] [Google Scholar]
- Soulet F, Yarar D, Leonard M, Schmid SL. SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Mol Biol Cell. 2005;16:2058–2067. doi: 10.1091/mbc.E04-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell MH, Marks B, Wigge P, McMahon HT. Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nat Cell Biol. 1999;1:27–32. doi: 10.1038/8997. [DOI] [PubMed] [Google Scholar]
- Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- Takei K, Haucke V, Slepnev V, Farsad K, Salazar M, Chen H, De Camilli P. Generation of coated intermediates of clathrin-mediated endocytosis on protein-free liposomes. Cell. 1998;94:131–141. doi: 10.1016/s0092-8674(00)81228-3. [DOI] [PubMed] [Google Scholar]
- Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- Warnock DE, Baba T, Schmid SL. Ubiquitously expressed dynamin-II has a higher intrinsic GTPase activity and a greater propensity for self-assembly than neuronal dynamin-I. Mol Biol Cell. 1997;8:2553–2562. doi: 10.1091/mbc.8.12.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamabhai M, Hoffman NG, Hardison NL, McPherson PS, Castagnoli L, Cesareni G, Kay BK. Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J Biol Chem. 1998;273:31401–31407. doi: 10.1074/jbc.273.47.31401. [DOI] [PubMed] [Google Scholar]
- Yamada H, Abe T, Li SA, Masuoka Y, Isoda M, Watanabe M, Nasu Y, Kumon H, Asai A, Takei K. Dynasore, a dynamin inhibitor, suppresses lamellipodia formation and cancer cell invasion by destabilizing actin filaments. Biochem Biophys Res Commun. 2009a;390:1142–1148. doi: 10.1016/j.bbrc.2009.10.105. [DOI] [PubMed] [Google Scholar]
- Yamada H, Padilla-Parra S, Park SJ, Itoh T, Chaineau M, Monaldi I, Cremona O, Benfenati F, De Camilli P, Coppey-Moisan M, Tramier M, Galli T, Takei K. Dynamic interaction of amphiphysin with N-WASP regulates actin assembly. J Biol Chem. 2009b;284:34244–34256. doi: 10.1074/jbc.M109.064204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Kinuta M, Abe T, Liang S, Araki K, Cremona O, Di Paolo G, Moriyama Y, Yasuda T, De Camilli P, Takei K. The stimulatory action of amphiphysin on dynamin function is dependent on lipid bilayer curvature. EMBO J. 2004;23:3483–3491. doi: 10.1038/sj.emboj.7600355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FQ, Cohan CS. Growth cone collapse through coincident loss of actin bundles and leading edge actin without actin depolymerization. J Cell Biol. 2001;153:1071–1084. doi: 10.1083/jcb.153.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhou K, Hao JJ, Liu J, Smith N, Zhan X. Regulation of cortactin/dynamin interaction by actin polymerization during the fission of clathrin-coated pits. J Cell Sci. 2005;118:807–817. doi: 10.1242/jcs.01668. [DOI] [PubMed] [Google Scholar]