Phylogenomics provides robust support for a two-domains tree of life (original) (raw)
. Author manuscript; available in PMC: 2020 Jun 9.
Published in final edited form as: Nat Ecol Evol. 2019 Dec 9;4(1):138–147. doi: 10.1038/s41559-019-1040-x
Abstract
Hypotheses about the origin of eukaryotic cells are classically framed within the context of a universal "tree of life" based upon conserved core genes. Vigorous ongoing debate about eukaryote origins is based upon assertions that the topology of the tree of life depends on the taxa included and the choice and quality of genomic data analysed. Here we have reanalysed the evidence underpinning those claims and bring more data to bear on the question by using supertree and coalescent methods to interrogate >3000 gene families in Archaea and eukaryotes. We find that eukaryotes consistently originate from within the Archaea in a two-domains tree when due consideration is given to the fit between model and data. Our analyses support a close relationship between eukaryotes and Asgard Archaea and identify the Heimdallarchaeota as the current best candidate for the closest archaeal relatives of the eukaryotic nuclear lineage.
Current hypotheses about eukaryotic origins generally propose at least two partners in that process: a bacterial endosymbiont that became the mitochondrion and a host cell for that endosymbiosis1–4. The identity of the host has been informed by analyses of conserved genes for the transcription and translation machinery that are considered essential for cellular life5. Traditionally, the host was considered to be a eukaryote based upon ribosomal RNA trees in either unrooted6,7 or rooted form8. In these trees, Archaea, Bacteria and Eukarya form three separate primary domains, with the rooted version suggesting that Archaea and Eukarya are more closely related to each other than to Bacteria8. A criticism of these three-domains (3D) trees is that they were constructed using overly simple phylogenetic models5,9,10. Phylogenetic analyses using models that better fit features of the data10–12, coupled with an expanded sampling of prokaryotic diversity13–15, have supported a two domains (2D) tree consistent with the eocyte16 hypothesis whereby the eukaryotic nuclear lineage - that is, the host for the mitochondrial endosymbiont - originated from within the Archaea (reviewed in5,17). The 2D tree has gained increasing traction in the field18, particularly with the discovery of the Asgard archaea19,20. The Asgard archaea branch together with eukaryotes in phylogenetic trees, and their genomes encode homologues of eukaryotic signature proteins - that is, proteins which underpin the defining cellular structures of eukaryotes, and which were previously thought7,21 to be unique to eukaryotes. However, the discoveries and analyses that support the 2D tree have been criticised from a variety of perspectives.
It has been suggested22,23 that the close relationship between eukaryotes and Asgard archaea in 2D trees19,20 is due to eukaryotic contamination of Asgard metagenomes combined with phylogenetic artifacts caused by the choice of genes analysed and the inclusion of fast evolving Archaea in tree reconstructions22–24; see also the comment25 and response24 to those analyses. The phenomenon of long branch attraction (LBA) due to the presence of fast-evolving sequences (FES) is a well-known artifact in phylogenetic analyses26–28. Indeed, it has previously been suggested that it is the 3D tree, rather than the 2D tree, that is an artifact of LBA5,9–11, both because analyses under better-fitting models have recovered a 2D tree, but also because the 3D topology is one in which the two longest branches in the tree of life - the stems leading to bacteria and to eukaryotes - are grouped together. Nevertheless, when putative FES were removed, Forterre and colleagues22,24 recovered a monophyletic Archaea within a three-domains tree, whether analysing 35 core genes, a particular subset of 6 genes, or RNA polymerases alone. Claims that the 2D tree is a product of unbalanced taxonomic sampling and inclusion of FES have also been made by others29.
In a more general criticism it has been suggested30–33 that protein sequences do not harbour sufficient signal to resolve the 2D/3D debate due to mutational saturation (but see11,12). One suggested solution is to analyse conserved structural motifs (folds) in proteins rather than primary sequence data31,33,34. Three-dimensional structures are thought to be more highly conserved than primary sequences. It has therefore been suggested that they should provide a more reliable indicator of ancient relationships, although it is not yet clear how best to analyse fold data for this purpose. Published unrooted trees based upon analyses of protein folds have recovered Archaea, Bacteria and Eukaryotes as separate groups34,35, a result that is consistent with the 3D, but not the 2D tree. Analyses of protein folds have recently been extended to use non-stationary models to infer a rooted tree of life31. In these analyses the inferred root separated cellular life into prokaryotes (Archaea plus Bacteria, termed akaryotes) and eukaryotes31,33. This tree is incompatible with the idea that Archaea and Eukaryotes share closer common ancestry, and recapitulates the hypothesis36 that the deepest division in cellular life is between prokaryotes and eukaryotes.
In this paper, we have evaluated the analyses and data that have led to conflicting hypotheses of relationships between the major groups of cellular life, and for the position of the eukaryotic nuclear lineage. We have also performed phylogenomic analyses using the best-available supermatrix, supertree, and coalescent methods on an expanded sample of genes and taxa, to further explore the deep structure of the tree of life and the relationship between archaea and eukaryotes.
Results and Discussion
Analysis of core genes consistently supports two primary domains, not three
It has recently been argued22–24 that the 2D tree is an artifact of data and taxon sampling, and that resolution of those issues provides support for a 3D tree. The molecular data at the core of this debate had first been used19 to support a 2D tree in which eukaryotes clustered within Archaea as the closest relatives of the Asgard Archaea. The original dataset19 comprised a concatenation of 36 "universal" genes for 104 taxa. In the initial critique, it was claimed that the close relationship reported19 between Asgard archaea and eukaryotes was caused by the inclusion in the data set of a contaminated Elongation Factor 2 (EF2) gene for Lokiarchaeum sample Loki322 (now Heimdallarchaeota20), and by the inclusion of fast-evolving archaeal lineages in the analysis. However, recent data suggest that the EF2 gene of Heimdallarchaeota is not contaminated with eukaryotic sequences because similar EF2 sequences have been found in additional Heimdallarchaeota metagenome-assembled genomes (MAGs) prepared from different environmental DNA (eDNA) samples in different laboratories20,37.
The claim22–24 that the presence of "fast evolving sequences" (FES) might be affecting the topology recovered could be seen as a reasonable challenge, since LBA can influence the tree topology recovered. A problem for this specific critique22 however, is that no single, clear and consistent criterion was used to identify the "fast evolving" sequences that were removed from the original dataset19 in order to recover the 3D tree. Long-branched archaea might result from either a fast evolutionary rate or a long period of time, and these possibilities are difficult to distinguish a priori. Moreover, the historical papers38,39 cited22 as providing topological evidence that some sequences are "fast evolving" used site- and time-homogenous phylogenetic models (that is, models in which the process of evolution is constant over the sites of the alignment and branches of the tree) which often fit data poorly5. To investigate further we ranked all of the taxa in the original dataset19 according to their root-to-tip distances for each species. This is equal to the summed branch length (expressed as expected number of substitutions/site) from the root of the tree (rooted between Bacteria and Archaea) to the relevant tip. We calculated distributions and 95% credibility intervals (Supplementary Table 1) for each of these root-to-tip distances from the samples drawn during an MCMC analysis under the best-fitting (see below) CAT+GTR+G4 model in PhyloBayes, in order to perform Bayesian relative rates tests (Supplementary Table 1). The 23 taxa previously identified as FES are not the 23 taxa with the longest root-to-tip distances; while some of the taxa chosen for exclusion (Parvarchaeum, Micrarchaeum, Nanoarchaeum Nst1, Nanosalinarum, and Korarchaeum) are indeed relatively long-branching, others (Iainarchaeum, Nanoarchaeum G17 and Aenigmaarchaeon) are in the bottom half of the branch length distribution, and many of the longest-branching Archaea (including the Thaumarchaeota) were retained. Nevertheless, analysis22 of the reduced dataset did recover a 3D tree, raising the question of why this result was obtained. In the following analyses we have followed the recent renaming20 of the 3 “Loki” MAGs originally analysed as Lokiarchaeum sp. GC14_75 (formerly Loki1), Heimdallarchaeota archaeon LC_2 (Loki2), and Heimdallarchaeota archaeon LC_3 (Loki3).
The published 3D tree22 was recovered from the 35-gene concatenated data set under the LG+G4+F model40 in PhyML 3.141, with moderate support (76% bootstrap) for monophyletic Archaea (Figure 5(b) in 22). In repeating this analysis, we noted that although PhyML returned a three-domains tree, analysis of the same alignment under the same substitution model (LG+G4+F) with IQ-Tree 1.6.242 and RAxML 8.2.443, two other maximum-likelihood phylogeny packages, instead yielded a 2D tree where Heimdallarchaeota and Lokiarchaeum were together the sister group to eukaryotes, with a better likelihood score (Supplementary Figure 1, Supplementary Table 2). To investigate further, we computed the log likelihoods of the 2D and 3D trees in all three packages, keeping the alignment and model constant (Supplementary Table 2). All three implementations accord the 2D tree a higher likelihood than the 3D tree (lnl ~= -684701.2, compared to ~= -684716.1 for the 3D tree). It thus appears that the recovery of a 3D tree reflects a failure of PhyML to find the more likely 2D tree, rather than to the removal of problematic sequences. The differences between the likelihoods are not significant according to an approximately-unbiased test (AU = 0.229 for the 3D tree, 0.771 for the 2D), meaning that analysis of the 35-gene dataset under LG+G4+F is equivocal with respect to the 2D and 3D trees; contrary to previous claims22, analysis of the 35-gene concatenation under the LG+G4+F model provides no unambiguous evidence to prefer the 3D tree.
A number of newer models accommodate particular features of empirical data better than the LG+G4+F, so we investigated which trees were produced from the 35-gene dataset using these models. We addressed three issues in particular: among-site compositional heterogeneity due to site-specific biochemical constraints44, changing composition in different lineages over time45, and variations in site- and lineage-specific evolutionary rates (heterotachous evolution)46.
The CAT+GTR+G4 model44,47 is an extension to the standard GTR model that allows compositions to vary across sites. Analysis of the 35-gene dataset using this model produced a 2D tree where eukaryotes group with Heimdallarchaeota and Lokiarchaeum with maximal support (Figure 1). It was previously reported22 that convergence in Bayesian analyses is a problem for this data set using the CAT+GTR+G422 model. In our analyses, we achieved good convergence between chains as assessed both by comparison of split frequencies and, for the continuous parameters of the model, means and effective sample sizes (Supplementary Table 4). As an additional check, we also carried out ML analyses using the LG+C60+G4+F model, which improves on the LG+G4+F model by modelling site-specific compositional heterogeneity using a mixture of 60 composition categories. This model fits the data much better than the LG+G4+F according to the BIC (Supplementary Table 3) and, like CAT+GTR+G4, it recovered a 2D tree with high bootstrap support (Supplementary Figure 1(c)). The 3D tree (AU = 0.036) could also be rejected at P < 0.05 using an AU test, based on the LG+C60+G4+F model and the 35-gene alignment.
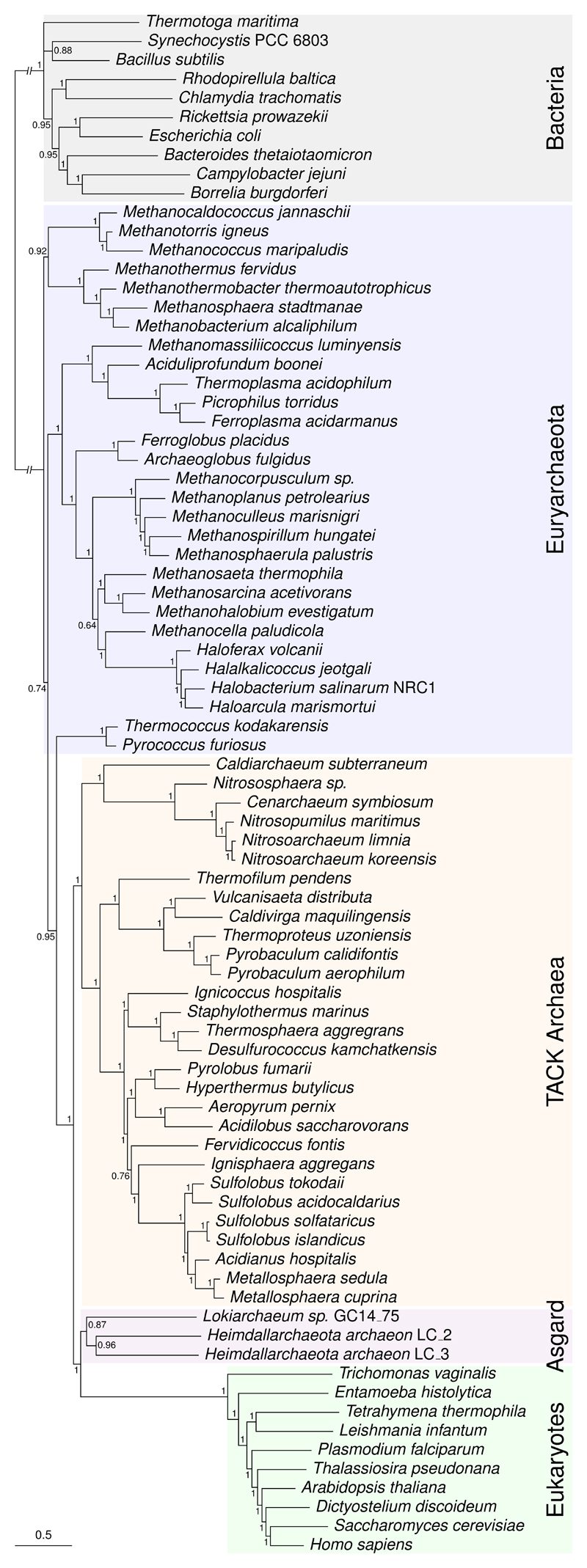
Figure 1. The 35-gene matrix of Da Cunha et al. favours a two-domains tree using the best-fitting models in both maximum likelihood and Bayesian analyses.
The eukaryotes (green) group with the sampled Asgard archaea (orange) with maximum posterior support. Bacteria are in grey, TACK Archaea in yellow, Euryarchaeota in blue. This is a consensus tree inferred under the CAT+GTR+G4 model in PhyloBayes-MPI; branch lengths are proportional to the expected number of substitutions per site, as indicated by the scale bar. A 2D topology was obtained under a variety of other models in ML analyses (LG+G4+F, LG+PMSF+G4, LG+C60+G4+F; Supplementary Figure 1), and also with 4-state Susko-Roger recoding under the CAT+GTR+G4 and NDCH2 models (Supplementary Figure 2).
Bayesian posterior predictive simulations48 provide a tool for evaluating the adequacy of models, by testing whether data simulated under a model is similar to the empirical data. Figure 2 plots the 2D tree (inferred under CAT+GTR+G4) and the 3D tree (inferred under LG+G4+F in PhyML) on the same scale (Figure 2(a)), revealing that --- from the same alignment --- CAT+GTR+G4 infers that many more substitutions have occurred in the core gene set during the evolutionary history of life. Model fit tests (Figure 2(b), Supplementary Table 4) indicate that LG+G4+F provided a much poorer fit to the data (larger Z-scores) than CAT+GTR+G4 in terms of across-site compositional heterogeneity (Z = 64.2 for LG+G4+F, Z = 6.9 for CAT+GTR+G4), and therefore systematically under-estimated the probability of convergent substitutions (Z = 19.7 for LG+G4+F; Z = 7.62 for CAT+GTR+G4). These differences arise because LG+G4+F assumes that amino acid frequencies are the same at all sites, whereas in empirical datasets different sites have different compositions, arising from distinct biochemical and selective constraints. Since this means the effective number of amino acids per site is in reality lower than that predicted by LG+G4+F, the probability of parallel convergence to the same amino acid in independent lineages is higher (Supplementary Table 5). CAT+GTR+G4 accounts for this across-site variation by incorporating site-specific compositions, and is therefore less prone to underestimating rates of convergent substitution. This is important because the longest branches in both the 2D and 3D trees are the lineages leading to the bacteria and eukaryotes. The lesser ability of LG+G4+F to detect convergent substitutions along these branches may favour inference of a 3D tree. While CAT+GTR+G4 provides a better fit than LG+G4+F, neither model completely fits the composition of the data (P = 0 for all tests; Supplementary Table 5). As a further data exploration step, we recoded49 the amino acid alignment into four categories of biochemically similar amino acids (AGNPST, CHWY, DEKQR, FILMV). Recoding has been shown to ameliorate sequence saturation and compositional heterogeneity49,50, and in this case it improved model fit (as judged by the magnitude of Z-scores; Supplementary Table 5). Analysis of this SR4-recoded alignment under CAT+GTR+G4 recovered a 2D tree where eukaryotes grouped with the Heimdallarchaeota (PP = 0.98, Supplementary Figure 2).
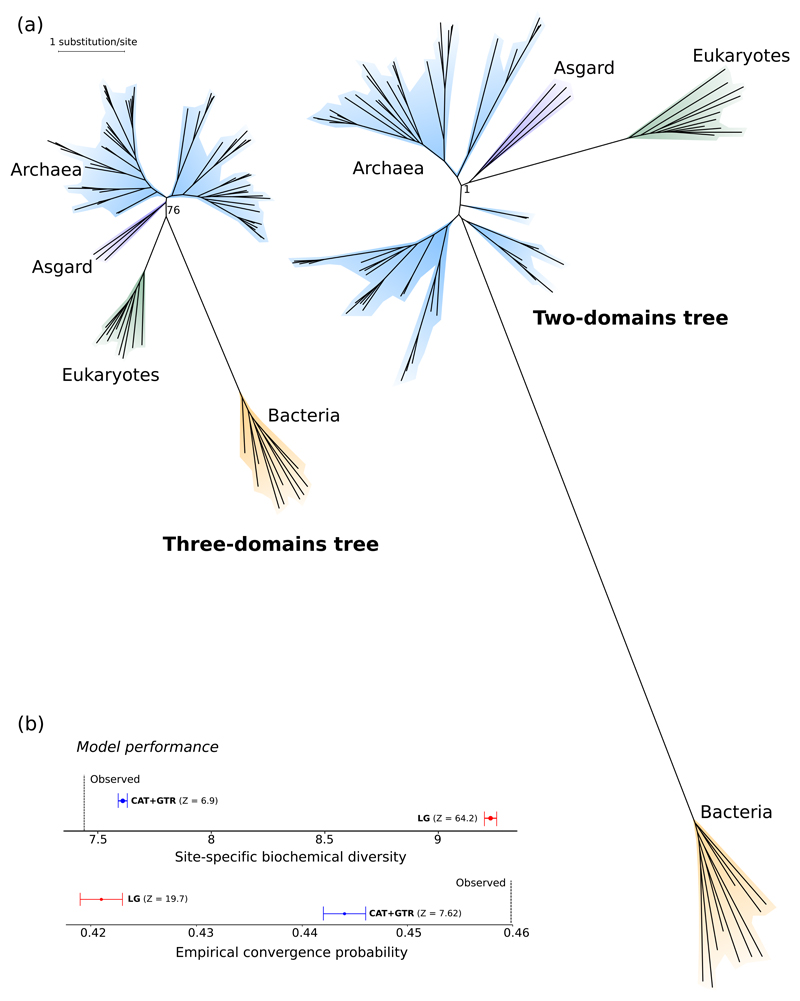
Figure 2. Evidence that the three-domains tree is an artifact of long branch attraction.
(a) Da Cunha et al. analysed a dataset of 35 core protein-coding genes under the LG+G4+F model and obtained a 3D tree; the better-fitting (Supplementary Table 4) CAT+GTR+G4 model recovers a 2D tree. Bootstrap support (a) and Bayesian posterior probability (b) are indicated for the key nodes defining the 3D and 2D trees. “Asgard” refers to a clade of Heimdallarchaeota and Lokiarchaeum. Plotting these trees to the same scale (in terms of substitutions per site) illustrates major differences in these analyses. The 3D/LG+G4+F analysis suggests that, on average, 30.77 changes have taken place per site; the two-domains/CAT+GTR+G4 analysis suggests that 47.4 changes per site have occurred. This difference amounts to ~128,511 additional substitutions in total inferred under the CAT+GTR+G4 model. (b) Posterior predictive tests indicate that CAT+GTR+G4 performs significantly better than LG+G4+F in capturing the site-specific evolutionary constraints reflected by lower biochemical diversity approaching that of the empirical data). This results in more realistic estimates of substitutional saturation and convergence found in the data. The longest branches on both the 3D and 2D tree are the stems leading to the bacteria and eukaryotes (in blue and green, respectively). CAT+GTR+G4 identifies many more convergent substitutions on these branches than does LG+G4+F, as can be seen by comparing the branch lengths in (a). This failure to detect convergent substitutions under LG+G4+F has the effect of drawing the bacterial and eukaryotic branches together, because convergences are mistaken for homologies (synapomorphies), resulting in a 3D tree.
Variation in sequence composition across the branches of the tree is also a pervasive feature of data that has been used to investigate the tree of life10,11. We tested each of the genes in the 35-gene dataset (see Methods), and found that 23/35 showed significant evidence of across-branch heterogeneity at P < 0.05 (Supplementary Table 6). Analysis of the concatenation of the 12 composition-homogeneous genes under CAT+GTR+G4 gave a 2D tree with maximal posterior support (PP = 1, Supplementary Figure 3), as did a partitioned analysis using the best-fitting homogeneous model for each of the 12 gene partitions (LG+G4+F in all cases; Supplementary Figure 3; PP = 1). We also inferred a phylogeny from the entire 35-gene dataset under the branch-heterogeneous node-discrete compositional heterogeneity (NDCH)2 model, which explicitly incorporates changing sequence compositions across the tree. NDCH2 is an extension of the NDCH model45; it has a separate composition vector for each tree node and is constrained via a sampled concentration parameter of a Dirichlet prior. Thus, the model adjusts to the level of across-branch compositional heterogeneity in the data during the MCMC analysis. For reasons of computational tractability, this analysis could only be run on the SR4-recoded version of the 35-gene alignment. NDCH2 obtained adequate model fit with respect to across-branch compositional heterogeneity (P = 0.7838), and recovered a 2D tree with Heimdallarchaeota as the sister group to eukaryotes (PP = 0.85; Supplementary Figure 2).
A failure to account for heterotachy, or rates of molecular evolution that are both site- and branch-specific, has been posited as a potential issue for phylogenomic analyses of ancient core genes51,52. We used the GHOST53 model of IQ-Tree to analyze the 35-gene alignment. GHOST is an edge-unlinked mixture model in which the sites of the alignment evolve along a shared tree topology, but are fit by a finite mixture of GTR exchangeabilities, sequence compositions and branch lengths. We fit a four component mixture model to both the original amino acid alignment (LG+G4+F components) and the SR4-recoded version (GTR+F components). The resulting trees were a weakly-supported (amino acids; 58% bootstrap support for eukaryotes plus Heimdallarchaeota and Lokiarchaeum) or strongly-supported (recoded data; 95% bootstrap support for eukaryotes plus Heimdallarchaeota) 2D tree (Supplementary Figure 5).
In summary, all of our analyses of the 35-gene alignment using better models recovered a 2D tree in which eukaryotes are either the sister group of Heimdallarchaeota plus Lokiarchaeum or Heimdallarchaeota alone, rather than the 3D tree which the data has previously been claimed22 to support.
Do some core genes have different histories?
Based upon AU tests under the LG+G4+F model for individual genes in the 35-gene dataset, it was suggested22 that the 35-gene dataset contains two subsets of genes with different evolutionary histories: a larger set supporting the 2D tree and a smaller set supporting the 3D tree. We used the better-fitting CAT+GTR+G4 model to analyse a concatenated dataset of the 6 genes which significantly favoured the 3D tree under LG+G4+F, and we also analysed a four-state recoded version of the same alignment. Analysis of the original amino acids recovered a moderately-supported 3D tree, while analysis of the recoded alignment recovered a weakly-supported 2D tree (Supplementary Figure 4); posterior predictive simulations indicated that model fit was improved by SR4 recoding (Supplementary Table 7), suggesting that support for the 3D tree from these 6 genes under LG+G4+F may be due to model misspecification.
It has also been suggested that phylogenetic analyses of RNA polymerase subunits22 provide robust support for a 3D tree. By contrast, other11 analyses of RNA polymerase subunits have already suggested that better fitting models prefer a 2D tree. We evaluated the fit of both models, LG+G4+F and CAT+GTR+G4, used22 to recover a 3D tree from RNA polymerase subunits, using posterior predictive simulations (Supplemental Text), and found that both models provide an inadequate fit to the data (Supplementary Table 8). Model fit was improved following SR4 recoding (Supplementary Table 8), and this analysis recovered a weakly-supported and poorly-resolved 2D tree (Supplementary Figure 6).
Expanded gene and taxon sampling supports a clade of eukaryotes and Asgard archaea
We took advantage of the recent dramatic improvements in genomic and transcriptomic sampling of free-living bacteria, archaea, and microbial eukaryotes to assemble a dataset of 125 species, including 53 eukaryotes, 39 archaea (including an expanded set of Asgard MAGs20 representing two new groups, Odinarchaeota and Thorarchaeota), and 33 bacteria, on the principle that improved sampling can sometimes help to resolve difficult phylogenetic problems54,55. We used free-living representatives of eukaryotic groups to avoid the well-documented problems for tree reconstruction caused by sequences from parasitic eukaryotes26. Our sampling of archaea and bacteria was also expanded to include representatives from the large number of uncultivated lineages that have recently been identified by single cell-genomics and metagenomics15,56,57.
To further investigate the claim22 that the tree inferred depends on the choice of universal marker genes, we used the Orthologous MAtrix (OMA58) algorithm to identify single-copy orthologues de novo on the 125 genome set. Benchmarks59 indicate that OMA is conservative, in that it returns a relatively low number of orthologues, but that these orthologues perform better than other methods at recovering the species tree. Combining OMA analysis with manual filtering to remove EF2 and genes of endosymbiotic origin (see Methods), we identified 21 broadly-conserved marker genes found in at least half of our set of bacteria, archaea, and eukaryotes, and 43 genes encoded by at least half of the archaea and eukaryotes (see Methods). We concatenated the 21 genes conserved in all three domains and inferred a tree under CAT+GTR+G4 (Figure 3a). Rooting on the branch separating bacteria and archaea resulted in a 2D tree, in which eukaryotes form a maximally-supported clade with Asgard archaea (Figure 3a); within Asgards, the closest relatives of eukaryotes was recovered as the Heimdallarchaeota, although with only modest support (PP = 0.79).
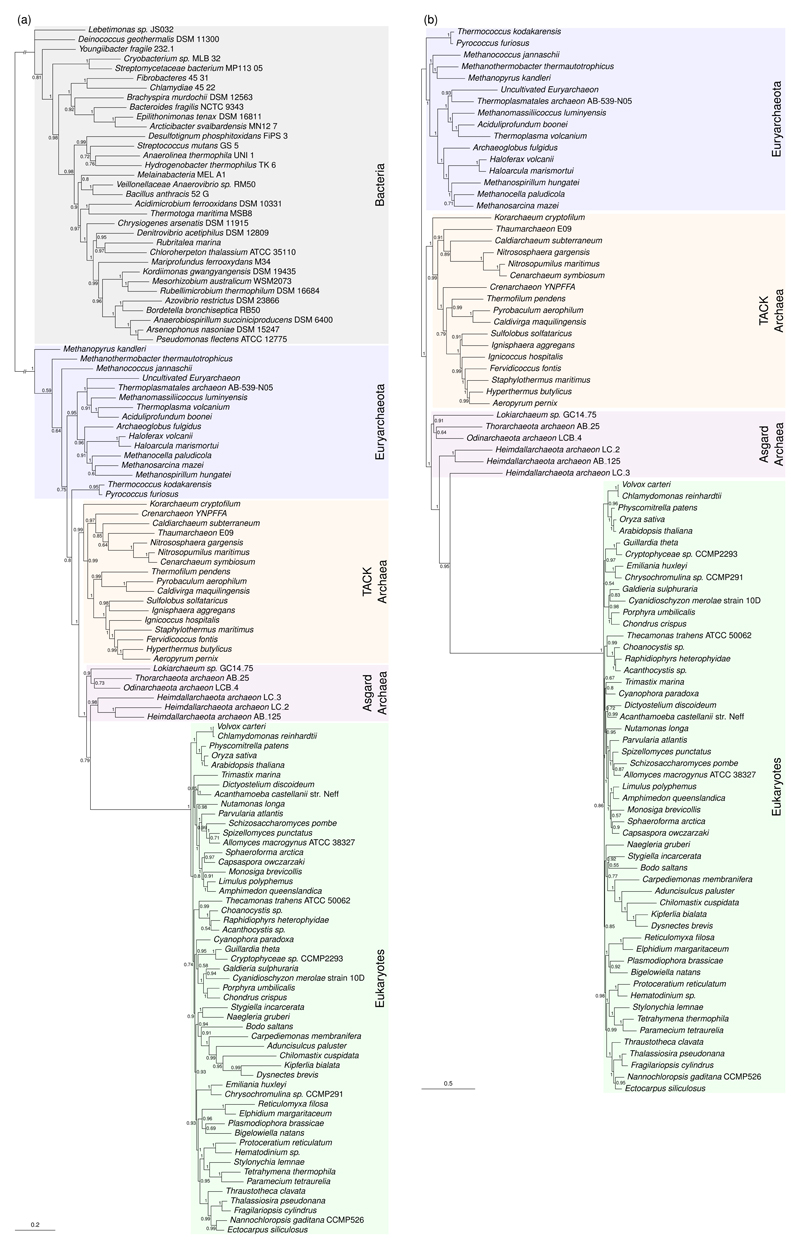
Figure 3. An expanded sampling of microbial diversity supports a two-domains tree.
(a) Bayesian phylogeny of 21 concatenated proteins conserved across bacteria, archaea and eukaryotes under the CAT+GTR+G4 model, rooted on the branch separating bacteria and archaea. Eukaryotes group with Asgard archaea with maximum posterior support. (b) Bayesian phylogeny of 43 genes conserved between Archaea and eukaryotes under CAT+GTR+G4. Eukaryotes group with, or within, Heimdallarchaeota. All support values are Bayesian posterior probabilities, and branch lengths are proportional to the expected number of substitutions per site, as indicated by the scale bars. The Euryarchaeota are paraphyletic in the consensus tree in (a), consistent with some recent analyses using bacterial outgroups11,12, although the relevant support values are low and the analysis does not robustly exclude the alternative hypothesis90 of a monophyletic Euryarchaeota. The tree in (b) is formally unrooted because it does not include a bacterial outgroup. Based on (a) and published analyses12,90, the root may lie between the Euryarchaeota and the other taxa, or within the Euryarchaeota. Amino acid data were recoded using the 4-state scheme of Susko and Roger, which our posterior predictive simulations (Supplementary Table 7) suggest improved model fit by ameliorating substitutional saturation and compositional heterogeneity; phylogenies inferred on the original amino acid data are provided in Supplementary Figure 7.
We next analyzed the expanded set of genes conserved between archaea and eukaryotes, placing the root outside the TACK/Asgard/eukaryote clade as suggested by the previous analysis including bacteria. The consensus tree under CAT+GTR+G4 (Figure 3b) resolves a clade of eukaryotes and Heimdallarchaeota with maximal posterior support; within that clade, eukaryotes group with one Heimdallarchaeota metagenome bin (LC3) with high (PP = 0.95) support.
Given ongoing debates about the impact of even single genes within concatenated datasets, we investigated in detail the overlap between the 35-gene set, the 21-genes selected by OMA, and a 29-gene set used in some previous analyses10,11,14,60,61 (Supplementary Table 10). After removing EF2, 7 genes are found in all three sets; 27 in at least two of the three, and 50 genes in total are present in at least one of the datasets. We obtained the orthologues for the 50 gene families from the 125 species dataset, and inferred trees using the best-fit ML model in IQ-Tree on the 7-, 27- and 50-gene concatenations (Supplementary Figure 8). We also expanded species sampling for the 35 genes to compare with the analyses described above. Analysis under the best-fitting ML model for all four concatenates resulted in a 2D tree, with either all Asgards (the 7- and 35-gene datasets) or Heimdallarchaeota (27 and 50 gene datasets) as sister to eukaryotes with moderate (7-gene set) to high (the other sets) bootstrap support. These results indicate that there is a congruent signal for a 2D tree, and a relationship between eukaryotes and Asgard archaea, that is robust to moderate differences in the choice of marker genes. The results of all our concatenation analyses are summarised in Supplementary Table 11.
Supertree and multispecies coalescent methods support the two-domains tree
Concatenation allows phylogenetic signal to be pooled and permits the use of complex, parameter-rich substitution models, but its assumptions are problematic in the context of microbial evolution. In particular, concatenation requires that all of the genes share a common phylogeny62,63, an assumption that is difficult to test because trees inferred from individual genes are often poorly supported. Some incongruence between single gene trees can be attributed to stochastic error or model misspecification14, but genuinely different evolutionary histories for different genes can arise from incomplete lineage sorting, gene duplication and loss, and horizontal gene transfer. We therefore investigated alternative methods for integrating phylogenetic signal from multigene datasets that account for gene tree incongruence in different ways. The probabilistic supertree method of Steel and Rodrigo (SR2008)64, and the Split Presence-Absence (SPA) method65, are supertree methods that model differences between gene trees as stochastic noise; ASTRAL is a supertree method that is consistent under the multispecies coalescent66. These methods have their own assumptions and limitations63, but these are distinct from --- and provide a useful contrast to --- concatenation. As these methods do not require genes to be broadly conserved across the species of interest, we analyzed a set of 3199 single-copy orthologues found in at least four of the taxa in our dataset (of these 3199 gene families, 479 included at least one archaeon and one eukaryote; see Supplementary Table 12 for the taxonomic distribution and phylogenetic relationships supported by the individual trees).
All of these analyses resolved a 2D tree including a clade of eukaryotes and Asgard archaea with high to maximal support (Supplementary Figures 9-10). Supertrees inferred under the SPA method and ASTRAL placed eukaryotes within the Asgard archaea as the sister lineage to the three Heimdallarchaeota metagenome bins (Supplementary Figures 9-10), while the SR2008 supertree recovered eukaryotes and Asgard archaea as monophyletic sister lineages (Supplementary Figure 10). To compare these supertrees independently of their models and assumptions, we calculated the summed quartet distances between the set of input trees and each supertree: that is, the total number of quartets (subtrees of four leaves) that differ between the input trees and each supertree (Table 1). The tree with the best score by this metric was the SPA supertree which, like the model-based ASTRAL analysis, recovered Heimdallarchaeota and eukaryotes as sister taxa. These results suggest that there is a congruent genome-wide signal for a specific relationship between eukaryotes and the Heimdallarchaeota, and that the 2D tree does not appear to be an artifact of concatenation.
Table 1. Summed quartet distances between the supertrees produced by several methods and the set of 3199 input trees.
All trees recover a clade of eukaryotes and Asgard archaea; in addition, the SPA and ASTRAL trees place eukaryotes within Asgard archaea, as the sister group to the Heimdallarchaeota.
| Supertree method | Summed quartet distance | Asgard-eukaryote relationship |
|---|---|---|
| SR2008 | 17287838 | Sister groups |
| MSC (ASTRAL) | 17213379 | Eukaryotes with Heimdallarchaeota (0.28 quadripartition support) |
| SPA | 17195042 | Eukaryotes with Heimdallarchaeota (BPP 1.0t) |
Is there support from protein folds for a root between prokaryotes and eukaryotes?
Debates about the 2D and 3D trees have typically assumed that the root of the tree lies on the branch separating bacteria and archaea67–69 or within the bacteria70–72. Recently, a non-stationary model of binary character evolution (the KVR73 model) was used31,33 to infer a rooted tree of life from a matrix of protein fold presence/absence data. Fold presence and absence were quantified by searching HMMs corresponding to Structural Classification of Proteins (SCOP) families against a set of bacterial, archaeal and eukaryotic genomes. The inferred trees are intrinsically rooted because the model is non-stationary: in this model there is one composition (probability of protein fold presence) at the root of the tree, and a second composition elsewhere. These analyses recovered a root between prokaryotes and eukaryotes31,33, suggesting this is the primary division within cellular life and rejecting both the 2D and 3D trees.
We performed simulations to evaluate the ability of the KVR model to recover the root of the tree from protein fold datasets. When data were simulated under the KVR model, the method recovered the true root of the simulation tree as might be expected. However, when protein fold compositions were allowed to vary over the tree, something which is observed in the empirical data31,33, the model fails to find the true root. Under these conditions, KVR finds a root on one of the branches with atypical sequence composition (see Supplementary Text). In the empirical data matrix, the eukaryotes encode significantly more protein folds than either bacteria or archaea (median of 871 folds per eukaryotic genome, compared to 521 for archaea and 615 for bacteria; P < 10-8 for the eukaryote-archaea and eukaryote-bacteria comparisons, P = 0.000278 comparing bacteria and archaea; n = 47 eukaryotes, 47 bacteria and 47 archaea, Wilcoxon rank-sum tests), but their higher compositions are in the minority because the matrix contains an equal number of genomes from each of the three domains. Thus, the inferred root between prokaryotes and eukaryotes may result from the model’s bias in placing the root on a branch with atypical composition; in simulations, the root inference can be controlled by varying which composition among tips - high or low - is in the majority (Supplementary Text). These results agree with recent work72,74 in suggesting that non-reversible models may provide reliable rooting information when the assumptions of the model are met, but that root inferences are sensitive to model misspecification. The KVR model is only one of the many possible non-stationary and non-homogeneous models, and does not appear to be well-suited to these data. Models that better describe the process by which fold (or sequence) compositions change through time, and across the tree --- or indeed those that make use of other sources of time information75,76 --- may perform better for rooting deep phylogenies. How best to root ancient radiations remains an open question, and method development is still at an early stage. A key challenge will be the development of methods that account for the heterogeneity of the evolutionary process across the data and through evolutionary time (that is, across the branches of the tree).
A potentially bigger problem than model misspecification for the published analyses31,33 is their assumption that the entire protein fold set evolves on a single underlying tree. This assumption is unlikely to be realistic because of the different histories generated by widespread horizontal gene transfer and, in eukaryotes, by endosymbiotic gene transfer from the bacterial progenitors of mitochondria and plastids77. The assumption of a single underlying tree to explain fold distributions also means that, despite claims to the contrary31, the published analyses cannot be used to reject the 2D tree because, as generally formulated5,16,78, it seeks to explain the inheritance of only a subset of the genes on cellular genomes.
To evaluate whether the protein folds in the published matrix31,33 share a common evolutionary tree, we inferred single-gene phylogenies for each fold (Supplementary Text). Although weakly supported, these trees are consistent with there being extensive disagreement between single fold-based topologies: only 22 of the protein folds supported the monophyly of eukaryotes, and none recovered all three domains as potentially monophyletic groups, even though this was the consensus topology obtained from analysis of the complete matrix. The trees contained signals for sister-group relationships between eukaryotes and Alphaproteobacteria (the most frequent sister-group among the protein folds shared between eukaryotes and bacteria) and for a relationship between eukaryotes and the TACKL archaea. These analyses are consistent with endosymbiotic theory2,79 and the ideas that underpin the 2D tree, namely that eukaryotes contain a mixture of genes from the archaeal host cell and the bacterial endosymbiont that became the mitochondrion2,3,5 (Supplemental Text).
Conclusions
Identifying the tree that best depicts the relationships between the major groups of life is important for understanding eukaryotic origins and the evolution of the complexity that distinguishes eukaryotic cells. It has recently been asserted that the tree recovered depends upon the species investigated and the choice and quality of the molecular data analysed22,23. In the present study we have investigated the data sets used to underpin these claims and find no compelling evidence to support them. Analyses using better-fitting phylogenetic models consistently recovered a 2D tree5,10,12,16,17,19,20 wherein eukaryotes are most closely related to members of the recently discovered Asgard archaea. These results are also supported by additional analyses of expanded concatenations and increased species sampling, and from large-scale genome-wide data sets analysed using supertree and coalescence methods.
We also investigated support from analyses of whole-genome protein folds for a rooted universal tree in which the deepest division is between prokaryotes and eukaryotes. Taken at face-value this tree would reject the 2D and 3D trees that are the focus of robust discussion in the current literature24,25. However, while protein structure is a useful guide to identifying homology when primary sequence similarity is weak, how best to analyse fold data to resolve deep phylogenetic relationships is still not clear. Published analyses31 do not account for the varied evolutionary histories of individual folds due to endosymbiosis and gene transfer, and our simulations suggest that root inference under existing models is unreliable and affected by variation in the abundance and distribution of folds across genomes. At present, the best supported root is on the branch separating bacteria and archaea67,68,80,81 or among the bacteria70,72, and the hypothesis that eukaryotes are younger than prokaryotes is supported by a range of phylogenetic, cell biological2,3 and palaeontological61,82–84 evidence.
Our analyses and published trees5,10,20 imply that the eukaryotic nuclear lineage evolved from within the Archaea. They provide robust phylogenomic support for a clade of eukaryotes and Asgard archaea, and identify the Heimdallarchaeota as the best candidate among sampled lineages19,20,85 for a sister group to eukaryotes. This sister group relationship will no doubt change with further sampling of the potentially vast archaeal diversity in nature still to be discovered. The prize will be ever more reliable inferences of the features that were in place in the last common ancestor of both groups and an improved evidence-based understanding of the building blocks that underpinned the transition from prokaryotic to eukaryotic cells.
Methods
Sequences and alignment
For the reanalyses of the Da Cunha et al. and Spang et al. datasets, alignments were obtained from the supplementary material of Da Cunha et al.22, and the EF2 gene removed according to the coordinates provided; the alignments from Spang et al. (2015) were generously provided by the authors. OMA 2.1.158 was used to identify putative single-copy orthologues among a dataset of 92 eukaryotic, archaeal and bacterial genomes. For putative orthologues present in at least half of the sampled species, single gene trees were inferred for each candidate under the LG+G4+F model in IQ-Tree, and the trees were manually inspected to filter out eukaryotic genes that were acquired from the mitochondrial or plastid endosymbionts. We also performed a BLASTP screen to identify organellar genes that might have been missed via the tree inspection approach. This procedure resulted in a set of 43 single-copy orthologues shared between archaea and eukaryotes, and 21 genes shared among all three domains, that were used for concatenation-based phylogenomic analyses. For all OMA gene families found in at least four species, we used a BLASTP-based screen to identify and filter out eukaryotic gene families of bacterial origin, resulting in 3261 gene families in four or more species that are either eukaryote-specific inventions, or shared between eukaryotes and archaea. For the comparisons of core gene sets, an iterative process of manual comparisons, similarity searches and tree building was used to identify common and distinct markers in the published sets, identify seed sequences for each marker in the genomes of Dictyostelium discoideum, Sulfolobus solfataricus and Escherichia coli K12, and build HMMs for each marker using the existing datasets. We used domain-specific HMM searches in HMMER386 followed by the reciprocal best hit criterion against our domain-specific reference genomes to identify candidate orthologues, followed by gene tree inference and manual curation to assemble final marker sets. Sequences were aligned using the L-INS-i mode in Mafft 787, and poorly aligning regions identified and removed using the BLOSUM30 model in BMGE 1.1288.
Phylogenetics
Maximum likelihood analyses were performed using IQ-Tree 1.6.242, and bootstrap supports were computed using UFBoot289, except where indicated in the main text. Model fitting was carried out using the MFP mode in IQ-Tree, adding the empirical site profile models (C20-C60) to the default candidate model set. Bayesian phylogenies were inferred under the CAT+GTR+G4 model in PhyloBayes-MPI 1.847, using the bpcomp and tracecomp programs to monitor convergence of two MCMC chains for each analysis. Posterior predictive simulations were performed using readpb_mpi in PhyloBayes. Tests for across-branch compositional heterogeneity were performed in p462: we inferred maximum-likelihood gene trees for each of the 35 genes in the concatenation, then simulated data for each gene under the LG+G4+F model. A Chi-square statistic reflecting compositional heterogeneity was calculated on the original and simulated datasets, and the values from the simulated data were used as a null distribution with which to evaluate the test statistic from the original data.
Supertrees
Supertrees were inferred from the maximum likelihood phylogenies for each single gene, with substitution models chosen as described above. MRP, SR2008 and SPA supertrees were inferred using p465. Multispecies coalescent trees were inferred using ASTRAL-III66.
Supplementary Material
Supplementary information
Acknowledgements
TAW is supported by a Royal Society University Research Fellowship and NERC grant NE/P00251X/1. GJSz received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 714774 as well from the grant GINOP-2.3.2.-15-2016-00057. PGF received funding from NERC grant NE/M015831/1. CJC received Portuguese national funds from Foundation for Science and Technology (FCT) through project UID/Multi/04326/2019 and the Portuguese node of ELIXIR, specifically BIODATA.PT ALG-01-0145-FEDER-022231. We thank Gareth Coleman for assistance with Figure 2.
Footnotes
Author contributions: All authors contributed to the conception and design of the project, and interpretation of results. TAW, CJC, PGF and GJSz performed analyses. TAW and TME wrote the manuscript, with input from all authors.
Competing interests statement: The authors declare they have no competing interests.
References
- 1.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 2.Martin WF, Garg S, Zimorski V. Endosymbiotic theories for eukaryote origin. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2014.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roger AJ, Muñoz-Gómez SA, Kamikawa R. The Origin and Diversification of Mitochondria. Curr Biol. 2017;27:R1177–R1192. doi: 10.1016/j.cub.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 4.Martijn J, Ettema TJG. From archaeon to eukaryote: the evolutionary dark ages of the eukaryotic cell. Biochem Soc Trans. 2013;41:451–457. doi: 10.1042/BST20120292. [DOI] [PubMed] [Google Scholar]
- 5.Williams Ta, Foster PG, Cox CJ, Embley TM. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504:231–236. doi: 10.1038/nature12779. [DOI] [PubMed] [Google Scholar]
- 6.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurland CG, Collins LJ, Penny D. Genomics and the irreducible nature of eukaryote cells. Science. 2006;312:1011–1014. doi: 10.1126/science.1121674. [DOI] [PubMed] [Google Scholar]
- 8.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tourasse NJ, Gouy M. Accounting for evolutionary rate variation among sequence sites consistently changes universal phylogenies deduced from rRNA and protein-coding genes. Mol Phylogenet Evol. 1999;13:159–168. doi: 10.1006/mpev.1999.0675. [DOI] [PubMed] [Google Scholar]
- 10.Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci USA. 2008;105:20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster PG, Cox CJ, Embley TM. The primary divisions of life: a phylogenomic approach employing composition-heterogeneous methods. Philos Trans R Soc Lond B Biol Sci. 2009;364:2197–2207. doi: 10.1098/rstb.2009.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raymann K, Brochier-Armanet C, Gribaldo S. The two-domain tree of life is linked to a new root for the Archaea. Proceedings of the National Academy of Sciences. 2015 doi: 10.1073/pnas.1420858112. 201420858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guy L, Ettema TJG. The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 2011;19:580–587. doi: 10.1016/j.tim.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Williams Ta, Foster PG, Nye TMW, Cox CJ, Embley TM. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc Biol Sci. 2012;279:4870–4879. doi: 10.1098/rspb.2012.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hug LA, et al. A new view of the tree of life. Nat Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.48. 16048. [DOI] [PubMed] [Google Scholar]
- 16.Lake Ja, Henderson E, Oakes M, Clark MW. Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci USA. 1984;81:3786–3790. doi: 10.1073/pnas.81.12.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eme L, Spang A, Lombard J, Stairs CW, Ettema TJG. Archaea and the origin of eukaryotes. Nat Rev Microbiol. 2017;15 doi: 10.1038/nrmicro.2017.133. nrmicro.2017.133. [DOI] [PubMed] [Google Scholar]
- 18.Williams TA, Embley TM, Williams TA, Embley TM. Changing ideas about eukaryotic origins. Philos Trans R Soc Lond B Biol Sci. 2015 doi: 10.1098/rstb.2014.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spang A, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015:173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaremba-Niedzwiedzka K, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 21.Hartman H, Fedorov A. The origin of the eukaryotic cell: a genomic investigation. Proc Natl Acad Sci USA. 2002;99:1420–1425. doi: 10.1073/pnas.032658599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Da Cunha V, Gaia M, Gadelle D, Nasir A, Forterre P. Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet. 2017;13:e1006810. doi: 10.1371/journal.pgen.1006810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaia M, Da Cunha V, Forterre P. The Tree of Life. In: Rampelotto PH, editor. Molecular Mechanisms of Microbial Evolution. Springer International Publishing; 2018. pp. 55–99. [Google Scholar]
- 24.Da Cunha V, Gaia M, Nasir A, Forterre P. Asgard archaea do not close the debate about the universal tree of life topology. PLoS genetics. 2018;14:e1007215. doi: 10.1371/journal.pgen.1007215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spang A, et al. Asgard archaea are the closest prokaryotic relatives of eukaryotes. PLoS genetics. 2018;14:e1007080. doi: 10.1371/journal.pgen.1007080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirt RP, et al. Microsporidia are related to Fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc Natl Acad Sci USA. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 2007;7(Suppl 1):S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–193. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- 29.Nasir A, Kim KM, Da Cunha V, Caetano-Anollés G. Arguments Reinforcing the Three-Domain View of Diversified Cellular Life. Archaea. 2016;2016 doi: 10.1155/2016/1851865. 1851865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penny D, McComish BJ, Charleston Ma, Hendy MD. Mathematical elegance with biochemical realism: the covarion model of molecular evolution. J Mol Evol. 2001;53:711–723. doi: 10.1007/s002390010258. [DOI] [PubMed] [Google Scholar]
- 31.Harish A, Kurland CG. Empirical genome evolution models root the tree of life. Biochimie. 2017;138:137–155. doi: 10.1016/j.biochi.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 32.Philippe H, Forterre P. The rooting of the universal tree of life is not reliable. J Mol Evol. 1999;49:509–523. doi: 10.1007/pl00006573. [DOI] [PubMed] [Google Scholar]
- 33.Harish A, Kurland CG. Akaryotes and Eukaryotes are independent descendants of a universal common ancestor. Biochimie. 2017;138:168–183. doi: 10.1016/j.biochi.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 34.Yang S, Doolittle RF, Bourne PE. Phylogeny determined by protein domain content. Proc Natl Acad Sci U S A. 2005;102:373–378. doi: 10.1073/pnas.0408810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caetano-Anolles G. An Evolutionarily Structured Universe of Protein Architecture. Genome Research. 2003;13:1563–1571. doi: 10.1101/gr.1161903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayr E. Two empires or three? Proc Natl Acad Sci U S A. 1998;95:9720–9723. doi: 10.1073/pnas.95.17.9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narrowe AB, et al. Complex evolutionary history of translation Elongation Factor 2 and diphthamide biosynthesis in Archaea and parabasalids. bioRxiv. 2018 doi: 10.1101/262600. 262600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brochier C, Forterre P, Gribaldo S. Archaeal phylogeny based on proteins of the transcription and translation machineries: tackling the Methanopyrus kandleri paradox. Genome Biol. 2004;5:R17. doi: 10.1186/gb-2004-5-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brochier C, Gribaldo S, Zivanovic Y, Confalonieri F, Forterre P. Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales? Genome Biol. 2005;6:R42. doi: 10.1186/gb-2005-6-5-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 41.Guindon S, et al. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 45.Foster P. Modeling Compositional Heterogeneity. Syst Biol. 2004;53:485–495. doi: 10.1080/10635150490445779. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Y, Brinkmann H, Rodrigue N, Lartillot N, Philippe H. A dirichlet process covarion mixture model and its assessments using posterior predictive discrepancy tests. Mol Biol Evol. 2010;27:371–384. doi: 10.1093/molbev/msp248. [DOI] [PubMed] [Google Scholar]
- 47.Lartillot NL, Odrigue NIR, Tubbs DAS, Icher JAR. PhyloBayes MPI : Phylogenetic Reconstruction with Infinite Mixtures of Profiles in a Parallel Environment. 2013;62:611–615. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
- 48.Bollback JP. Bayesian model adequacy and choice in phylogenetics. Mol Biol Evol. 2002;19:1171–1180. doi: 10.1093/oxfordjournals.molbev.a004175. [DOI] [PubMed] [Google Scholar]
- 49.Susko E, Roger AJ. On reduced amino acid alphabets for phylogenetic inference. Mol Biol Evol. 2007;24:2139–2150. doi: 10.1093/molbev/msm144. [DOI] [PubMed] [Google Scholar]
- 50.Hrdy I, et al. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- 51.Whelan S. Spatial and temporal heterogeneity in nucleotide sequence evolution. Mol Biol Evol. 2008;25:1683–1694. doi: 10.1093/molbev/msn119. [DOI] [PubMed] [Google Scholar]
- 52.Gouy R, Baurain D, Philippe H. Rooting the tree of life: the phylogenetic jury is still out. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2014.0329. 20140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crotty SM, et al. GHOST: Recovering Historical Signal from Heterotachously-evolved Sequence Alignments. bioRxiv. 2019 doi: 10.1101/174789. 174789. [DOI] [PubMed] [Google Scholar]
- 54.Graybeal A. Is it better to add taxa or characters to a difficult phylogenetic problem? Syst Biol. 1998;47:9–17. doi: 10.1080/106351598260996. [DOI] [PubMed] [Google Scholar]
- 55.Hedtke SM, Townsend TM, Hillis DM. Resolution of phylogenetic conflict in large data sets by increased taxon sampling. Syst Biol. 2006;55:522–529. doi: 10.1080/10635150600697358. [DOI] [PubMed] [Google Scholar]
- 56.Castelle CJ, Banfield JF. Major New Microbial Groups Expand Diversity and Alter our Understanding of the Tree of Life. Cell. 2018;172:1181–1197. doi: 10.1016/j.cell.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 57.Parks DH, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 58.Roth ACJ, Gonnet GH, Dessimoz C. Algorithm of OMA for large-scale orthology inference. BMC Bioinformatics. 2008;9:518. doi: 10.1186/1471-2105-9-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Altenhoff AM, et al. Standardized benchmarking in the quest for orthologs. Nat Methods. 2016;13:425–430. doi: 10.1038/nmeth.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams Ta, Embley TM. Archaeal ‘dark matter’ and the origin of eukaryotes. Genome Biol Evol. 2014;6:474–481. doi: 10.1093/gbe/evu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Betts HC, et al. Integrated genomic and fossil evidence illuminates life’s early evolution and eukaryote origin. Nat Ecol Evol. 2018;2:1556–1562. doi: 10.1038/s41559-018-0644-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roch S, Steel M. Likelihood-based tree reconstruction on a concatenation of aligned sequence data sets can be statistically inconsistent. Theor Popul Biol. 2015;100C:56–62. doi: 10.1016/j.tpb.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 63.Roch S, Nute M, Warnow T. Long-Branch Attraction in Species Tree Estimation: Inconsistency of Partitioned Likelihood and Topology-Based Summary Methods. Syst Biol. 2019;68:281–297. doi: 10.1093/sysbio/syy061. [DOI] [PubMed] [Google Scholar]
- 64.Steel M, Rodrigo A. Maximum likelihood supertrees. Syst Biol. 2008;57:243–250. doi: 10.1080/10635150802033014. [DOI] [PubMed] [Google Scholar]
- 65.Akanni WA, Wilkinson M, Creevey CJ, Foster PG, Pisani D. Implementing and testing Bayesian and maximum-likelihood supertree methods in phylogenetics. R Soc Open Sci. 2015;2 doi: 10.1098/rsos.140436. 140436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang C, Sayyari E, Mirarab S. Comparative Genomics. Springer, cham; 2017. ASTRAL-III: Increased Scalability and Impacts of Contracting Low Support Branches; pp. 53–75. [Google Scholar]
- 67.Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci U S A. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gogarten JP, et al. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proc Natl Acad Sci U S A. 1989;86:6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fournier GP, Gogarten JP. Rooting the ribosomal tree of life. Mol Biol Evol. 2010;27:1792–1801. doi: 10.1093/molbev/msq057. [DOI] [PubMed] [Google Scholar]
- 70.Lake Ja, Skophammer RG, Herbold CW, Servin Ja. Genome beginnings: rooting the tree of life. Philos Trans R Soc Lond B Biol Sci. 2009;364:2177–2185. doi: 10.1098/rstb.2009.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cavalier-Smith T. Rooting the tree of life by transition analyses. Biol Direct. 2006;1:19. doi: 10.1186/1745-6150-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams TA, et al. New substitution models for rooting phylogenetic trees. Philos Trans R Soc Lond B Biol Sci. 2015 doi: 10.1098/rstb.2014.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klopfstein S, Vilhelmsen L, Ronquist F. A Nonstationary Markov Model Detects Directional Evolution in Hymenopteran Morphology. Syst Biol. 2015;64:1089–1103. doi: 10.1093/sysbio/syv052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cherlin S, et al. The effect of non-reversibility on inferring rooted phylogenies. Molecular Biology and Evolution. 2018;35:984–1002. doi: 10.1093/molbev/msx294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tria FDK, Landan G, Dagan T. Phylogenetic rooting using minimal ancestor deviation. Nature Ecology & Evolution. 2017;1 doi: 10.1038/s41559-017-0193. s41559–017–0193. [DOI] [PubMed] [Google Scholar]
- 76.Szöllõsi GJ, Rosikiewicz W, Boussau B, Tannier E, Daubin V. Efficient exploration of the space of reconciled gene trees. Syst Biol. 2013;62:901–912. doi: 10.1093/sysbio/syt054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Timmis JN, Ayliffe Ma, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 78.McInerney JO, O’Connell MJ, Pisani D. The hybrid nature of the Eukaryota and a consilient view of life on Earth. Nat Rev Microbiol. 2014;12:449–455. doi: 10.1038/nrmicro3271. [DOI] [PubMed] [Google Scholar]
- 79.Gray MW, Doolittle WF. Has the endosymbiont hypothesis been proven? Microbiol Rev. 1982;46:1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brown JR, Doolittle WF. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci U S A. 1995;92:2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhaxybayeva O, Lapierre P, Gogarten JP. Ancient gene duplications and the root(s) of the tree of life. Protoplasma. 2005;227:53–64. doi: 10.1007/s00709-005-0135-1. [DOI] [PubMed] [Google Scholar]
- 82.Knoll AH. Paleobiological perspectives on early eukaryotic evolution. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Butterfield NJ. Early evolution of the Eukaryota. Palaeontology. 2015;58:5–17. [Google Scholar]
- 84.Parfrey LW, Lahr DJG, Knoll AH, Katz La. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Spang A, et al. Proposal of the reverse flow model for the origin of the eukaryotic cell based on comparative analyses of Asgard archaeal metabolism. Nature Microbiology. 2019 doi: 10.1038/s41564-019-0406-9. [DOI] [PubMed] [Google Scholar]
- 86.Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Williams TA, et al. Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc Natl Acad Sci U S A. 2017;114:E4602–E4611. doi: 10.1073/pnas.1618463114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Williams, et al. Data from: Phylogenomics provides robust support for a two-domains tree of life. Figshare. doi: 10.6084/m9.figshare.8950859.v2. fileset. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information