Prochlorococcus, a Marine Photosynthetic Prokaryote of Global Significance (original) (raw)
Abstract
The minute photosynthetic prokaryote Prochlorococcus, which was discovered about 10 years ago, has proven exceptional from several standpoints. Its tiny size (0.5 to 0.7 μm in diameter) makes it the smallest known photosynthetic organism. Its ubiquity within the 40°S to 40°N latitudinal band of oceans and its occurrence at high density from the surface down to depths of 200 m make it presumably the most abundant photosynthetic organism on Earth. Prochlorococcus typically divides once a day in the subsurface layer of oligotrophic areas, where it dominates the photosynthetic biomass. It also possesses a remarkable pigment complement which includes divinyl derivatives of chlorophyll a (Chl a) and Chl b, the so-called Chl a2 and Chl b2, and, in some strains, small amounts of a new type of phycoerythrin. Phylogenetically, Prochlorococcus has also proven fascinating. Recent studies suggest that it evolved from an ancestral cyanobacterium by reducing its cell and genome sizes and by recruiting a protein originally synthesized under conditions of iron depletion to build a reduced antenna system as a replacement for large phycobilisomes. Environmental constraints clearly played a predominant role in Prochlorococcus evolution. Its tiny size is an advantage for its adaptation to nutrient-deprived environments. Furthermore, genetically distinct ecotypes, with different antenna systems and ecophysiological characteristics, are present at depth and in surface waters. This vertical species variation has allowed Prochlorococcus to adapt to the natural light gradient occurring in the upper layer of oceans. The present review critically assesses the basic knowledge acquired about Prochlorococcus both in the ocean and in the laboratory.
The large body of work achieved since the discovery about 10 years ago of the minute and ubiquitous photosynthetic prokaryote Prochlorococcus (20, 21) has changed our view of the community structure of oceanic picoplankton. It also has implications in fields as different as the phylogeny of cyanobacteria and photosynthesis. Prochlorococcus has proven exceptional from several standpoints.
(i) The tiny size of Prochlorococcus (equivalent spherical diameter in culture, 0.5 to 0.7 μm [110]) makes it the smallest known photosynthetic organism, having the lowest predictable size for an O2 evolver (136). The discovery and first field studies of this organism were made possible only by the use of sensitive flow cytometers onboard research vessels (21, 88, 116). Since then, the development of procedures to fix and preserve picoplanktonic cells has allowed cells to be transported to the laboratory for analysis (104, 173). As a result, the number of studies of picoplankton including Prochlorococcus has steadily increased. The ubiquity of this organism within the 40°S to 40°N latitudinal band, its high density, and its occupation of a 100- to 200-m-deep layer make it the most abundant photosynthetic organism in the ocean and presumably on Earth.
(ii) Prochlorococcus possesses a remarkable pigment complement, which includes divinyl derivatives of chlorophyll a (Chl a) and Chl b, the so-called Chl _a_2 and Chl _b_2 (41), that are unique to this genus. Recent developments of high-performance liquid chromatography (HPLC) (42) and spectrofluorimetric techniques (114) have made it possible to identify these pigments in natural assemblages and therefore to assess precisely the contribution of Prochlorococcus to the total planktonic photosynthetic biomass. Another peculiarity of its photosynthetic apparatus is the presence of light-harvesting complexes which are similar in function but not in structure to those of higher plants or green algae (77). In some strains, there are also small amounts of a particular type of phycoerythrin (55). The combination of Chl a, Chl b, and at least one phycobiliprotein is a unique trait among oxygen-evolving phototrophs.
(iii) In subtropical oligotrophic areas of the Atlantic and Pacific Oceans, the vertical distribution of Prochlorococcus often exceeds the boundaries of the euphotic layer (i.e., the part of the water column extending from the surface to the depth that receives 1% of the surface irradiance). Thus, cells of this genus seem to be able to sustain growth and photosynthesis over an irradiance range extending for more than 3 orders of magnitude. This raises the question whether natural Prochlorococcus populations exhibit an outstanding ability for photoacclimation or whether other processes such as the occurrence of different Prochlorococcus species or pigment types along the vertical light gradient are implicated in this intriguing phenomenon. Recent biochemical studies (132) suggest that different Prochlorococcus strains may have different antenna systems specifically adapted to the light environment from which they have been isolated.
(iv) Prochlorococcus typically divides once a day in the subsurface layer of oligotrophic areas, such as the central oceanic gyres (94), where it dominates the photosynthetic biomass (16). In these environments, where nutrients such as nitrogen are extremely scarce, Prochlorococcus has an obvious advantage with respect to uptake because of its high surface/volume ratio resulting from its very small cell size (19). However, it must also possess either unusually small nutrient requirements (at least compared to other phytoplankton) or an extremely efficient capacity to scavenge elements recycled by heterotrophic bacteria.
(v) The remarkable diversity of Prochlorococcus observed both among laboratory isolates (145, 166) and within field populations (124) raises questions about the role of genetic variability in the success of Prochlorococcus in the field. One may wonder how environmental constraints, such as the absence of iron in oligotrophic areas, conditioned the evolution of Prochlorococcus over geologic timescales from the ancestor it shares with the cyanobacteria.
This review critically assesses the basic knowledge acquired about Prochlorococcus both in the ocean and in the laboratory since its recent discovery. It clearly indicates that some research areas, such as the ecological distribution, photosynthetic capacities, and phylogeny of Prochlorococcus, are well ahead of others, in particular nutrient acquisition, which need to be developed.
CHARACTERIZATION AND CULTIVATION
Methods of Characterization
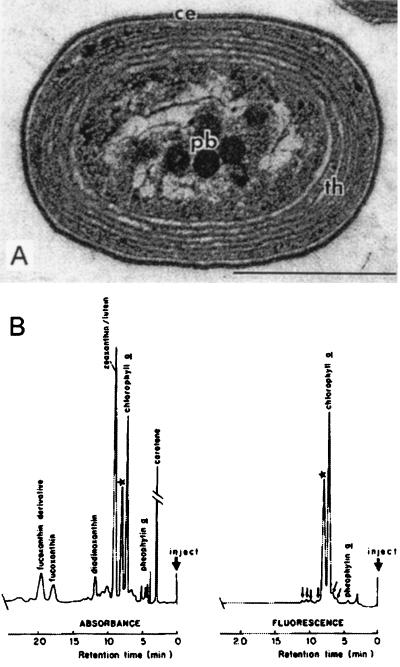
The first published records of Prochlorococcus in nature are the electron microscopy sections of “type II” cells from marine samples reported in the paper by Johnson and Sieburth (63). They revealed what is now known as the typical ultrastructure of Prochlorococcus (Fig. 1A). Although these authors clearly suggested that the cells they observed were probably devoid of phycoerythrin (63), Guillard et al. mistakenly identified them as Synechococcus (47), the other coccoid photosynthetic prokaryote discovered in abundance in marine waters around that time (184). The second published historical record came indirectly in 1983 from the discovery of an unknown “red-shifted” Chl a derivative in subtropical Atlantic waters (37) (Fig. 1B). In fact, years later, Gieskes (36) revealed that in 1977 he had found both red-shifted Chl a and Chl b in these waters by using thin layer chromatography. Prochlorococcus was clearly identified only once flow cytometry was used (21). During a cruise out of Barbados in 1985, Rob Olson and Ginger Armbrust were the first to visualize in a deep sample a new population of red-fluorescing particles, dimmer than Synechococcus (117). The finding that these cells possessed an unusual divinyl derivative of Chl a and the visualization of their ultrastructure led to the announcement in 1988 of the discovery of Prochlorococcus by Chisholm et al. (21). By then, other workers had began to record Prochlorococcus either by flow cytometry or by epifluorescence microscopy (88, 116).
FIG. 1.
First records of the occurrence of Prochlorococcus. (A) Electron microscope photograph of “type II cells” from deep samples of the North Atlantic ocean. Reprinted from reference 63 with permission of the publisher. ce, cell envelope; pb, polyhedral bodies; th, thylakoids. Scale bar, 0.5 μm. (B) Analysis of a pigment extract obtained by normal-phase HPLC showing the “unknown Chl a derivative,” indicated by a star. Reprinted from reference 37 with permission of the publisher.
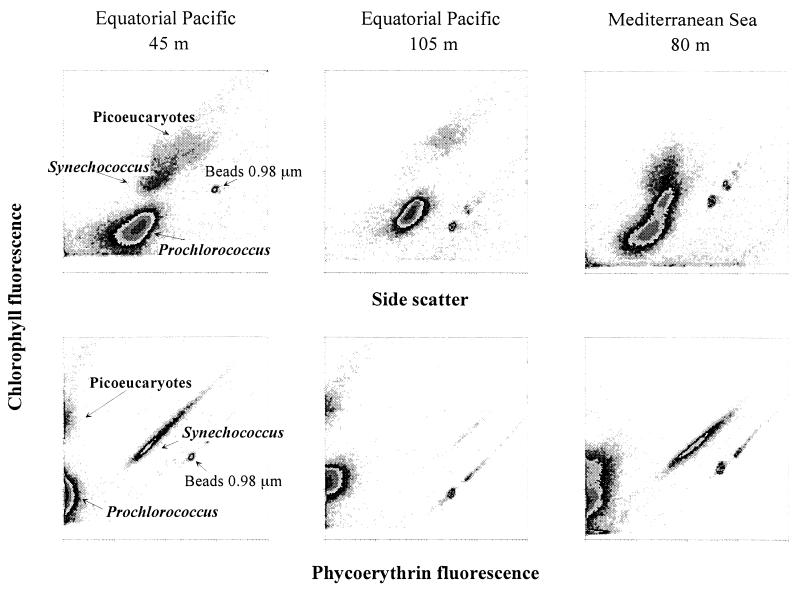
Since the late 1980s, many observations on Prochlorococcus in natural waters have been made. Most of these observations were obtained by flow cytometry on either live (Fig. 2) or preserved samples (99, 129, 173). One disadvantage of most commercially available flow cytometers is their inability to completely resolve surface populations from background noise in very oligotrophic surface waters, due to the very low Chl fluorescence of surface Prochlorococcus. This remains a serious problem for water column studies in areas where Prochlorococcus dominates the phytoplankton community. Two possible solutions to this problem have been put forward. The first requires the use of very high laser power (above 1 W) and modified optics on old instruments equipped with water-cooled lasers (119). Such bulky instruments are not very convenient for field work. The second possibility involves modifying the optics of the small instruments equipped with air-cooled lasers to focus laser beam more narrowly and increase the excitation power (28).
FIG. 2.
Analysis of natural Prochlorococcus populations by flow cytometry. (Top) Side scatter (a function of cell size) plotted against red fluorescence of chlorophyll. (Bottom) Orange fluorescence of phycoerythrin plotted against red fluorescence of chlorophyll. All scales are 4-decades logarithmic from 1 to 10,000 (arbitrary units). (Left) Typical surface-layer sample (45 m, deep Equatorial Pacific, 7°S, 150°W, collected 10 November 94). Synechococcus is easily distinguished from Prochlorococcus by its orange phycoerythrin fluorescence. (Middle) Typical deep sample (105 m deep, Equatorial Pacific, 7°S, 150°W, collected 10 November 94). Synechococcus is virtually absent, and the chlorophyll fluorescence of Prochlorococcus is much higher than near the surface (compare the fluorescence of Prochlorococcus and that of the standard beads). Note also the weak orange fluorescence displayed by Prochlorococcus at this depth. (Right) Example of two Prochlorococcus populations coexisting at the same depth (80 m deep, Mediterranean Sea, 37°5′N, 16°52′E, collected 20 June 1996).
Alternative methods to quantify the Prochlorococcus cell population in the field include epifluorescence microscopy, electron microscopy, and pigment analysis. Although it has been claimed that Prochlorococcus abundance can be recorded by ordinary epifluorescence microscopy (60, 61, 88, 92), sophisticated image recording with, for example, a cooled charge-coupled device camera (158) is necessary to avoid severe underestimates of cell abundance, especially in surface waters, where the fluorescence is very low. For example, concentrations of Prochlorococcus in the Central Pacific estimated by epifluorescence microscopy (60) are twofold lower than those estimated by flow cytometry in the same region. Epifluorescence microscopy can, however, provide direct estimates of cell size, a key characteristic of oceanic biomass budgets (158). Because of its technical difficulty, electron microscopy has very seldom been used with marine samples (1, 21, 63). Finally, pigment analysis can be used to routinely detect Prochlorococcus in the field. Although it has been possible to discriminate Chl a2 from its monovinyl counterpart by direct-phase HPLC analyses for quite a long time (37), a major step forward came with the ability to separate these pigments by reverse-phase HPLC, the most commonly used oceanographic HPLC technique, which also allows reliable separation of Chl _b_1 and _b_2 (see, e.g., references 3, 42, 170, and 182). Moreover, HPLC analysis offers the ability to measure the growth rate of Prochlorococcus by monitoring the incorporation of 14C into Chl a2 (see “Growth rates and loss processes in the ocean” below). Spectrofluorometry, a technique that is hardly ever used in oceanography, offers a very attractive alternative to detect divinyl Chls, especially because of its sensitivity, speed, and ease of implementation (114).
Cultivation
Available cultures and isolation methods.
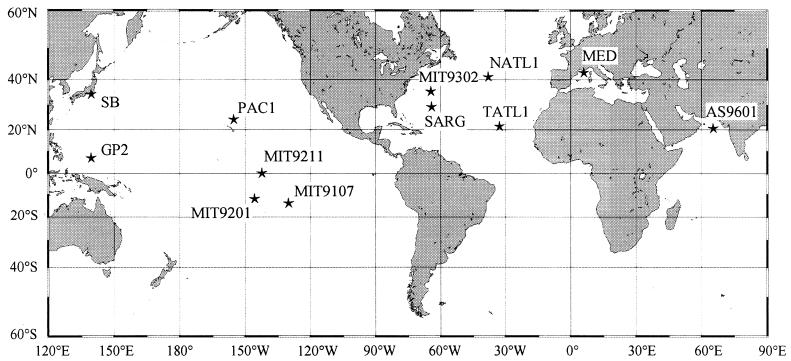
The first Prochlorococcus strain was isolated by Palenik in May 1988 from the bottom of the euphotic zone in the Sargasso Sea (depth, 120 m). It was initially dubbed LG (125), for “Little Greens,” but was then renamed SARG for its geographical origin (131). Since then, several strains have been isolated both at the surface and at depth from various sites in oceans around the world (Table 1 and Fig. 3). Strains are now available from most areas where Prochlorococcus populations have been sampled.
TABLE 1.
Prochlorococcus strains isolated in culture for which published data are available
| Strain | Derived clone | Other name | Origin | Datea | Latitude | Longitude | Depth (m) | Isolator | Remarks | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|---|
| SARG | LG | Sargasso Sea | 30/5/88 | 28°59′N | 64°21′W | 120 | B. Palenik | First strain to be isolated | 131 | |
| SS120 | CCMP 1375 | L. R. Moore | Obtained by serial dilution | 20, 107 | ||||||
| PCC 9511 | R. Rippka | First axenic strain | 143 | |||||||
| MED | DV | NW Mediterranean | 8/1/89 | 43°12′N | 6°52′E | 5 | D. Vaulot, F. Partensky | 131 | ||
| MED4 | CCMP 1378 | L. R. Moore | Obtained by serial dilution | 20, 107 | ||||||
| NATL1 | FP12 | N Atlantic | 1/4/90 | 37°39′N | 40°1′W | 30 | F. Partensky | 131 | ||
| NATL2 | FP5 | N Atlantic | 1/4/90 | 38°59′N | 40°33′W | 30 | F. Partensky | 145, 166 | ||
| MIT9107 | S Pacific | 8/8/91 | 14°60′S | 134°60′W | 25 | J. A. Dusenberry | 166 | |||
| TATL 1 | EUM11 | E Tropical Atlantic | 19/10/91 | 20°57′N | 31°6′W | 20 | F. Partensky | 145, 166 | ||
| TATL 2 | EUM17 | E Tropical Atlantic | 21/10/91 | 20°25′N | 31°8′W | 30 | F. Partensky | 145, 166 | ||
| GP2 | W Pacific | 10/9/92 | 8°32′N | 136°31′E | 150 | A. Shimada | 155, 156 | |||
| SB | Sugura Bay, Japan | 21/10/92 | 35°0′N | 138°30′E | 40 | A. Shimada | 156, 157 | |||
| PAC 1 | N Tropical Pacific | 1/4/92 | 22°45′N | 158°0′W | 100 | L. Campbell | 145, 166 | |||
| MIT9201 | S Pacific | 26/9/92 | 11°60′S | 145°25′W | 0 | B. Binder | 106 | |||
| MIT9202 | S Pacific | 26/9/92 | 11°60′S | 145°25′W | 79 | B. Binder | 106 | |||
| MIT9211 | Equatorial Pacific | 10/4/92 | 0 | 140°W | 83 | R. J. Olson | 106 | |||
| MIT9215 | Equatorial Pacific | 3/9/92 | 0 | 140°W | 0 | B. Binder | 106 | |||
| MIT9302 | Sargasso Sea | 15/7/93 | 34°45′N | 66°11′W | 100 | L. Moore | Sorted by flow cytometry | 108 | ||
| MIT9303 | Sargasso Sea | 15/7/93 | 34°45′N | 66°11′W | 100 | L. Moore | Sorted by flow cytometry | 108, 166 | ||
| MIT9312 | N Atlantic | 17/7/93 | 37°30′N | 68°14′W | 135 | L. Moore | Sorted by flow cytometry | 108 | ||
| MIT9313 | N Atlantic | 17/7/93 | 37°30′N | 68°14′W | 135 | L. Moore | Sorted by flow cytometry | 108 | ||
| AS9601 | Arabian Sea | 1/11/95 | 19°12′N | 67°10′E | 50 | R. J. Olson | 150 |
FIG. 3.
Locations of some of the available Prochlorococcus strains (see Table 1).
For most of the strains reported in Table 1, samples were collected and processed by trace-metal clean techniques (30), although some other strains (e.g., MED) were obtained without special precautions. Isolation steps usually consist of a gentle filtration through two stacked 0.6-μm-pore-size filters followed by enrichment of the filtrate with sterile stock solutions of phosphate, nitrogen, and chelated trace metals (20, 105). Another elegant way of obtaining isolates is flow cytometric cell sorting, which can be directly applied to natural samples. For example, this technique has been used to obtain separate strains from subpopulations coexisting at the same depth in the Gulf Stream and the Sargasso Sea (108).
Optimal growth medium.
Chisholm et al. (20) showed that among the various media initially tested, seawater enriched with urea, β-glycerophosphate, a minimum trace metal mix, and 100 μM CPTC (cis,cis,cis,_cis_-1,2,3,4-cyclopentanetetracarboxylic acid), a chelator, led to sustained growth of Prochlorococcus (Table 2). However, Prochlorococcus can also be adapted to a seawater-based medium containing only inorganic additions (20). To date, the most widely used culture media (PC, PRO2, and modified K/10-Cu [Table 2]) are derived from the K medium used for marine microalgae (69), with EDTA as the chelator, a 10-fold-diluted trace metal stock solution, and no copper (Table 2). The PRO2 medium proved to be very efficient for isolation purposes (105). Other media, such as PC with trace metals as in f/20 (48) or PCR-S11 (142) (Table 2), which use a modified “Gaffron” metal stock solution (141) have also been successful in tests for culturing. Maximum cell yields with these media are 2 × 108 to 3 × 108 cells ml−1, corresponding to a Chl _a_2 yield of ca. 0.2 to 0.4 mg liter−1.
TABLE 2.
Composition of media used for growing Prochlorococcusa
| Component | Concn of component inb: | ||||
|---|---|---|---|---|---|
| CPTC-based (A) | K/10-Cu (B) | PC (C) | PRO2 (D) | PCR-S11 (E) | |
| Nutrients | |||||
| Urea | 20 μM | 50 μM | 100 μM | ||
| NH4ClB,C,D or (NH4)2SO4E | 50 μM | 50 μM | 50 μM | 400 μM | |
| β-glycerol-phosphateA,C (or NaH2PO4)B,D,E | 10 μM | 10 μM | 10 μM | 10 μM | 50 μM |
| Buffer | |||||
| HEPES | 1 mM | ||||
| Chelator/trace metals | |||||
| CPTC | 100 μM | ||||
| EDTA-Na2 | 11.7 μM | 11.7 μM | 1.2 μM | 8 μM | |
| FeSO4A or FeCl3B,C,D,E | 0.1 μM | 1.2 μM | 1.2 μM | 1.2 μM | 8 μM |
| MnCl2A,B,C,D or MnSO4E | 10 nM | 90 nM | 90 nM | 90 nM | 30 nM |
| ZnCl2B,C,D or ZnSO4E | 8 nM | 8 nM | 8 nM | 3 nM | |
| CoCl2B,C,D [or Co(NO3)2]E | 5 nM | 5 nM | 5 nM | 1.5 nM | |
| Na2MoO4A,B,C,D or (NH4)6 Mo7O24E | 10 nM | 3 nM | 3 nM | 3 nM | 1.5 nM Mo |
| Na2SeO3B,C,D or SeO2E | 10 nM | 10 nM | 10 nM | 1.5 nM | |
| NiSO4 or NiCl2D,E | 10 nM | 10 nM | 1.5 nM | ||
| Na2WO4 | 0.3 nM | ||||
| KBr | 3 nM | ||||
| KI | 1.5 nM | ||||
| Cd(NO3)2 | 1.5 nM | ||||
| CuSO4 | 1.5 nM | ||||
| Cr(NO3)3 | 0.3 nM | ||||
| VOSO4 | 0.3 nM | ||||
| KaI(SO4)2 | 3 nM | ||||
| H3BO3 | 150 nM | ||||
| Vitamins | |||||
| Thiamine-HCl | 10 μM | ||||
| Biotin | 50 nM | ||||
| B12 | 50 nM | 7 nM |
Growth on solid medium, despite repeated attempts, has not been successful to date, restraining the possible use of Prochlorococcus for genetic manipulations such as site-directed mutagenesis. This is one of the most critical bottlenecks for future research on Prochlorococcus, and it is the focus of much investigation. At present, cloning is possible only by using extinction serial dilutions, leading to “statistical” clones. Clones SS120 and MED4 (Table 2) have been obtained in this way. The same technique, combined with centrifugation to eliminate contaminant heterotrophic bacteria larger than Prochlorococcus, has been used to isolate the first axenic strain of Prochlorococcus, PCC 9511 (143).
PHYSIOLOGY
Basic Cellular Features
Ultrastructure.
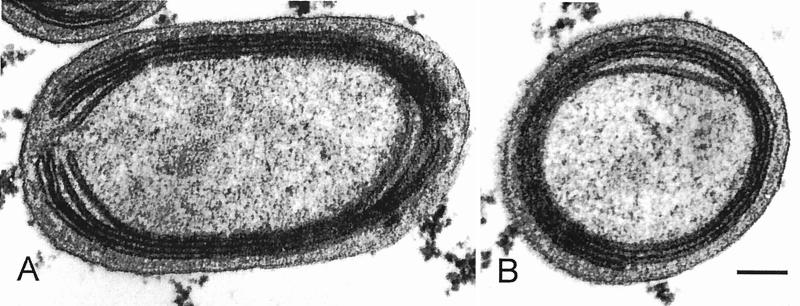
Because of their tiny size, Prochlorococcus cells are very difficult to identify by optical microscopy. They are hardly distinguishable from small heterotrophic bacteria, except for their very weak Chl fluorescence. Transmission electron microscopy reveals typical cyanobacterial architecture (Fig. 4), which is best compared to that of the other abundant oceanic photosynthetic prokaryote, Synechococcus. However, the Prochlorococcus cell is distinctly elongated (Fig. 4A) whereas the Synechococcus cell is much more spherical (see 21, 63, 158). In some cases, the membrane appears fairly electron dense, but it is not known whether this is a consequence of environmental conditions or if it is strain specific. The cytoplasm contains DNA fibrils, carboxysomes that can be labeled with an antibody against ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco), and glycogen granules, located near or between thylakoids (90). In cross-sections, there are generally between two and four thylakoids, and there are sometimes up to six (157). They run parallel to the cell membrane (Fig. 4) and are much more appressed than in Synechococcus. In Prochlorococcus marinus type strain (SARG) and in most cells from natural populations, they are closed. In contrast, in the surface isolate Prochlorococcus sp. strain MED (90) and in the deep isolate MIT 9313 (Fig. 4A), they are horseshoe shaped. Interestingly, Johnson and Sieburth (63) described a “type III” cell with noncircular thylakoids that looks somewhat like MED. No phycobilisomes are visible, but since phycoerythrin has been found in strains such as CCMP 1375 (55), immunocytochemistry would be useful to investigate its intracellular localization.
FIG. 4.
Electron micrographs of longitudinal and cross sections of Prochlorococcus strain MIT9313 showing tightly appressed thylakoids at the periphery of the cell. Scale bar, 0.1 μm. Unpublished photographs courtesy of C. Ting, J. King, and S. W. Chisholm.
Size and carbon content.
Most methods used for cell sizing, such as electronic (Coulter) sizing and optical and electronic microscopy, exhibit a number of biases for such tiny objects as Prochlorococcus cells. The best estimates made on cultures give a range of 0.5 to 0.8 μm for length and 0.4 to 0.6 μm for width (90, 110). The cell size appears to vary with environmental conditions. For example, it was shown to increase from 0.45 to 0.75 μm between the surface and a depth of 150 m in the Sargasso Sea (158). Moreover, forward scatter measured by flow cytometry, a function of both size and refractive index, increases from dusk to dawn at the equator, a corollary of synchronized cell division (7, 174).
Determination of the carbon content of Prochlorococcus is important to assess its relative contribution to the oceanic biomass (16, 87). A commonly used method is to assume a given cell size and to rely on some universal carbon-to-volume ratio derived from the literature. Depending on the hypotheses made, this yields values ranging between 50 and 60 fg of C cell−1 (8, 16, 87), although higher values have been proposed (124 fg of C cell−1 [181]). The only direct measurements made on cultures (13) provided an average value of 49 fg of C cell−1, surprisingly close to those indirect estimates. The recent availability of an axenic strain (143) should allow us to further refine this value.
Genome size and base composition.
Existing data obtained on the axenic strain PCC 9511 either by DNA renaturation kinetics (143) or by flow cytometry (97) suggest a genome size of 1.9 to 2.0 Mbp. This value is within the size range of genomes of free-living eubacteria, which extends from 1.55 Mbp for Aquifex aeolicus (25) to 4.21 Mbp for Bacillus subtilis (71). Genome sizes have also been estimated for a variety of cyanobacteria (mainly freshwater) by renaturation kinetics (49). These range from 2.55 to 13.2 Mbp (1.66 × 109 to 8.58 × 109 Da (49; Catalogue of the Pasteur Culture Collection at http://www.pasteur.fr). For instance, the only sequenced cyanobacterial genome, Synechocystis strain PCC 6803 (67), is about twice as large (3.6 Mbp) as that of Prochlorococcus whereas Prochlorothrix hollandica has a genome size of 5.5 Mbp (149). Although more genome size data are required for marine cyanobacteria, it seems that Prochlorococcus possesses the smallest genome of all prokaryotes evolving oxygen (or oxyphotobacteria; see “Phylogeny” below). Some of the genetic characteristics of P. marinus SS120, such as the presence of only one copy of psbA (57), overlapping genes in the dihydrodipicolinate synthase operon (95), or overlapping promoter regions (53), are consistent with the finding of a relatively compact genome in this microorganism. The genes sequenced to date in Prochlorococcus are listed in Table 3.
TABLE 3.
Genes sequenced in Prochlorococcus
| Category | Gene name | Molecule encoded | Localization or function | EMBL/GenBank accession no. | Reference(s) |
|---|---|---|---|---|---|
| Photosynthesis | cpeY | Phycoerythrobilin, phycourobilin lyase? | Light harvesting? | AJ001230 | 52 |
| cpeZ | Phycoerythrobilin, phycourobilin lyase? | Light harvesting? | AJ001230 | 52 | |
| mpeX | Bile pigment biosynthesis, coupling? | Light harvesting? | AJ001230 | 52 | |
| orf463 | Assembly of light-harvesting structures? | Light harvesting? | AJ001230 | 52 | |
| pcb | Chl _a_2 and Chl _b_2 antenna | PS II antenna | U57660, U57661 | 77 | |
| petB | _b_-type cytochrome | Cytochrome _b_6-f complex | AF001487–AF001489 | 166 | |
| petD | Subunit IV | Cytochrome _b_6-f complex | AF001487–AF001489 | 166 | |
| cpeA | Phycoerythrin III α subunit | Light harvesting? | Z68890, AJ001230 | 55 | |
| cpeB | Phycoerythrin III β subunit | Light harvesting? | Z68890, AJ001230 | 55 | |
| ppeC | Linker polypeptide of phycoerythrin | Light harvesting? | Z92525, AJ001230 | 54 | |
| psaA | PsaA | PS I core | 167 | ||
| psaB | PsaB | PS I core | 167 | ||
| psaF | Subunit PsaF of PS I | Binding of plastocyanin? | 167 | ||
| psaI | Subunit PsaI of PS I | Trimerization of PS I | Z98595, AJ002427 | 168 | |
| psaJ | Subunit PsaF of PS I | Binding of plastocyanin? | 167 | ||
| psaL | Subunit PsaL of PS I | Trimerization of PS I | Z98595, AJ002427 | 168 | |
| psbA | D1 | PS II core | Z49201 | 57 | |
| psbB | CP47 | Internal antenna of PS II | AF001481–AF001485 | 166 | |
| rbcL | Large subunit of Rubisco | Carbon assimilation | U93857, U93858 | 52, 133, 155 | |
| General metabolism | aroC | Chorismate synthase | Amino acid metabolism | Z49201 | 51 |
| aspA | Aspartoacylase | Amino acid metabolism | Z80110 | 51 | |
| cpn60 | Chaperonin-60 | Protein folding | Z37730 | 52 | |
| dapA | Dihydrodipicolinate synthase | Amino acid metabolism | Z37733, Z68126 | 95 | |
| rpoC1 | DNA-dependent RNA polymerase | Biosynthesis of RNA | Z11160 | 125 | |
| rnpB | RNase P ribozyme | RNA processing | Y12789, AJ001135 | 53 | |
| rnc | RNase III | RNA processing | AJ001135 | 52 | |
| rrn | 16S rRNA | Protein synthesis | X63140, AF001466– AF001476 | 165, 166 | |
| trnR | Transfer RNA arginine (CCU) | Translation | Y12789, AJ001135 | 53 | |
| Nutrient uptake | pstS | Phosphate-binding protein | Phosphorus assimilation | U75514 | 146 |
| Cell cycle | dnaA | DnaA | DNA replication | U44977 | 139 |
| ftsZ | FtsZ | Cell division | AJ011025 | 58 | |
| uvrD | DNA helicase | DNA replication and DNA repair processes | AJ001230 | 52 |
In contrast to marine Synechococcus cyanobacteria, characterized by a high G+C content (47.4 to 69.5%; [185]), most Prochlorococcus strains have a low G+C content. This was suggested first from the relatively high cell DNA fluorescence when cells were stained with DAPI (4′,6-diamino-2-phenylindole), which is AT specific (9), and then from the analysis of rpoC1 gene sequences of cultivated strains (125). When the whole set of sequences available for P. marinus SS120, the strain which at present has the best-known genome, is examined (Table 4), the global G+C content is 36.82% over an accumulated length of 25,083 nucleotides. This G+C content is comparable to the ones estimated for a few freshwater Synechocystis strains (34.7 to 36.7%), which have the lowest G+C scores among 176 cyanobacteria strains examined by Herdman et al. (50). Since the G+C content of P. marinus is an absolute number based on sequences of both genes and noncoding intergenic regions, it can be compared to the notably higher absolute G+C value of 43% obtained for the total genome of Synechocystis strain PCC 6803 (68). In accordance with its low G+C content, the codon usage of P. marinus SS120 (Table 4) is shifted towards A or T at the third base position (T > A > C > G), suggesting mutational biases as the most likely cause (65, 153, 161). From a practical point of view, it is important to take these factors into account, e.g., when oligonucleotide primers or gene probes are constructed. Noncoding regions are frequently even more enriched in A and T. Lorenz et al. (96) found G+C scores as low as 29% for one noncoding DNA fragment in P. marinus SS120. When individual genes are considered, ppeC has the lowest G+C content (33.7%) whereas the highly expressed psbA gene is less biased, with 44.6% G+C. Similar differences in G+C content in strongly versus weakly expressed genes are known for other bacteria. In Escherichia coli genes, for instance, the codon usage is biased towards higher G+C contents with increasing gene expression (59, 152). The evolutionary forces causing the compositional bias toward a low G+C content in most investigated Prochlorococcus strains are not known. It might be related to higher mutation rates relative to other prokaryotes, including Synechococcus (124), perhaps as a result of the higher sensitivity of AT-rich DNA sequences to UV light-induced oxidative DNA damage (187).
TABLE 4.
Codon usage in P. marinus CCMP 1375 (6,365 codons)a
| Amino acid | First and second positions | No. of occurrences at third position | |||
|---|---|---|---|---|---|
| A | C | G | T | ||
| Ala | GC | 247 | 70 | 34 | 255 |
| Arg | CG | 27 | 16 | 5 | 52 |
| Arg | AG | 121 | 41 | ||
| Asn | AA | 89 | 222 | ||
| Asp | GA | 86 | 259 | ||
| Cys | TG | 21 | 43 | ||
| Gln | CA | 158 | 77 | ||
| Glu | GA | 244 | 101 | ||
| Gly | GG | 152 | 107 | 66 | 167 |
| His | CA | 44 | 131 | ||
| Ile | AT | 145 | 72 | 294 | |
| Leu | TT | 236 | 106 | ||
| Leu | CT | 119 | 36 | 29 | 199 |
| Lys | AA | 239 | 100 | ||
| Phe | TT | 123 | 190 | ||
| Pro | CC | 103 | 22 | 10 | 123 |
| Ser | TC | 124 | 27 | 20 | 150 |
| Ser | AG | 65 | 103 | ||
| Thr | AC | 111 | 65 | 11 | 137 |
| Tyr | TA | 50 | 111 | ||
| Val | GT | 115 | 40 | 36 | 203 |
| Stop | TA | 6 | 4 | ||
| Stop | TG | 6 | |||
| Total | 2,153 | 933 | 640 | 2,639 |
Although it is a salient feature for most available Prochlorococcus strains, a low G+C content is not a general characteristics within this genus. One strain, MIT9303, has a significantly higher G+C content than all other Prochlorococcus strains available in culture (55% G+C at third codon positions [164]). Interestingly, molecular studies with the 16S rRNA gene (166) place MIT9303 as the closest known relative of the high-G+C marine Synechococcus strains, whereas strain MED4, which has a very low G+C content of 35.7% (when considering concatenated sequences for three regions: pcb, rpoC1, and the psbB-petB/D intergenic region) belongs to the most recently evolved clade within Prochlorococcus (see “Genetic diversity” below). Therefore, there may be a correlation between the global G+C content of a particular genotype and its phylogenetic position. A systematic analysis of the G+C content in all isolates should allow us to verify this interesting hypothesis.
Photosynthesis
Pigment composition.
The presence of Chl _a_2 and Chl _b_2 is a trait common to all Prochlorococcus strains characterized to date (41, 107, 131, 166). Besides Prochlorococcus, Chl _a_2 (but not Chl _b_2) has been observed only in a mutant of corn (5), although divinyl derivatives of chlorophyll(ide) are probable intermediates in the biosynthesis of Chl in higher plants (126). Both Chl _a_2 and Chl _b_2 have absorption and fluorescence excitation maxima in the blue part of the visible spectrum red-shifted by 8 to 10 nm compared to their monovinyl counterparts (107, 110). The other pigments of Prochlorococcus, which include zeaxanthin, α-carotene, and small amounts of a Chl _c_-like pigment (Mg,3-8 divinyl phaeoporphyrin a5), are shared with a limited number of other phytoplanktonic groups, including chlorophytes, cryptomonads, and cyanobacteria (41). It is noteworthy that natural populations of Prochlorococcus from suboxic waters of the Arabian Sea also possess a novel 7,8-dihydro derivative of zeaxanthin, probably parasiloxanthin, which is absent in cultured strains (40). One notable peculiarity of Prochlorococcus is the dramatic difference in pigment ratios among different isolates. For example, several isolates, notably SARG, have a Chl _b_2/Chl _a_2 ratio equal to or higher than 1 whereas other isolates display much lower ratios, with the MED strain exhibiting the lowest (0.13 [105–107, 131]). At least three isolates (MIT9302, MIT9312, and SARG and its clonal derivative SS120 [Table 1]) also synthesize normal (monovinyl) Chl b (or Chl _b_1) when grown under high light conditions (105, 107, 131), suggesting that this light condition may trigger the expression of enzymes which can transform, probably in a single step, Chl _b_2 into Chl _b_1 (5, 131, 144). Surprisingly, although these enzymes should also be able to allow the transformation of Chl _a_2 in Chl _a_1, no Chl _a_1 has been detected in Prochlorococcus. Another difference between these strains is the presence in SS120 but not in MED4 of a novel type of phycoerythrin (55). Because of its low concentration and the fact that classical pigment HPLC analyses do not detect phycobiliproteins, this pigment remained undetected for a long time. It also was overlooked by researchers making absorption measurements, because the major phycobilin associated with this phycoerythrin is phycourobilin. Its absorption maximum at 495 nm is very close to that of the very abundant Chl _b_2 (480 nm), and thus they cannot be discriminated by their absorption spectra. It was only after the discovery of the phycoerythrin gene that Hess et al. (55) examined water-soluble fractions by spectrofluorimetry and found evidence of phycobilins.
Photosynthetic performances.
In a study with the MED and SARG isolates (131), the observed ranges of assimilation rates, expressed per Chl unit, at various growth irradiances were similar between these strains (1.5 to 4.8 and 1.4 to 5.6 fg of C fg of Chl−1 h−1 for MED and SARG, respectively), but expressed per cell, they were almost constant for MED (4.9 to 5.8 fg of C cell−1 h−1) and more variable for SARG (2.8 to 6.2 fg of C cell−1 h−1). However, light-saturated carbon fixation rates (P _m_Chl or P _m_cell [m stands for maximum]) were found to vary significantly between strains grown under similar irradiances (131). A more recent study confirmed that differences in pigmentation among isolates correlate with differing photosynthetic efficiencies: when grown at 9 μmol quanta m−2 s−1, two Prochlorococcus isolates (MIT9303 and MIT9313) with high ratios of Chl _b_2 to Chl a_2 (>1.1) had a significantly higher PChl_m than did two other isolates (MIT9302 and MIT9312) with Chl _b_2/Chl _a_2 ratios lower by a factor of 2 (2.4 and 1.8 fg of C fg of Chl−1 h−1) (105). Laboratory values also compare fairly well with data on natural Prochlorococcus populations from the Moroccan upwelling (0.6 to 4 fg of C cell−1 h−1), obtained by labeling a natural seawater sample with 14C and sorting individual Prochlorococcus cells by flow cytometry (82). However, photosynthetic rates measured by the same method for Prochlorococcus cells at the base of the euphotic zone in the open ocean were significantly lower (0.03 to 0.3 fg of C cell−1 h−1). This method also allowed us to estimate the fraction of the total phytoplanktonic production attributable to Prochlorococcus, which varied from 11 to 57% (82).
Some of its photosynthetic properties may give Prochlorococcus a definite selective advantage for growth at depth in oligotrophic areas, particularly compared to Synechococcus. These properties include (i) its absorption characteristics with maxima (for the SARG-like pigment type) in the range from 430 to 490 nm, which are more suitable than those of oceanic Synechococcus for collecting photons in the blue part of the spectrum, the wavelengths which penetrate the deepest in the ocean (107, 131); (ii) its higher photosynthetic yield than Synechococcus at all wavelengths (156); and (iii) its higher probability of absorbing rather than scattering incident photons (in contrast to Synechococcus), especially in the blue part of the spectrum (110).
In contrast, all Prochlorococcus isolates studied to date appear fairly sensitive to high irradiances, such as those available in near surface waters. In particular, curves of photosynthesis versus irradiance (108, 131, 156) saturate at relatively low irradiances (typically around 200 μmol quanta m−2 s−1) and display a strong inhibition at high irradiances, in a way reminiscent of that observed for deep natural populations (82). This is true even for Prochlorococcus strains belonging to the so-called high light clade, such as MED (see “Genetic diversity” below). One may wonder, therefore, whether researchers have been successful at obtaining isolates that are truly representative of populations thriving in the uppermost layers of the ocean.
Photoacclimation versus photoadaptation.
One remarkable characteristic of the distribution of Prochlorococcus is that viable cells can be found over a very thick (150 to over 200 m) layer when the hydrologic conditions are stable enough to allow the water column to stratify, either quasi-permanently, such as in the tropical Atlantic and Pacific Oceans, or seasonally, such as in the Sargasso Sea or the Mediterranean Sea in summer. Over this natural light gradient, ranging from ca. 1,500 μmol quanta m−2 s−1 near the surface to less than 1 μmol quanta m−2 s−1 below 150 m, cells display a variety of differences. The most obvious ones are concomitant increases in cell size (see “Size and carbon content” above) and pigment content, as well as variations of the ratios of accessory pigments to Chl _a_2, which generally occur below the depth of the mixed layer.
Goericke and Repeta (42) were among the first to measure Chl _a_2 and Chl _b_2 simultaneously in field populations and thus to provide direct information on the response of wild Prochlorococcus cells to changes in light intensity. The ratio of Chl _b_2 to Chl _a_2 they measured ranged from 0.15 in the surface layer to 2.9 below the deep Chl maximum. Similar ranges were observed in the Red Sea by using normal phase HPLC (180) and in the tropical Atlantic by using spectrofluorometry (129). In these two studies, Prochlorococcus cell concentrations were also enumerated by flow cytometry, which allowed variations of the content of Prochlorococcus cells in divinyl Chls to be computed. The Chl _b_2 content varied 45-fold from 0.1 to 4.5 fg cell−1, and the Chl _a_2 content varied only 12-fold, from 0.23 to 2.7 fg cell−1. In samples dominated by Prochlorococcus, such large variations in Prochlorococcus divinyl Chl content and in the ratio of Chl _b_2 to Chl _a_2 are also reflected by conspicuous changes in the absorption and fluorescence excitation spectra of total phytoplankton pigments, which at depth display a marked peak at 480 nm, specific of Chl _b_2 (10, 78).
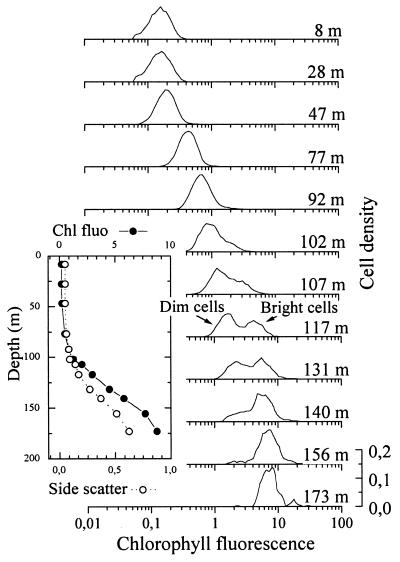
All these variations concern the whole Prochlorococcus population, which is considered homogeneous. In fact, the ranges of size, Chl _a_2 or Chl _b_2 content per cell, ratio of Chl _b_2 to Chl _a_2, or red Chl fluorescence determined by flow cytometry measured in the field largely exceed those measured for individual Prochlorococcus isolates (43, 107, 131). A close look at flow cytometric red fluorescence histograms obtained from field samples reveals that several populations may be present over the vertical light gradient (Fig. 5). Bimodal red-fluorescence distributions of Prochlorococcus have very often been observed around the deep chlorophyll maximum layer in oligotrophic areas (7, 8, 17, 102, 121, 129). In such bimodal distributions, the fluorescence of the dim population is ca. 2 to 3 times lower than that of the bright population. Only the dim population seems to occur in the upper layer, while the bright population is the sole representative below the deep chlorophyll maximum. Each population, considered separately, does exhibit photoacclimation, in the physiological sense, as suggested by the increase of its modal red fluorescence (and scatter) with depth (Fig. 5). However, the bright and dim populations seem to be genetically distinct and have significantly different irradiance optima for growth. This hypothesis is supported by the fact that these populations also have genome sizes that differ by 14%, as determined by flow cytometry (17). Recently, Reckermann and Veldhuis (137) showed that if a sample with two populations is incubated at 20 m deep, only the dim population, i.e., the one that is acclimated to the prevailing light at 20 m, can grow, while the bright one rapidly disappears, probably consumed by grazers. Finally, using flow cytometric cell sorting, Moore et al. (108) were able to sort cooccurring bright and dim populations from natural samples at two sites and grow coisolates separately. Even after 2 years of culture, these coisolates maintained their physiological distinctness. Isolates with high red fluorescence had a higher ratio of Chl _b_2 to Chl _a_2 and were adapted to growth and photosynthesis at lower light levels relative to the dim populations. In addition, the high- and low-red-fluorescence coisolates were more than 2% different in their 16S rRNA sequence (see “Genetic diversity” below), confirming that genetically and physiologically different populations of Prochlorococcus can coexist. The relative differences in the light-dependent physiological parameters between the high- and low-red-fluorescence coisolates are similar to the differences observed between SS120 and MED4, leading to the hypothesis that Prochlorococcus isolates can be distinguished as low- or high-light-adapted ecotypes (108). The low-light-adapted isolates (SS120-like) have ratios of Chl _b_2 to Chl _a_2 that are 2- to 10-fold higher than those of the high-light-adapted isolates (MED4-like) over all irradiances (105). Furthermore, these two ecotypes display shifted optimal growth irradiances. Culture studies have also shown that no single isolate of Prochlorococcus is able to thrive over a range greater than 2.5 orders of magnitude of light irradiance (107). Thus, the ability of wild Prochlorococcus strains to stand a very wide range of light conditions in the field apparently results from both physiological and genetic diversity among species or groups within this genus.
FIG. 5.
Vertical distributions from bright and dim Prochlorococcus populations in the subtropical Pacific Ocean off Hawaii. The insert represents the vertical profile of side scatter and red chlorophyll fluorescence of the total Prochlorococcus population measured by flow cytometry. Adapted from reference 17 with permission of the publisher.
Photosynthetic apparatus.
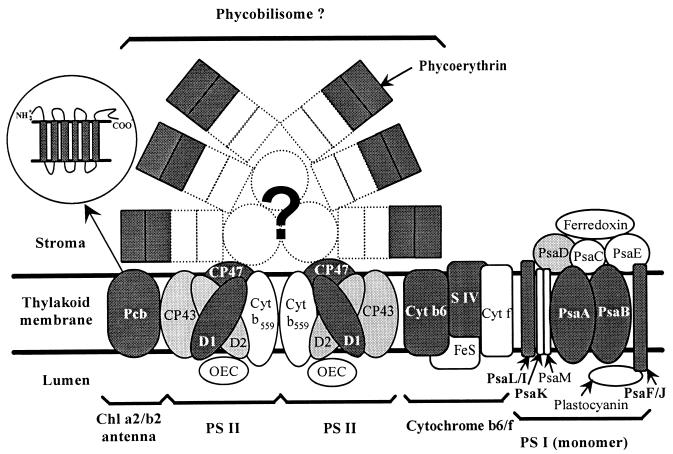
One basic question raised by the observation of different Prochlorococcus pigment types both in the field (17, 129) and in culture (107, 110, 131) is whether strains or populations representative of those found near the surface and those found at the bottom of the euphotic zone have not only different pigmentation and ecophysiological features but also structurally different photosynthetic apparatuses (Fig. 6). This question was addressed through the biochemical characterization of the pigment complexes of clones MED4 and SS120 (132). Since these strains differed mainly in their ratios of Chl _b_2 to Chl _a_2 (even at a given growth irradiance; see “Pigment composition” above) and since the majority of Chl b is located in antenna systems (46), the most striking structural difference between strains was expected to be found at the level of their major light-harvesting complexes. Proteins constituting these antenna complexes have apparent molecular masses on denaturing electrophoresis gels of 32.5 and 34 to 38 kDa in MED4 and SS120, respectively. Moreover, they are more abundant (at low light intensities), their relative amount varies more with irradiance, and they bind ca. 7 times as much Chl _b_2 in SS120 as in MED4 (132). Sequencing of the pcb genes, which encode these antenna proteins, confirmed the large differences between strains, since polypeptides deduced from gene sequences have only 76% identity at the amino acid level (77). These polypeptides are, however, similar between MED4 and SS120 in length (352 and 351 amino acids, respectively), mass (44.9 and 44.6 kDa, respectively), and the presence of six putative transmembrane helices (insert in Fig. 6), suggesting that the discrepancy noticed between their apparent molecular masses resulted from posttranslational modifications, e.g., different levels of phosphorylation, affecting their migration properties in denaturing gels (132). The antenna proteins of Prochlorococcus (Pcb), as well as those of the two other “prochlorophytes” (Prochlorothrix and Prochloron; see “Phylogeny” below), are closely related to the IsiA proteins found in iron-stressed freshwater cyanobacteria, such as Synechococcus strain PCC 7942, and are probably derived from them (77). All these proteins belong to the same family of Chl-binding proteins as the psbC and the psbB gene products (CP43 and CP47), which are major constituents of the photosystem II (PS II) internal antenna in all oxygenic photosynthesizing organisms (77). Despite the progress made recently in the identification of “prochlorophyte” antenna proteins, major questions still remain to be answered. The presence in both Prochlorothrix and Prochloron of several pcb genes (77, 169) raises the question whether some or all Prochlorococcus strains might not also possess multiple pcb genes, since these genes could play an important role in the photoacclimation capacity of these organisms. Other important topics are the exact localization of Chl-binding residues within the Pcb amino acid sequences and their total Chl-binding capacity per molecule. Furthermore, state transitions, a characteristic of photosynthesis in many cyanobacteria (see reference 147 and references therein), and the possible role of Pcb-antenna complexes in this phenomenon, if it takes place in Prochlorococcus, might be specifically targeted in future studies.
FIG. 6.
Diagram showing the thylakoid proteins in Prochlorococcus and their putative organization by homology to the photosynthetic apparatus of cyanobacteria. The proteins whose genes have been sequenced partially or totally are shown in dark gray (see Table 3), and those which have been characterized only by immunoblotting (35, 90, 132) are shown in light gray. Although phycoerythrin is present in some strains such as SS120, it is not clear whether it is integrated in phycobilisomes, which, if present, would be very scarce. PS I is probably organized in trimers (35), but only one monomer is shown. The insert shows a detail of the Pcb protein which includes six transmembrane hydrophobic domains. For the electron transport chain, only the cytochrome b6-f complex is shown because the existence of other components (such as NADH dehydrogenase and plastoquinone) has not been demonstrated yet. Other probable components of the photosynthetic apparatus such as cytochrome oxidase or ATP synthase are not shown either, since they are still uncharacterized in Prochlorococcus. Abbreviations: CP, chlorophyll-protein complex; Cyt, cytochrome; OEC, oxygen evolving complex.
Although in both Prochlorococcus strains, most Chl _b_2 is found associated with these major antenna complexes, a significant quantity of this pigment is also found in PS I fractions (35). This suggests that Chl _b_2 may be associated either with some minor PS I-specific Chl _a_2-Chl _b_2-protein complexes or with the PS I core itself. Both hypotheses imply an unusual organization of the Prochlorococcus PS I with regard to other phototrophs since (i) cyanobacteria do not have any PS I-specific antenna and (ii) the PS I core of all photosynthetic organisms studied to date does not include any Chl b. Additionally, two proteins found in purified PS I fractions have unusual apparent molecular masses of 21 and 25 kDa, whereas no PS I proteins with such masses are known in cyanobacteria (35). Sequence data show that these proteins are related to PsaF and PsaL, respectively. Their anomalous sizes, compared to the sizes of the equivalent molecules known in other phototrophs, are caused by an insertion in the central part of the molecule in PsaF (167) and an N-terminal extension in PsaL (168). As in most cyanobacteria, the _psaL_-like gene is found in an operon with the psaI gene, which is located upstream of psaL, and the two genes are cotranscribed (168). In cyanobacteria, PsaL-PsaI complexes are known to be implicated in the formation or stabilization of PS I trimers (44), and the presence of such trimers in Prochlorococcus (35) suggests that despite its anomalous length, the PsaL-like protein of Prochlorococcus and its PsaI-like companion must have maintained similar functions. Knockout mutants of Synechococcus strain PCC 7002 with psaL mutation revealed that transitions from state 2 to state 1 (resulting from excess excitation of PS II and PS I, respectively) proceeded approximately three times more rapidly than in the wild type, possibly linked to the missing ability to form PS I trimers and the consequently enhanced mobility of PS I particles in the mutant (147). An interference of the N-terminal extension of Prochlorococcus PsaL with similar processes would be intriguing. The function of the PsaF-like protein, which in cyanobacteria also makes complexes with another protein, PsaJ, is even less clear so far. It has been suggested that the PsaF-PsaJ complex is active in docking negatively charged donors such as plastocyanin (44). Further genetic studies are required to know whether these modifications in PS I proteins have some consequences in the structural organization and function of this photosystem.
Despite their originality, the PS I components appear fairly similar between SS120 and MED4. However, there is one striking difference between the two Prochlorococcus pigment types (beside their antenna proteins). Hess et al. (55) found a functional operon encoding the two subunits (α and β) of a phycoerythrin in SS120. Attempts to detect phycoerythrin genes in MED4 by hybridization and PCR did not provide any evidence for their presence in this strain (55). With a homologous antiserum to the SS120 phycoerythrin β subunit, it was not possible to detect even traces of this protein in MED4 or PCC 9511, whereas the same serum showed a strong reaction with total proteins from SS120 (56). The phycoerythrin found in SS120 was characterized as “type III” (55), based on major differences with regard to type I, which is widespread in cyanobacteria, and type II, which is present only in some marine Synechococcus (122). Characteristics of type III phycoerythrin include the presence of a single chromophore-binding site in the α subunit (as opposed to two or three in other phycoerythrin α subunits) and the absence of a few amino acids in the central part of the molecule (55). Future comparative and functional studies are required to elucidate whether these modifications play a functional role. Preliminary immunocytochemical data suggest an association of phycoerythrin with thylakoid membranes in SS120 (56). However, whether this phycoerythrin is included in structures comparable to the phycobilisomes of cyanobacteria (Fig. 6) or is present in the thylakoid lumen as in cryptophytes (32) is not known. In this context, it is noteworthy that the newly discovered Chl _d_-containing prokaryote Acaryochloris marina, a species similar in many aspects to Prochloron sp. (see “Phylogeny” below), contains phycobiliprotein aggregates that are not organized in the form of phycobilisomes (100, 103). The phycoerythrin genes in SS120 are part of a larger gene cluster. The latter includes ppeC, a gene encoding a putative gamma phycoerythrin-like protein which may serve as linker polypeptide (54), and two genes homologous to cpeY and cpeZ (56). These two genes are thought to encode lyases involved in chromophore attachment to the α or β phycoerythrin subunits in Fremyella diplosiphon (66), and a similar role may be attributed to their gene products in Prochlorococcus. The presence of at least one linker polypeptide and two putative lyases in SS120 suggests that its phycoerythrin is chromophorylated and may participate in light harvesting. In contrast, this finding, together with the relatively low concentration of phycoerythrin in the cell and its probable intracellular localization within the thylakoids, constitute evidence against a role for Prochlorococcus phycoerythrins as nitrogen storage molecules, a function which was proposed for marine nitrogen-replete Synechococcus (190). Future studies are clearly needed to clarify the functional relevance of Prochlorococcus phycoerythrin.
Most of the other photosynthetic genes investigated so far in one or several Prochlorococcus strains (Table 1) show few unique characteristics compared to known cyanobacterial, algal, or plant models. However, the psbA gene raises some interesting questions. It is present as a single copy in both the Prochlorococcus strains SS120 (57) and MED4 genomes (145). This contrasts with other cyanobacteria, such as Synechococcus strain PCC 7942, which generally possess two isoforms of the D1 protein, D1:1 and D1:2, that are differentially regulated by light (45, 123). D1:1 (encoded by psbAI) is the only type detectable in the thylakoid membrane at low light intensities. Upon a shift to high light intensities, the psbAII/III genes encoding the D1:2 form are induced, the psbAI mRNA is actively degraded, and D1:2 is substituted for D1:1 in the PS II reaction center (70). However, following this transient photoinhibition phase, the process reverses and D1:1 dominates again. Thus, D1:1 corresponds to long-term photoacclimation whereas D1:2 is expressed only transiently. The latter form provides a higher photochemical efficiency to PS II, thus dissipating excess energy via photochemical quenching and preventing photodamage to the photosynthetic machinery (123, 160). The two Synechococcus strain PCC 7942 D1 isoforms differ by only 25 amino acids. Among the few internal differences, residue 130 is a prime candidate for being responsible for the respective photochemical properties of each D1 type (38): D1:1 has a glutamine at this site, whereas D1:2 has a glutamate. Because of its single psbA gene copy, Prochlorococcus cannot use a similar regulation mechanism. The D1 protein of Prochlorococcus strain SS120 is phylogenetically closer to D1:1 of Synechococcus strain PCC 7942 than to D1:2 (57), and a glutamine is present at position 130 as in D1:1. In steady-state cultures of strains SS120 and MED4 acclimated to different irradiances, psbA transcript levels are proportional to light irradiances (34). They increase when cultures are shifted from low to high light intensities, more quickly in MED4 than in SS120, and decrease during the opposite shift. Thus, the D1 protein from both strains seems to have a mode of regulation closer (but not identical) to that of D1:1. Although the highly developed antenna system of Prochlorococcus strain SS120 is particularly efficient at harvesting low photon fluxes (132), it may become detrimental in the setting of abrupt light changes, because it conveys excess energy to the reaction center that cannot be completely dissipated by the turnover of D1 because its transcription is too slow.
Nutrient Assimilation
One of the most intriguing ecological characteristics of Prochlorococcus, besides its capacity to grow over a very wide range of irradiances in nature, is its ability to colonize extremely oligotrophic areas. For example, Prochlorococcus represents on average 73% of the photosynthetic carbon in the surface mixed layer off Hawaii (16). Under these conditions, its very small cell size and the resulting high surface-to-volume ratio are obvious adaptative advantages for nutrient uptake (19). However, very little is known about its basic physiological characteristics with respect to nutrient assimilation, partly because of the unavailability of axenic cultures, a gap only recently filled (143).
Most oceanic areas are assumed to be limited by nitrogen. In tropical areas, Prochlorococcus is present both in the surface layer, where presumably only reduced nitrogen forms are available, and in the deep chlorophyll maximum, where nitrates are present. However, in the Mediterranean Sea in winter, NO3− addition was able to stimulate Prochlorococcus cell cycling (177), suggesting that some populations might be able to take up oxidized forms of nitrogen. These data raise the possibility that there are different physiological types of Prochlorococcus adapted to grow on different nutrient sources, a hypothesis consistent with the available information for marine Synechococus (185) and with the existence of genetically different Prochlorococcus (see “Genetic diversity” below). Nutrient uptake experiments must be performed with the axenic strain now available (143) and other forthcoming axenic strains more representative of deep ecotypes. It will also be very interesting to investigate whether all Prochlorococcus strains possess the components necessary for NO3− uptake, such as the ntcA gene, whose product is involved in the activation of transcription of a set of genes required for the use of oxidized nitrogen sources (91).
Although nitrogen is the primary limiting nutrient in many oceanic areas, recent evidence points to phosphorus as limiting, either permanently, in areas such as the Mediterranean Sea, or transiently, in areas such as the Sargasso Sea (23, 162). Prochlorococcus is well represented in both areas, suggesting that it also has the ability to thrive under very low P concentrations. Parpais et al. (127) showed that Prochlorococcus becomes limited only at P-PO43− concentrations of the order of 30 nM. Although the initial medium used to isolate Prochlorococcus contained organic instead of mineral phosphorus (see “Optimal growth medium” above), Prochlorococcus can grow very well on the latter form. In fact, organic phosphorus is rapidly mineralized by the heterotrophic bacteria present in the culture (127). It is possible that a similar mechanism occurs in situ and provides natural Prochlorococcus populations with mineral P, which is consumed so quickly that it remains undetectable. It is worth noting that Prochlorococcus possesses a pstS gene encoding a phosphate-binding protein, that is expressed only under P depletion and may play a role in its adaptation to oligotrophy; however, this gene is not unique to this organism (146).
Iron is the third major nutrient recently implicated in the limitation of primary productivity in remote areas of the ocean not subjected to eolian inputs, such as the equatorial Pacific (101), as well as in more coastal areas (76). The low iron requirement of Prochlorococcus could be a key factor in its success in central oceanic areas. This is suggested in particular by the fact that the Prochlorococcus PS II antenna is similar to IsiA, a protein induced under iron stress in Synechococcus sp. strain PCC 7942 (77). However, field experiments suggest that natural populations of Prochlorococcus are somewhat iron limited (although much less so than larger cells such as diatoms), since the addition of iron induces increases in cell size and chlorophyll fluorescence (192). Clearly, future laboratory experiments should aim at a better understanding of the regulation of cellular processes by iron in Prochlorococcus and should search for the eventual presence of siderophores, as evidenced previously in a variety of marine and freshwater cyanobacteria (189).
Cell Cycle
Although little studied in photosynthetic prokaryotes, cell cycling, a key cellular process that coordinates growth, DNA replication, and cell division, has received some attention in Prochlorococcus because of its application to assessment of growth rate and population status with respect to nutrient limitation in the field (see “Growth rates and loss processes in the ocean” below). The Prochlorococcus cell cycle can be easily studied by flow cytometry after staining with a DNA-binding dye (98, 104). The Prochlorococcus cell cycle resembles that of eukaryotes, with a discrete DNA synthesis phase (S phase) and two well-defined G1 and G2 phases, in contrast to some strains of marine Synechococcus, which may have more than two genome copies (6). As in most phytoplankton species (171), light or nutrient deprivation arrests cells in the G1 phase of the cell cycle. A notable exception occurs when phosphorus is depleted. In this case, cells are blocked in all phases of the cell cycle, including the DNA synthesis S phase. Cells arrested in S are not able to recover upon addition of fresh phosphorus, in contrast to cells arrested in G1 and G2 (127). Light-dark entrainment induces a very strong synchronization of the cell cycle both in cultures (150) and in nature (94, 129, 175). The S phase usually takes place in late afternoon, and division occurs during the early part of the night. Natural Prochlorococcus populations from the Arabian Sea, as well as two cultured strains, can divide more than once per photocycle while remaining highly synchronized (150). In such cases, two cohorts of dividing cells occur in rapid succession, the second with a G1 duration as short as 1 h. It would be interesting to investigate whether Prochlorococcus can display behaviors typical of circadian clock control (e.g., free running in continuous light), as demonstrated for Synechococcus strain PCC 7942 (62). Indeed, S-phase initiation appears to be linked to a light-triggered timer (150).
The only gene related to the cell cycle that has fully been characterized in Prochlorococcus so far is dnaA. In most bacteria, the encoded protein, DnaA, is essential for initiating chromosomal replication recognizing short, asymmetric sequence elements, the DnaA boxes, near the origin of replication, oriC (159). In P. marinus CCMP 1375 (139), DnaA consists of 461 amino acids and has 63% identity in a 315-residue overlap to Synechocystis strain PCC 6803 DnaA (140). Among the more than 35 bacterial DnaA sequences available in data banks, those of Prochlorococcus and Synechocystis form a distinguishable subfamily. The C-terminal DNA-binding domain of the Prochlorococcus recombinant DnaA protein expressed in vitro recognizes and binds specifically the heterologous oriC from Bacillus subtilis and E. coli, suggesting that the primary structure of the DnaA boxes might be evolutionarily conserved in Prochlorococcus. In many bacteria, oriC is physically near the dnaA gene and DnaA boxes are frequently present in its promoter region, providing a basis for binding and autoregulation (159). Surprisingly, no putative DnaA box could be identified in the analyzed genomic region of 5,304 bp containing Prochlorococcus dnaA (139). However, this is consistent with a unique gene organization in Prochlorococcus, which is very different from the highly conserved gene order, rnpA-rpmH-dnaA-recF-gyrB, found in a wide variety of bacteria. It is worth noting that the cyanobacterium Synechocystis strain PCC 6803 also lacks a DnaA-binding box near dnaA and displays an unusual gene arrangement in this region (140). Surprisingly, knockout mutants of Synechocystis strain PCC 6803 grow as well as the wild type, suggesting that DnaA is not essential for this strain, in contrast to all known eubacteria (138).
ECOLOGY
Oceanic Distribution
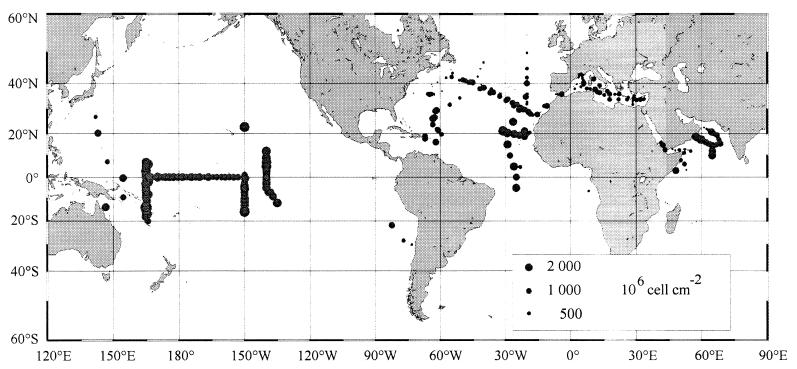
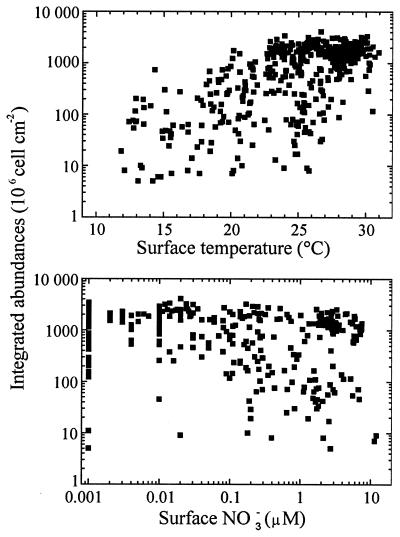
Prochlorococcus appears to have a very wide oceanic distribution. This is shown in Fig. 7, which is based on the analysis of more than 8,400 field measurements of Prochlorococcus made throughout the world oceans by flow cytometry (Table 5). Prochlorococcus is virtually ubiquitous in the latitudinal band extending from 40°N to 40°S (Fig. 7). It can still be found beyond 40°, but its concentrations decline fairly rapidly. The highest latitude where it has been recorded is 60°N off Iceland in the North Atlantic (11). This oceanic distribution suggests that low temperatures are lethal to Prochlorococcus, and this is confirmed by culture data (107). In the field, the lowest surface temperature at which Prochlorococcus is recorded is about 10°C (Fig. 8A). This contrasts strongly with that for Synechococcus, the other major marine unicellular cyanobacterium, which can be encountered, albeit at low concentrations, at temperatures as low as 2°C (151). There does not appear to be an upper temperature limit for Prochlorococcus distribution, since it is found in warm equatorial waters that reach 30°C at the surface. Still, the maximum integrated concentrations occur between 26 and 29°C and decrease above that temperature (Fig. 8A).
FIG. 7.
Concentrations Prochlorococcus integrated over the water column throughout the world oceans. Based on data from Table 5.
TABLE 5.
Oceanographic data used to construct Fig. 7 to 10
| Ocean or sea | Area | Cruise | Ship | Yr(s) | Months | Reference |
|---|---|---|---|---|---|---|
| Atlantic Ocean | North Atlantic | 87022 | CSS Hudson | 1987 | June | 88 |
| 88026 | CSS Hudson | 1988 | Sept | 87 | ||
| 89003 | CSS Baffin | 1989 | Apr | 86 | ||
| NIOZ Natl 89 | RV Tyro | 1989 | Aug–Sept | 179 | ||
| 91001 | CSS Hudson | 1991 | Apr | 81 | ||
| 92037 | CSS Hudson | 1992 | Sept | 83 | ||
| 93002 | CSS Hudson | 1993 | May | 83 | ||
| NOAA 93 | NOAA Malcolm Baldbridge | 1993 | July–Aug | 11 | ||
| Delaware 95 | RV Cape Henlopen | 1995 | Apr | 84 | ||
| 95016 | CSS Hudson | 1995 | July | 84 | ||
| Sargasso Sea | CHLOMAX | NO Suroit | 1987 | Sept–Oct | 116 | |
| Lopez 96 | Launch | 1995 | June | 85 | ||
| Tropical Atlantic | EUMELI 3 | NO Atalante | 1991 | Oct | 129 | |
| EUMELI 4 | NO Atalante | 1992 | June | 129 | ||
| EUMELI 5 | NO Atalante | 1992 | Dec | 129 | ||
| Indian Ocean | Arabian Sea | Arabian TTN043 | RV Thomson | 1995 | Jan | 14 |
| Arabian TTN045 | RV Thomson | 1995 | Mar–Apr | 14 | ||
| Arabian TTN049 | RV Thomson | 1995 | July–Aug | 117 | ||
| Indian Ocean and Red Sea | NIOZ Indian | 1992–1993 | May–Feb | 181 | ||
| Pacific Ocean | Equatorial Pacific | EQPAC TT007 | RV Thomson | 1992 | Feb–Mar | 74 |
| EQPAC TT008 | RV Thomson | 1992 | Mar–Apr | 7 | ||
| EQPAC TT008D | RV Thomson | 1992 | Mar–Apr | 27 | ||
| EQPAC TT011 | RV Thomson | 1992 | Aug–Sept | 74 | ||
| EQPAC TT012 | RV Thomson | 1992 | Sept–Oct | 27 | ||
| SURTROPAC 17 | NO Noroit | 1992 | Aug | 8 | ||
| OLIPAC | NO Atalante | 1994 | Nov | 115 | ||
| FLUPAC | NO Suroit | 1994 | Sept–Oct | 115 | ||
| Tropical Pacific | HOT | Moana Wave | 1990–1994 | 15 | ||
| Coast of Japan | Suruga Bay | Boat | 1992–1993 | May–Oct | 157 | |
| South Pacific | Chile 95 | RV Sonne | 1995 | June | 85 | |
| West Tropical Pacific | Australia | RV Sohgen-Maru | 1990 | Nov–Dec | 154 | |
| Mediterranean Sea | EROS Discovery 89 | HMS Discovery | 1989 | Jan | 178 | |
| EROS Bannock 89 | Bannock | 1989 | July | 128 | ||
| POEM 91 | RV Bilim/RV Shikmona | 1991 | Oct | 89 | ||
| Eddy 92 | RV Shikmona | 1992 | Mar | 193 | ||
| EROS Valdivia 92 | Valdivia | 1992 | Mar | 97 | ||
| EROS Discovery 93 | HMS Discovery | 1993 | July | 4 | ||
| Malaga 93 | Launch | 1993 | Jan | 33 | ||
| MINOS | NO Suroit | 1995 | Jun | 176 |
FIG. 8.
Relationship between Prochlorococcus integrated concentrations and surface temperature (A) and surface nitrate concentrations (B). Based on data from Table 5.
Although Prochlorococcus is most abundant in oligotrophic waters, both in absolute terms and relative to the other photosynthetic populations, it is by no means restricted to nutrient-depleted waters (Fig. 8B). In particular, it can be found in relatively coastal areas, such as the plume of the Rhone river in the Mediterranean Sea (178) or the Japanese waters of Suruga Bay (157). It has also been observed in the inner lagoons of Pacific atolls (18). Whether Prochlorococcus grows actively in such environments or is simply advected is an open question. In contrast, although it is often present in offshore temperate waters, it has never been observed in some temperate, permanently mixed shallow seas such as the English Channel (175). Very recently, it has been discovered in yet another niche, in secondary deep chlorophyll maxima situated below the oxycline in the Arabian Sea (40, 64). Still, vast areas of the world oceans remain uncharted (Fig. 7), especially in the southern hemisphere, although more data are now available from regions such as the Indian Ocean (Table 5), where large oceanographic surveys have been recently completed as part of the Joint Global Ocean Flux Study.
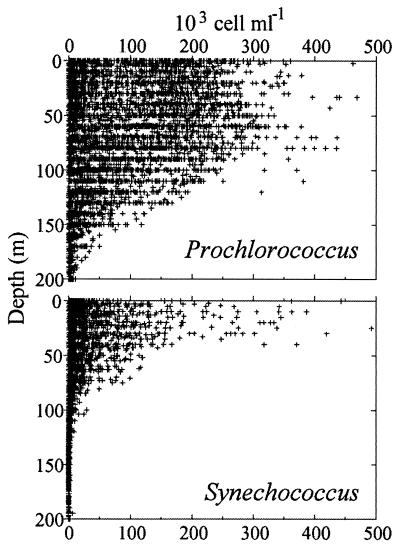
Vertical plots of the available measurements of Prochlorococcus and Synechococcus concentration by flow cytometry (Fig. 9) reveal the broad features that distinguish these two photosynthetic prokaryotes. On average Synechococcus concentrations are about 1 order of magnitude lower than Prochlorococcus concentrations, although their abundance maxima are very comparable. Maximum Prochlorococcus concentrations on the order of 700,000 cells ml−1 have been recorded in the Arabian Sea (14). Clearly, Prochlorococcus extends much deeper than Synechococcus, since the latter disappears virtually beyond 100 m deep. Three major types of vertical distributions are observed (Fig. 10). The first type is encountered mostly in nearshore waters (Fig. 10A and B). Prochlorococcus is restricted to the surface mixed layer, and its abundance drops abruptly below the thermocline (Fig. 10A), where a sharp and narrow maximum can be localized (Fig. 10B). In such cases, Synechococcus has a vertical distribution that parallels that of Prochlorococcus and its abundance is similar or slightly higher. In the second type of distribution (Fig. 10C and D), Prochlorococcus presents a very sharp maximum concentration on the order of 105 cells ml−1 near the bottom of the euphotic zone, which decreases by at least 1 order of magnitude at the surface. The third type of vertical distribution (Fig. 10E and F), with Prochlorococcus extending from the surface to the bottom of the euphotic zone at nearly constant concentrations on the order of 1 × 105 to 3 × 105 cells ml−1, is the most widespread in oceanic waters. Under these circumstances, the Synechococcus concentration is 1 to 2 orders of magnitude lower. Very often, a slight Prochlorococcus maximum occurs just above the depth at which concentrations begin to drop off (e.g., at 70 m in Fig. 10E). Generally, two different types of populations are found in the surface mixed layer and near the bottom of the euphotic zone (Fig. 5). This type of vertical profile is typical of oligotrophic waters. As oligotrophy increases (e.g., in the Pacific, going from the equator to the south gyre [Fig. 10E and F]), the Prochlorococcus layer deepens and Synechococcus concentrations decrease. Under these circumstances, the chlorophyll fluorescence of Prochlorococcus is very weak near the surface and cells are difficult to detect. This may lead to underestimates of cell concentrations at the surface and to creation of artifactual abundance maxima at depth.
FIG. 9.
Vertical distributions of Prochlorococcus (A) and Synechococcus (B). Based on data from Table 5.
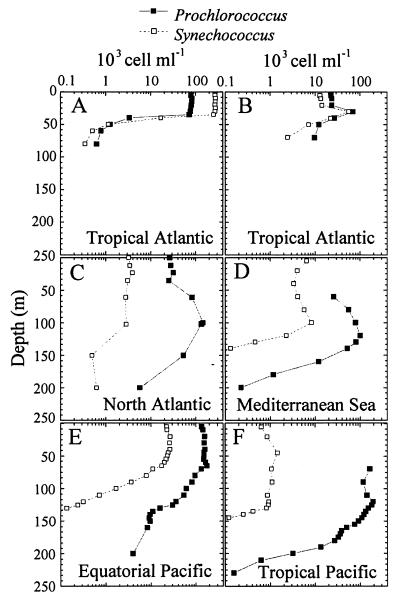
FIG. 10.
Typical vertical distributions of Prochlorococcus and Synechococcus. (A and B) Surface layer maximum. (C and D) Deep maximum. (E and F) Uniform distribution over the euphotic zone. (A and B) North tropical Atlantic, EUMELI3 cruise (129). (A) EU site, off the coast of Mauritania (20°N, 18°W). (B) MESO site (18°N, 21°W). (C) North Atlantic 30°N, 23°W (11). (D) Eastern Mediterranean Sea, 34°N 18°E, MINOS cruise (176). (E) Equatorial Pacific 150°W, 5°S (174). (F) Tropical Pacific 150°W, 16°S (174). (D and F) Prochlorococcus chlorophyll fluorescence was too weak to be detected at the surface.
In oceanic areas, where the water column is never mixed beyond 100 to 150 m deep (roughly between 30°S and 30°N), Prochlorococcus depth profiles vary only slightly throughout the year and are of the third type, as observed, for example, off Hawaii (15). In contrast, when the water column is mixed seasonally to depths below the euphotic zone, which usually happens in winter in more temperate waters, Prochlorococcus completely disappears during that period of the year (92). In spring, when the water column begins to restratify and abundant nutrients are found at the surface, Synechococcus may start to bloom in the mixed layer, followed by Prochlorococcus, yielding vertical profiles of the first type (119). As the bloom develops and nutrients become depleted, a well-defined Prochlorococcus layer deepens, resulting in a vertical profile of the second type (119). At these sites, profiles of the third type are not observed, at least to our knowledge, suggesting that there is not enough time between the end of the spring bloom and the next winter mixing event for a Prochlorococcus population to recolonize fully the oligotrophic surface layer. However, such recolonization may happen at locations where deep mixing does not occur every winter.
Growth Rates and Loss Processes in the Ocean
In culture, maximal steady-state division rates reported for Prochlorococcus are in general slightly below (or, more rarely, above [150]) one division per day, which is equivalent to a population growth rate of 0.5 to 0.6 day−1 (107, 131, 156). Besides being under the control of parameters such as light and nutrients, the division rate is under the strict control of temperature. For example, the MED4 and SS120 strains both have optimal growth rates at 24°C and cannot grow at or above 28°C, while their minimum growth temperatures are, respectively, 15 and 12.5°C (107).
In the field, division rates are very difficult to estimate directly from changes in cell concentrations because loss and growth processes balance each other and maintain populations at near constant levels. Initially, average estimates of Prochlorococcus growth rates of 0.2 to 0.3 day−1 in the top, euphotic layer of the Sargasso Sea off Bermuda were provided by the labeling of Chl a2 with 14C (43). However, this method yields strong over- or underestimates for populations subjected to photoacclimation (13), a common occurrence in the field. Use of the dilution method (72) to measure division rates in Equatorial Pacific waters gave variable and in some cases unrealistically low estimates for the Prochlorococcus division rate (0 to 0.5 day−1 at 10 m deep in reference 73), while its use to measure rates in the western Arabian Sea off Somalia suggested that Prochlorococcus can undergo more than two divisions per day in the field (137), i.e., higher than ever observed in culture.
Using cell cycle analysis, a method avoiding all artifacts linked to sample incubation, several groups demonstrated that the Prochlorococcus cell cycle is highly synchronized in the field, with DNA synthesis taking place in the afternoon and division taking place after dusk (94, 174, 175). Until recently, maximal growth rates recorded in areas such as the equatorial and subtropical Pacific Ocean were around 0.8 day−1, i.e., slightly above 1 division per day. However, populations from the Arabian Sea seem to be able to sustain growth rates of almost 1 day−1, i.e., about 1.4 divisions per day (150). Surprisingly, maximal rates were observed not at the surface but between 30 and 70 m deep. Above this depth, DNA synthesis is retarded by up to 4 h (a possible deleterious consequence of UV radiation that affects the top 20 to 30 m of the ocean) and, concomitantly, division rate decreases by up to 30% at the surface (174, 175).
Since Prochlorococcus cells reach division rates of the order of 1 division per day in marine waters and population abundance remain very stable over large timescales and spatial scales (15), loss processes must precisely balance division. This has been established in field experiments (93). However, the exact nature of the major loss processes has not been elucidated yet. Grazing by microzooplankton is probably important, as it is for heterotrophic bacteria (188). The single experimental study devoted to this question (22) revealed that ciliates exhibit a marked preference for Synechococcus over Prochlorococcus when given both together, leading to the speculation that the micrograzer community could be very different in _Prochlorococcus_- and _Synechococcus_-dominated marine ecosystems. Large filtering organisms, such as sponges in coastal systems (134) or filter feeders in reefs (191), could also have a major impact on Prochlorococcus. Viral lysis of Prochlorococcus has not been studied, but it has been shown not to be a significant cause of mortality of Synechococcus natural populations (183). Finally, near the surface, UV-induced lysis probably affects Prochlorococcus as strongly as it does heterotrophic bacteria (111).
DIVERSITY AND PHYLOGENY
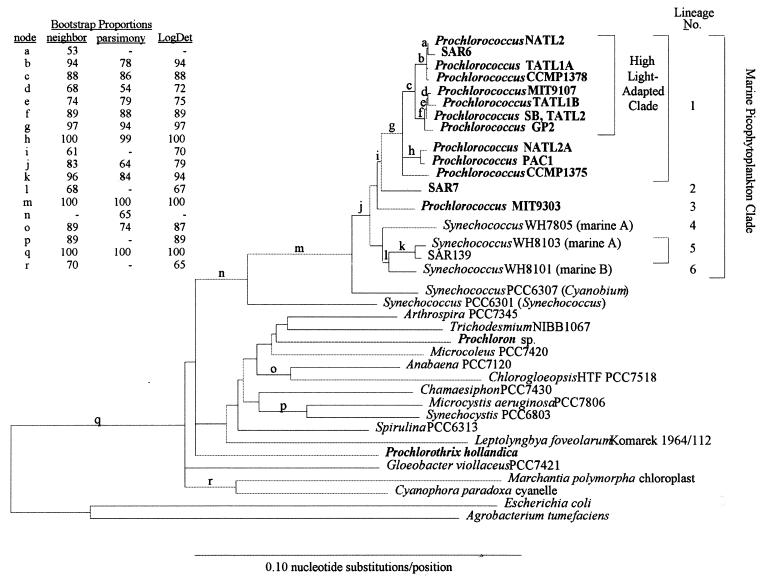
Genetic Diversity
Although in the previous sections we mainly discussed data obtained for two strains, MED (or its clonal derivative MED4) and SARG (or its clonal derivative SS120), which are representative of surface and deep oceanic layers, respectively, many more strains have been isolated (Table 1). Molecular studies of this set of isolates suggest that there is no correlation between their genetic distance, as assessed by using either restriction fragment length polymorphism patterns (145) or 16S rRNA gene sequence comparisons (166), and the geographic distance between sites from which these strains were isolated. For example, the genetic distance between strains isolated from the mixed layer of distant sites such as the Mediterranean Sea and the northwestern Atlantic Ocean seems to be much shorter than that between the latter isolates and others from deep waters (≥100 m) of the Sargasso Sea. Thus, the natural vertical gradients of light and nutrients may provide more stringent conditions for speciation within the Prochlorococcus genus than does geographic distance. For isolates whose pigmentation has been determined, genetic differentiation seems to be related to differences in the ratios of Chl _b_2 to Chl _a_2. The “high-light-adapted clade” (Fig. 11), as defined by Urbach et al. (166), groups strains from distant sites, all having low ratios of Chl _b_2 to Chl _a_2 (from 0.1 to 0.6). The well-studied clone MED4 is typical of this clade. Strains with low ratios of Chl _b_2 to Chl_a_2, such as strain GP2, _a_2 have sometimes been isolated from deeper layers (156), and the converse is also true. This is probably the main cause of exceptions in strain clustering, when the isolation depth is the only factor considered, as in the work by Scanlan et al. (145). It is most likely that strains with such deviant pigment signatures did not constitute the dominant genotype in the original seawater sample but were selected from a genetically mixed population by the conditions of culture following isolation (29, 108). A much larger genetic variation seems to exist among strains with high ratios of Chl _b_2 to Chl _a_2 than among those with low ratios (166), which is consistent with the more intense physical mixing rates experienced by surface oceanic populations. Moreover, their position in the phylogenetic tree suggests that members of the high-light clade are the most recently evolved of the Prochlorococcus strains.
FIG. 11.
Phylogenetic relationship between Prochlorococcus isolates and cyanobacteria inferred from 16S rRNA gene sequences. Reprinted from reference 166 with permission of the publisher.
As mentioned above, MED4 and SS120 differ by the presence in the latter strain but not in the former of a large gene cluster encoding phycoerythrin and related proteins (see “Photosynthetic apparatus” above). It would be interesting to know if this distinctive peculiarity extends to all strains, with those from the high-light-adapted clade being deprived of this gene cluster and others possessing it. This cluster is an example of the role that environmental parameters (in that case mainly light) may play in driving genetic diversity within the genus Prochlorococcus, and it is expected that other genes or operons exclusive to one of the groups might soon be discovered. Furthermore, it would be very informative to compare whole genome regions between different Prochlorococcus genotypes to determine whether there are hot spots for recombination that change more rapidly than do other regions.
Most of the conclusions drawn from culture studies are confirmed by molecular studies on the genetic diversity of Prochlorococcus in the field (124, 163). In particular, they also suggest a very high genetic variability among individuals of _Prochlorococcus_-like populations. The first sequences from natural Prochlorococcus populations were determined in studies investigating the diversity of the whole prokaryotic community in oceanic waters by using the 16S rRNA gene as a marker (31, 39, 148). Since these sequences were obtained prior to the sequencing of this gene in cultured Prochlorococcus cells (165), they were first misinterpreted as belonging to organisms from the Synechococcus marine A cluster. However, a later reanalysis of a number of sequences of shotgun clones obtained from the field, i.e., SAR6 (39), ALO7, ALO23, and ALO37 (148), and NH16-15 (31), showed their closer relatedness to Prochlorococcus, with a sequence similarity of around 98% (112).
The rpoC1 gene (encoding the RNA polymerase γ subunit) has also been used as a marker for more specific studies of natural populations of photosynthetic prokaryotes from the Sargasso Sea and the California Current (29, 124). In neither of these field studies was any gene sequence retrieved by amplifying rpoC1 absolutely identical to that in cultured isolates. Environmental clones could be separated into two genetically distant clades, A and B, with the former being genetically close to the MED4 clone. In contrast, the SS120 clone did not fall into either clade. For samples obtained under well-established stratified conditions (e.g., in the California Current), surface and deep environmental clones fell neatly into the A and B clades, respectively. However, clones from recently or weakly stratified water columns fell into both clades, suggesting that surface and deep populations probably occur together in the water column following mixing and become separated only once stratification is sufficiently (or permanently) established. The A clade is probably equivalent to the “high-light clade” identified by using 16S rRNA (166). The variability of the rpoC1 gene allows us to further subdivide this A clade into two deep branches (A1 and A2), which may differ in their temperature or nutrient preferences (29).
Another important outcome of the rpoC1 studies (29, 124) is that the genetic diversity among clones of a cluster is significantly higher for Prochlorococcus than for the phylogenetically related cyanobacterial genus Synechococcus, which generally occurs together with Prochlorococcus in the field. In contrast, Synechococcus seems to display a larger number of deeply branching clades (29), which may correspond to the wider oceanic distribution of Synechococcus compared to Prochlorococcus. It must be stressed that, as for Prochlorococcus, several pigment types of Synechococcus may be identified by flow cytometry or spectrofluorometry in the ocean, owing to their different ratios of phycourobilin to phycoerythrobilin. High-phycoerythrobilin Synechococcus strains dominate in mesotrophic environments, whereas only high-phycourobilin populations are found in oligotrophic waters (75, 118, 120). Thus, in an oligotrophic environment, the genetic variability of Synechococcus is expected to be lower than that of Prochlorococcus. Moreover, because of the lower population density of Synechococcus than Prochlorococcus in oligotrophic environments, extinction of lineages due to selective pressures and random genetic drift may occur more frequently for the former (124). More work is needed to check whether the reverse is true in mesotrophic environments, where Synechococcus may outnumber Prochlorococcus.
In another study, Urbach and Chisholm (164) gathered an extended data set, consisting of sequences from Prochlorococcus cells that had been sorted by flow cytometry from different depths at two stratified sites in the North Atlantic ocean. They used the variable intergenic region separating petB and petD genes and part of these genes as a marker. Of 68 Prochlorococcus alleles analyzed, only 16 were found more than once, and of these 16, only 12 appeared in more than one sample. The results of this study were consistent with the hypothesis that populations from different depths of stratified water columns are derived from a single gene pool but that gene frequencies vary at different depths, resulting in populations with different ratios of Chl _b_2 to Chl _a_2. The main difference between this study and the previous ones based on rpoC1 (29, 124) is that in the latter, DNA sequences were classified as Prochlorococcus or Synechococcus on the basis of the G+C content of their third codon position and their phylogenetic proximity to the two Prochlorococcus isolates MED and SS120 or to Synechococcus isolates WH7803, WH7805, and 8103. However, it is possible that a number of the sequences classified as “_Synechococcus_-like” by this method (e.g., clones 1CNB, 1CNI, and 1CNJ in reference 124) were in fact from Prochlorococcus cells. This inference can be drawn from the fact that Prochlorococcus strain MIT9303 has 55% G+C at third codon posi tions and is more closely related to Synechococcus strain WH8103 than to any known Prochlorococcus strains (166). There is also the (wholly theoretical) possibility that some of the “_Prochlorococcus_-like” rpoC1 clones were in fact from Synechococcus cells. Urbach and Chisholm (164) safely classified as Prochlorococcus a large number of clones, which could have been classified as Synechococcus because of their high G+C content; the correct classification occurred because the cells were initially purified by flow cytometric cell sorting. If some of _Synechococcus_-like clones in the study by Palenik (124) were indeed Prochlorococcus and none of the _Prochlorococcus_-like clones were Synechococcus, this strengthens the conclusion that the Prochlorococcus population in the sample analyzed was more genetically diverse than the Synechococcus population.
The apparently limited genetic difference observed between near surface Prochlorococcus from remote areas, a conclusion which comes mainly from culture studies (145, 166) and remains to be confirmed in the field, raises a number of questions. For instance, surface populations appear to be physiologically different between mesotrophic areas, such as the northwestern Mediterranean Sea in winter, and very oligotrophic subtropical areas. Growth of the former populations seems to be limited by inorganic nitrogen (177), whereas the latter populations are able to sustain high growth rates in the nutrient-depleted mixed layer (94). It is surprising that these physiologically different populations may correspond to the same genotype (or species), since they probably have very different nutrient acquisition systems and metabolisms. Along the same lines, temporal changes in the dominant Prochlorococcus species may also occur at a given site and depth, even over the short term, as suggested by observations of changes in scatter or fluorescence characteristics of natural Prochlorococcus populations from the Equatorial Pacific following iron addition (192) or the passage of a tropical instability wave (7). In both cases, authors have put forward the hypothesis that these artificial or natural events induced physiological changes within a homogeneous population (i.e., growth and/or photosynthesis), but a change in the Prochlorococcus population structure is also possible. Although never applied in such studies, use of molecular markers should be able to answer a number of such questions on population dynamics in the field.
Phylogeny
The original pigment complement found in Prochlorococcus cells led Chisholm et al. (21) to call this organism a prochlorophyte, which was the name previously given to the only two other known oxyphototrophic prokaryotes possessing Chl b but no phycobilins. The first of these organisms to be discovered was Prochloron sp., which is an obligate symbiont of marine ascidians (80). The second was Prochlorothrix hollandica, a free-living, filamentous prokaryote found in some freshwater lakes (12). Hence, the three known “prochlorophytes” appear to differ by many aspects. Although they were considered for some time to belong to a distinct phylum, the Prochlorophyta (79), or order, the Prochlorales (113), molecular studies involving, e.g., 16S rRNA (165), rpoC1 (125), or psbA (57) gene alignments have clearly demonstrated the very distinct origins of these organisms. For example, Prochlorothrix lacks seven amino acids near the C terminus of the otherwise very conserved D1 protein, a characteristic it shares with green plastids (109), whereas Prochlorococcus SS120 does not lack this region (57). All three “prochlorophytes” are emerging within the cyanobacteria but on clearly separate branches. Thus, there is no phylogenetic unity within “prochlorophytes” (165), and these should soon be reclassified as Oxyphotobacteria (135), together with the Chl _d_-containing Acaryochloris marina (103) and all classical cyanobacteria. Prochlorococcus is phylogenetically nearest to Synechococcus of the marine A cluster, such as WH 8103, a typical oceanic species (165). In fact, the fuzzy phylogenetic limits between Prochlorococcus and marine Synechococcus suggest a near simultaneous diversification of these groups from a common ancestor (166). Among Prochlorococcus strains, those belonging to the “high-light clade” (Fig. 11), which inhabit the nutrient-depleted surface layer, appear to be the most recent to diverge (166). This observation is consistent with the idea that the ancient oceans might have been less nutrient limited than they are nowadays.
The discovery of phycobilins and phycobiliproteins in Prochlorococcus strain SS120 (55) invalidates even more strongly the original definition of the term “prochlorophytes” as being oxyphototrophic prokaryotes possessing Chl b but no phycobilins. The phylogenetic origin of these phycobilins has not yet been fully resolved. There is no evidence that these genes were acquired through a lateral gene transfer. Instead, the characteristics of Prochlorococcus phycoerythrins are consistent with the supposed evolution of the organism based on 16S rRNA or rpoC1 phylogenies. The two structural genes encoding the α and β subunits of phycoerythrin are part of a larger gene cluster that also contains a putative linker polypeptide as well as several open reading frames and genes frequently found clustered in cyanobacterial phycobiliprotein operons (54). This gene arrangement resembles that of cyanobacteria such as Synechococcus strain WH8020, and phylogenetic analyses place Prochlorococcus type III phycoerythrin slightly closer to the marine type II phycoerythrin than to type I (54). That these genes are absent in at least one member of the phylogenetically recent high-light clade indicates that these strains may have eliminated from their genome any genes that had become useless, a process consistent with the relatively small size of Prochlorococcus genome.
The assertion of a common origin of Prochlorococcus and Synechococcus has been challenged, however, by one study that used the rbcL gene, encoding the large subunit of Rubisco (155). In this analysis, rbcL of strain GP2 was found to resemble more closely the Rubisco form IA of the γ subdivision of the Proteobacteria than that of most cyanobacteria (form IB). From these data, the authors concluded that Prochlorococcus was the most primitive oxygenic prokaryote. However, later analysis of the rbcL gene in two other Prochlorococcus strains (MED and PAC) did not support this conclusion, since they were found to group with form IB (133). Furthermore, the marine Synechococcus strain WH7803 possesses a Rubisco form IA (186), as does Prochlorococcus strain GP2. Thus, the Rubisco form is probably not a useful marker for the phylogeny of Prochlorococcus and Synechococcus. Since the rbcL gene is often transferred horizontally, such an event probably occurred between ancestors of Prochlorococcus strain GP2 and purple bacteria in the γ subdivision of the Proteobacteria (26). The close clustering between the GP2 and MED strains based on 16S rRNA (Fig. 11) suggests that this transfer could be fairly recent. It is interesting that the GP2 Rubisco is very similar to the corresponding protein of Hydrogenovibrio marinus, an aerobic chemolithotrophic bacterium which is present in similar environments to those favored by Prochlorococcus and Synechococcus and thus would be an example of a possible donor for such a lateral gene transfer to recent ancestors of GP2 and WH7803. A remaining question to be answered by a more detailed study is whether in Prochlorococcus strains such as GP2, Rubisco genes are organized in a tricistronic operon together with a ccmK homologue that encodes a component of the inorganic carbon-concentrating mechanism (186), as is the case in Synechococcus strain WH7803.
CONCLUSION
In the decade since its discovery, Prochlorococcus has revealed itself to be a truly fascinating organism. It is unambiguously one of the most important components of marine phytoplankton in terms of biomass, although its real importance in terms of global production is still only very approximately estimated (130). The analysis of its natural distribution provides many clues to interpret the physiological and genetic features it displays in the laboratory and vice versa. Its most spectacular traits include its extremely small size, its unique pigmentation, and its ability to proliferate in nutrient-depleted areas. From a phylogenetic viewpoint, it is unfortunately not the long-sought-after direct ancestor of the green chloroplast. Still, the evolution of Prochlorococcus, Prochloron, and Prochlorothrix, which have evolved independently from different cyanobacterial ancestors and converged to recruit the same protein to build a novel antenna to replace the more complex phycobilisomes (77), is a very exciting topic. Much remains to be discovered about Prochlorococcus, and this review has pointed out some directions for future research. In particular, there is a clear need for more axenic strains isolated from a variety of ecosystems where Prochlorococcus proliferates. Because of its many intriguing features, but even more importantly because of its true significance from a global environmental perspective, Prochlorococcus could establish itself in the near future as a standard model in photosynthetic prokaryote research, warranting a concerted effort to sequence its genome.
ACKNOWLEDGMENTS
We thank all the scientists who communicated both published and unpublished results, in particular those used to compile Fig. 7 through 10. D. Marie’s contribution was essential since he performed many of the flow cytometric measurements leading to Fig. 7 through 10. Special thanks are due to Ena Urbach, who kindly provided an electronic copy of Fig. 11; Claire Ting, who provided unpublished electron micrographs of Prochlorococcus strain MIT 9313; and R. Rippka, who gave us the recipe of the PCRS11 medium as well as her precious axenic strain. Susan de Goër, Jean Houmard, Penny Chisholm, Ena Urbach, Gabrielle Rocap, Lisa Moore, and Brian Palenik are gratefully acknowledged for their very useful comments on earlier versions of this paper and for communicating submitted manuscripts. Anonymous referees helped improve the structure of the final version.
Some of the work presented in this review was supported by the following sources: CNRS-SDU Action Incitative, JGOFS-France PROSOPE, Deutsche Forschungsgemeinschaft (to W.R.H.), CNRS Poste Rouge (to Georg W. M. van der Staay), EU BASIC program (BIO4-CT96-0256), and EU MAST III programs MEDEA (MAS3-CT96-015) and PROMOLEC (MAS3-CT97-0128).
REFERENCES
- 1.Andersen R A, Bidigare R R, Keller M D, Latasa M. A comparison of HPLC pigment signatures and electron microscopic observations for oligotrophic waters of the North Atlantic and Pacific Oceans. Deep-Sea Res II. 1996;43:517–537. [Google Scholar]
- 2.Andersen R A, Morton S L, Sexton J P. CCMP-Provasoli-Guillard National Center for culture of marine phytoplankton. J Phycol. 1997;33:1–75. [Google Scholar]
- 3.Barlow R G, Cummings D G, Gibb S W. Improved resolution of mono- and divinyl chlorophylls a and b and zeaxanthin and lutein in phytoplankton extracts using reverse phase C-8 HPLC. Mar Ecol Prog Ser. 1997;161:303–307. [Google Scholar]
- 4.Barlow R G, Mantoura R F C, Cummings D G, Fileman T W. Pigment chemotaxonomic distributions of phytoplankton during summer in the western Mediterranean. Deep-Sea Res II. 1997;44:833–850. [Google Scholar]
- 5.Bazzaz M B, Brereton R G. 4-Vinyl-4-desetyl chlorophyll a: a new naturally occurring chlorophyll. FEBS Lett. 1982;138:104–108. [Google Scholar]
- 6.Binder B J, Chisholm S W. Cell cycle regulation in marine Synechococcus sp. strains. Appl Environ Microbiol. 1995;61:708–717. doi: 10.1128/aem.61.2.708-717.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binder B J, Chisholm S W, Olson R J, Frankel S L, Worden A Z. Dynamics of picophytoplankton, ultraphytoplankton and bacteria in the central equatorial Pacific. Deep-Sea Res II. 1996;43:907–931. [Google Scholar]
- 8.Blanchot J, Rodier M. Picophytoplankton abundance and biomass in the western tropical Pacific Ocean during the 1992 El Niño year: results from flow cytometry. Deep-Sea Res I. 1996;43:877–895. [Google Scholar]
- 9.Boucher N, Vaulot D, Partensky F. Flow cytometric determination of phytoplankton DNA in cultures and oceanic populations. Mar Ecol Prog Ser. 1991;71:75–84. [Google Scholar]
- 10.Bricaud A, Stramski D. Spectral absorption coefficients of living phytoplankton and nonalgal biogenous matter: a comparison between the Peru upwelling area and the Sargasso Sea. Limnol Oceanogr. 1990;35:562–582. [Google Scholar]
- 11.Buck K R, Chavez F P, Campbell L. Basin-wide distributions of living carbon components and the inverted trophic pyramid of the central gyre of the North Atlantic Ocean, summer 1993. Aquat Microb Ecol. 1996;10:283–298. [Google Scholar]
- 12.Burger-Wiersma T, Veenhuis M, Korthals H J, Van de Wiel C C M, Mur L R. A new prokaryote containing chlorophylls a and b. Nature (London) 1986;320:262–264. [Google Scholar]
- 13.Cailliau C, Claustre H, Vidussi F, Marie D, Vaulot D. Carbon biomass, and gross growth rates as estimated from C-14 pigment labelling, during photoacclimation in Prochlorococcus CCMP 1378. Mar Ecol Prog Ser. 1996;145:209–221. [Google Scholar]
- 14.Campbell L, Landry M R, Constantinou J, Nolla H A, Brown S L, Liu H. Response of microbial community structure to environmental forcing in the Arabian Sea. Deep-Sea Res II. 1998;45:2301–2325. [Google Scholar]
- 15.Campbell L, Liu H B, Nolla H A, Vaulot D. Annual variability of phytoplankton and bacteria in the subtropical North Pacific Ocean at Station ALOHA during the 1991–1994 ENSO event. Deep-Sea Res I. 1997;44:167–192. [Google Scholar]
- 16.Campbell L, Nolla H A, Vaulot D. The importance of Prochlorococcus to community structure in the central North Pacific Ocean. Limnol Oceanogr. 1994;39:954–961. [Google Scholar]
- 17.Campbell L, Vaulot D. Photosynthetic picoplankton community structure in the subtropical North Pacific Ocean near Hawaii (station ALOHA) Deep-Sea Res I. 1993;40:2043–2060. [Google Scholar]
- 18.Charpy L, Blanchot J. Prochlorococcus contribution to phytoplankton biomass and production of Takapoto atoll (Tuamotu archipelago) C R Acad Sci (Paris) Ser III. 1996;319:131–137. [Google Scholar]
- 19.Chisholm S W. Phytoplankton size. In: Falkowski P G, Woodhead A D, editors. Primary productivity and biogeochemical cycles in the sea. New York, N.Y: Plenum Press; 1992. pp. 213–237. [Google Scholar]
- 20.Chisholm S W, Frankel S L, Goericke R, Olson R J, Palenik B, Waterbury J B, West-Johnsrud L, Zettler E R. Prochlorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b. Arch Microbiol. 1992;157:297–300. [Google Scholar]
- 21.Chisholm S W, Olson R J, Zettler E R, Waterbury J, Goericke R, Welschmeyer N. A novel free-living prochlorophyte occurs at high cell concentrations in the oceanic euphotic zone. Nature (London) 1988;334:340–343. [Google Scholar]
- 22.Christaki U, Jacquet S, Dolan J R, Vaulot D, Rassoulzadegan F. Growth and grazing on Prochlorococcus and Synechococcus by two marine ciliates. Limnol Oceanogr, 1999;44:52–61. [Google Scholar]
- 23.Cotner J B, Ammerman J W, Peele E R, Bentzen E. Phosphorus-limited bacterioplankton growth in the Sargasso Sea. Aquat Microb Ecol. 1997;13:141–149. [Google Scholar]
- 24.Davey H M, Kell D B. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol Rev. 1996;60:641. doi: 10.1128/mr.60.4.641-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature (London) 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 26.Delwiche C F, Palmer J D. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol Biol Evol. 1996;13:873–882. doi: 10.1093/oxfordjournals.molbev.a025647. [DOI] [PubMed] [Google Scholar]
- 27.DuRand M D, Olson R J. Contributions of phytoplankton light scattering and cell concentration changes to diel variations in beam attenuation in the equatorial Pacific from flow cytometric measurements of pico-, ultra- and nanoplankton. Deep-Sea Res II. 1996;43:891–906. [Google Scholar]
- 28.Dusenberry J A, Frankel S L. Increasing the sensitivity of a FACScan flow cytometer to study oceanic picoplankton. Limnol Oceanogr. 1994;39:206–209. [Google Scholar]
- 29.Ferris M J, Palenik B. Niche adaptation in ocean cyanobacteria. Nature (London) 1998;396:226–228. [Google Scholar]
- 30.Fitzwater S E, Knauer G A, Martin J H. Metal contamination and its effect on primary production measurements. Limnol Oceanogr. 1982;27:544–551. [Google Scholar]
- 31.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gantt E. Pigment protein complexes and the concept of the photosynthetic unit: Chlorophyll complexes and phycobilisomes. Photosynth Res. 1996;48:47–53. doi: 10.1007/BF00040995. [DOI] [PubMed] [Google Scholar]
- 33.García C M, Jiménez-Gómez F, Rodríguez J, Bautista B, Estrada M, García Braun J, Gasol J M, Gómez Figueiras F, Guerrero F, Jiménez Montes F, Li W K W, Lopez Díaz J M, Santiago G, Varel M. The size structure and functional composition of ultraplankton and nanoplankton at a frontal station in the Alboran Sea. Working groups 2 and 3 report. Sci Mar. 1994;58:43–52. [Google Scholar]
- 34.Garcia-Fernandez J M, Hess W R, Houmard J, Partensky F. Expression of the psbA gene in the marine oxyphotobacteria Prochlorococcus spp. Arch Biochem Biophys. 1998;359:17–23. doi: 10.1006/abbi.1998.0862. [DOI] [PubMed] [Google Scholar]
- 35.Garczarek L, Van der Staay G W M, Thomas J C, Partensky F. Isolation and characterization of photosystem I from two strains of the marine oxychlorobacterium Prochlorococcus. Photosynth Res. 1998;56:131–141. [Google Scholar]
- 36.Gieskes W W C. Algal pigment fingerprints: clue to taxon-specific abundance, productivity and degradation of phytoplankton in seas and oceans. NATO ASI Ser. 1991;G27:61–99. [Google Scholar]
- 37.Gieskes W W C, Kraay G W. Unknown chlorophyll a derivatives in the North Sea and the tropical Atlantic Ocean revealed by HPLC analysis. Limnol Oceanogr. 1983;28:757–766. [Google Scholar]
- 38.Giorgi L B, Nixon P J, Merry S A, Joseph D M, Durrant J R, de Las Rivas J, Barber J, Porter G, Klug D R. Comparison of primary charge separation in the photosystem II reaction center complex isolated from wild-type and D1-130 mutants of the cyanobacterium Synechocystis PCC 6803. J Biol Chem. 1996;271:2093–2101. doi: 10.1074/jbc.271.4.2093. [DOI] [PubMed] [Google Scholar]
- 39.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature (London) 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 40.Goericke, R., R. J. Olson, and A. Shalapyonok. A novel niche for Prochlorococcus sp. in low-light suboxic environments in the Arabian Sea and the Eastern Tropical North Pacific. Submitted for publication.
- 41.Goericke R, Repeta D J. The pigments of Prochlorococcus marinus: the presence of divinyl chlorophyll a and b in a marine prochlorophyte. Limnol Oceanogr. 1992;37:425–433. [Google Scholar]
- 42.Goericke R, Repeta D J. Chlorophylls a and b and divinyl chlorophylls a and b in the open subtropical North Atlantic Ocean. Mar Ecol Prog Ser. 1993;101:307–313. [Google Scholar]
- 43.Goericke R, Welschmeyer N A. The marine prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary production in the Sargasso Sea. Deep-Sea Res I. 1993;40:2283–2294. [Google Scholar]
- 44.Golbeck J H. Photosystem I in cyanobacteria. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 319–360. [Google Scholar]
- 45.Golden S S. Light-responsive gene expression in cyanobacteria. J Bacteriol. 1995;177:1651–1654. doi: 10.1128/jb.177.7.1651-1654.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green B R. The chlorophyll-protein complexes of higher plant photosynthetic membranes, or just what green band is that? Photosynth Res. 1988;15:3–32. doi: 10.1007/BF00054985. [DOI] [PubMed] [Google Scholar]
- 47.Guillard R R L, Murphy L S, Foss P, Liaaen-Jensen S. Synechococcus spp. as likely zeaxanthin-dominant ultraphytoplankton in the North-Atlantic. Limnol Oceanogr. 1985;30:412–414. [Google Scholar]
- 48.Guillard R R, Ryther J H. Studies of marine diatoms. I. Cyclotella nana Husdedt and Detonula confervacea Gran. Can J Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- 49.Herdman M, Janvier M, Rippka R, Stanier R Y. Genome size of cyanobacteria. J Gen Microbiol. 1979;111:73–85. [Google Scholar]
- 50.Herdman M, Janvier M, Waterbury J B, Rippka R, Stanier R Y. Deoxyribonucleic acid base composition of cyanobacteria. J Gen Microbiol. 1979;111:63–71. [Google Scholar]
- 51.Hess W R. Localization of an open reading frame with homology to human aspartoacylase upstream from psbA in the prokaryote Prochlorococcus marinus CCMP 1375. DNA Seq. 1997;7:301–306. doi: 10.3109/10425179709034049. [DOI] [PubMed] [Google Scholar]
- 52.Hess, W. R. Unpublished data.
- 53.Hess W R, Fingerhut C, Schön A. RNase P RNA from Prochlorococcus marinus: contribution of substrate domains to recognition by a cyanobacterial ribozyme. FEBS Lett. 1998;431:138–142. doi: 10.1016/s0014-5793(98)00729-7. [DOI] [PubMed] [Google Scholar]
- 54.Hess, W. R., and F. Partensky. Identification of a putative gamma linker polypeptide gene in the marine oxyphotobacterium _Prochlorococcus marinus_—Implications for the phylogeny of Prochlorococcus phycoerytrins. In G. A. Peschek, W. Löffelhardt, and G. Schmetterer (ed.), The phototrophic procaryotes, in press. Plenum Press, New York, N.Y.
- 55.Hess W R, Partensky F, Van der Staay G W M, Garcia-Fernandez J M, Börner T, Vaulot D. Coexistence of phycoerythrin and a chlorophyll a/b antenna in a marine prokaryote. Proc Natl Acad Sci USA. 1996;93:11126–11130. doi: 10.1073/pnas.93.20.11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hess, W. R., C. Steglich, C. Lichtlé, and F. Partensky. The phycoerythrins of Prochlorococcus marinus are associated to the thylakoid membrane and are encoded by a single large gene cluster. Submitted for publication. [DOI] [PubMed]
- 57.Hess W R, Weihe A, Loiseaux-de Goër S, Partensky F, Vaulot D. Characterization of the single psbA gene of Prochlorococcus marinus CCMP 1375 (Prochlorophyta) Plant Mol Biol. 1995;27:1189–1196. doi: 10.1007/BF00020892. [DOI] [PubMed] [Google Scholar]
- 58.Holtzendorff, J., and W. R. Hess. Unpublished data.
- 59.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:12–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 60.Ishizaka J, Kiyosawa H, Ishida K, Ishikawa K, Takahashi M. Meridional distribution and carbon biomass of autotrophic picoplankton in the Central North Pacific during late Northern summer 1990. Deep-Sea Res I. 1994;41:1745–1766. [Google Scholar]
- 61.Jochem F J. Phototrophic picoplankton structure in three different pelagic regimes in the Arabian Sea. Mar Ecol Prog Ser. 1995;117:307–314. [Google Scholar]
- 62.Johnson C H, Golden S S, Ishiura M, Kondo T. Circadian clocks in prokaryotes. Mol Microbiol. 1996;21:5–11. doi: 10.1046/j.1365-2958.1996.00613.x. [DOI] [PubMed] [Google Scholar]
- 63.Johnson P W, Sieburth J M. Chroococcoid cyanobacteria in the sea: a ubiquitous and diverse phototrophic biomass. Limnol Oceanogr. 1979;24:928–935. [Google Scholar]
- 64.Johnson, Z., M. L. Landry, R. R. Bidigare, S. L. Brown, L. Campbell, J. Gunderson, J. Marra, R. Olson, A. Shalapyonok, and C. Trees. Energetics and growth of a deep Prochlorococcus population in the Arabian Sea. Deep-Sea Res. II, in press.
- 65.Jukes T H, Bushan V. Silent nucleotide substitutions and G+C content of some mitochondrial and bacterial genes. J Mol Evol. 1986;24:39–44. doi: 10.1007/BF02099949. [DOI] [PubMed] [Google Scholar]
- 66.Kahn K, Mazel D, Houmard J, Tandeau de Marsac N, Schaefer M R. A role for cpeYZ in cyanobacterial phycoerythrin biosynthesis. J Bacteriol. 1997;179:998–1006. doi: 10.1128/jb.179.4.998-1006.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 68.Kaneko T, Tabata S. Complete genome structure of the unicellular cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 1997;38:1171–1176. doi: 10.1093/oxfordjournals.pcp.a029103. [DOI] [PubMed] [Google Scholar]
- 69.Keller M D, Selvin R C, Claus W, Guillard R R L. Media for the culture of oceanic ultraphytoplankton. J Phycol. 1987;23:633–638. [Google Scholar]
- 70.Kulkarni R D, Golden S S. Form II of D1 is important during transition from standard to high light intensity in Synechococcus sp strain PCC 7942. Photosynth Res. 1995;46:435–443. doi: 10.1007/BF00032298. [DOI] [PubMed] [Google Scholar]
- 71.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature (London) 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 72.Landry M R, Hassett R P. Estimating the grazing impact of marine microzooplankton. Mar Biol. 1982;67:283–288. [Google Scholar]
- 73.Landry M R, Kirshtein J, Constantinou J. A refined dilution technique for measuring the community grazing impact of microzooplankton, with experimental tests in the central equatorial Pacific. Mar Ecol Prog Ser. 1995;120:53–63. [Google Scholar]
- 74.Landry M R, Kirshtein J, Constantinou J. Abundances and distributions of picoplankton populations in the central equatorial Pacific from 12 degrees N to 12 degrees S, 140 degrees W. Deep-Sea Res II. 1996;43:871–890. [Google Scholar]
- 75.Lantoine F, Neveux J. Spatial and seasonal variations in abundance and spectral characteristics of phycoerythrins in the tropical northeastern Atlantic Ocean. Deep-Sea Res I. 1997;44:223–246. [Google Scholar]
- 76.La Roche J, Murray H, Orellana M, Newton J. Flavodoxin expression as an indicator of iron limitation in marine diatoms. J Phycol. 1995;31:520–530. [Google Scholar]
- 77.La Roche J, van der Staay G W M, Partensky F, Ducret A, Aebersold R, Li R, Golden S S, Hiller R G, Wrench P M, Larkum A W D, Green B R. Independent evolution of the prochlorophyte and green plant chlorophyll a/b light-harvesting proteins. Proc Natl Acad Sci USA. 1996;93:15244–15248. doi: 10.1073/pnas.93.26.15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lazzara L, Bricaud A, Claustre H. Spectral absorption and fluorescence excitation properties of phytoplanktonic populations at a mesotrophic and an oligotrophic site in the tropical North Atlantic (EUMELI program) Deep-Sea Res I. 1996;43:1215–1240. [Google Scholar]
- 79.Lewin R A. Prochlorophyta as a proposed new division of algae. Nature (London) 1976;261:697–698. doi: 10.1038/261697b0. [DOI] [PubMed] [Google Scholar]
- 80.Lewin R A, Withers N W. Extraordinary pigment composition of a prokaryotic alga. Nature (London) 1975;256:735–737. [Google Scholar]
- 81.Li W K W. Estimation of primary production by flow cytometry. ICES Mar Sci Symp. 1993;197:79–91. [Google Scholar]
- 82.Li W K W. Primary productivity of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol Oceanogr. 1994;39:169–175. [Google Scholar]
- 83.Li W K W. Composition of ultraphytoplankton in the central North Atlantic. Mar Ecol Prog Ser. 1995;122:1–8. [Google Scholar]
- 84.Li W K W. Cytometric diversity in marine ultraphytoplankton. Limnol Oceanogr. 1997;42:874–880. [Google Scholar]
- 85.Li, W. K. W. Unpublished data.
- 86.Li W K W, Dickie P M, Harrison W G, Irwin B D. Biomass and production of bacteria and phytoplankton during the spring bloom in the western North Atlantic Ocean. Deep-Sea Res I. 1993;40:307–327. [Google Scholar]
- 87.Li W K W, Dickie P M, Irwin B D, Wood A M. Biomass of bacteria, cyanobacteria, prochlorophytes and photosynthetic eukaryotes in the Sargasso Sea. Deep-Sea Res I. 1992;39:501–519. [Google Scholar]
- 88.Li W K W, Wood A M. Vertical distribution of North Atlantic ultraphytoplankton: analysis by flow cytometry and epifluorescence microscopy. Deep-Sea Res I. 1988;35:1615–1638. [Google Scholar]
- 89.Li W K W, Zohary T, Yacobi Y Z, Wood A M. Ultraphytoplankton in the eastern Mediterranean Sea: towards deriving phytoplankton biomass from flow cytometric measurements of abundance, fluorescence and light scatter. Mar Ecol Prog Ser. 1993;102:79–87. [Google Scholar]
- 90.Lichtlé C, Thomas J C, Spilar A, Partensky F. Immunological and ultrastructural characterization of the photosynthetic complexes of the prochlorophyte Prochlorococcus (Oxychlorobacteria) J Phycol. 1995;31:934–941. [Google Scholar]
- 91.Lindell D, Padan E, Post A F. Regulation of ntcA expression and nitrite uptake in the marine Synechococcus sp. strain WH 7803. J Bacteriol. 1998;180:1878–1886. doi: 10.1128/jb.180.7.1878-1886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lindell D, Post A F. Ultraphytoplankton succession is triggered by deep winter mixing in the Gulf of Aqaba (Eilat), Red Sea. Limnol Oceanogr. 1995;40:1130–1141. [Google Scholar]
- 93.Liu H, Campbell L, Landry M. Growth and mortality rates of Prochlorococcus and Synechococcus measured with a selective inhibitor technique. Mar Ecol Prog Ser. 1995;116:277–287. [Google Scholar]
- 94.Liu H B, Nolla H A, Campbell L. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquat Microb Ecol. 1997;12:39–47. [Google Scholar]
- 95.Lorenz M, Börner T, Hess W R. Molecular cloning and characterization of a dihydrodipicolinate synthase (DHDPS) gene from the photoautotrophic prokaryote Prochlorococcus marinus CCMP 1375 (prochlorophyta) Endo Cell Res. 1995;11:59–68. [Google Scholar]
- 96.Lorenz M, Partensky F, Börner T, Hess W R. Sequencing of RAPD fragments amplified from the genome of the prokaryote Prochlorococcus marinus (Prochlorophyta) Biochem Mol Biol Int. 1995;36:705–713. [PubMed] [Google Scholar]
- 97.Marie D, Jacquet S, Vaulot D. Proceedings of the International Symposium on Marine Cyanobacteria and Related Organisms. 1997. DNA and cell cycle analysis of marine Synechococcus and Prochlorococcus by flow cytometry; p. 34. [Google Scholar]
- 98.Marie D, Partensky F, Jacquet S, Vaulot D. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl Environ Microbiol. 1997;63:186–193. doi: 10.1128/aem.63.1.186-193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marie, D., F. Partensky, N. Simon, L. Guillou, and D. Vaulot. Flow cytometry analysis of marine picoplankton. In R. A. Diamond and S. DeMaggio (ed.), In living colors: protocols in flow cytometry and cell sorting, in press. Springer-Verlag, Heidelberg, Germany.
- 100.Marquardt J, Senger H, Miyashita S, Morschel E. Isolation and characterization of biliprotein aggregates from Acaryochloris marina, a Prochloron-like prokaryote containing mainly chlorophyll d. FEBS Lett. 1997;410:428–432. doi: 10.1016/s0014-5793(97)00631-5. [DOI] [PubMed] [Google Scholar]
- 101.Martin J H, Coale K H, Johnson K S, Fitzwater S E, Gordon R M, Tanner S J, Hunter C N, Elrod V A, Nowicki J L, Coley T L, Barber R T, Lindley S, Watson A J, Van Scoy K, Law C S, Liddicoat M I, Ling R, Stanton T, Stockel J, Collins C, Anderson A, Bidigare R, Ondrusek M, Latasa M, Millero F J, Lee K, Yao W, Zhang J Z, Friederich G, Sakamoto C, Chavez F, Buck K, Kolber Z, Greene R, Falkowski P, Chisholm S W, Hoge F, Swift R, Yungel J, Turner S, Nightingale P, Hatton A, Liss P, Tindale N W. Testing the iron hypothesis in ecosystems of the equatorial Pacific Ocean. Nature (London) 1994;371:123–129. [Google Scholar]
- 102.McManus G B, Dawson R. Phytoplankton pigments in the deep chlorophyll maximum of the Caribbean Sea and the western tropical Atlantic Ocean. Mar Ecol Prog Ser. 1994;113:199–206. [Google Scholar]
- 103.Miyashita H, Ikemoto H, Kurano N, Adachi K, Chihara M, Miyachi S. Chlorophyll d as a major pigment. Nature (London) 1996;383:402. [Google Scholar]
- 104.Monger B C, Landry M R. Flow cytometric analysis of marine bacteria with Hoechst 33342. Appl Environ Microbiol. 1993;59:905–911. doi: 10.1128/aem.59.3.905-911.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moore L R. Physiological ecology of Prochlorococcus: a comparison of isolates from diverse oceanographic regimes. Ph.D. thesis. Cambridge: Massachusetts Institute of Technology; 1997. [Google Scholar]
- 106.Moore, L. R., and S. W. Chisholm. Photophysiology of the marine cyanobacterium Prochlorococcus: ecotypic differences among cultured isolates. Limnol. Oceanogr., in press.
- 107.Moore L R, Goericke R, Chisholm S W. Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar Ecol Prog Ser. 1995;116:259–275. [Google Scholar]
- 108.Moore L R, Rocap G, Chisholm S W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature (London) 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 109.Morden C W, Golden S S. psbA genes indicate common ancestry of prochlorophytes and chloroplasts. Nature (London) 1989;337:382–385. doi: 10.1038/337382a0. [DOI] [PubMed] [Google Scholar]
- 110.Morel A, Ahn Y-W, Partensky F, Vaulot D, Claustre H. Prochlorococcus and Synechococcus: a comparative study of their size, pigmentation and related optical properties. J Mar Res. 1993;51:617–649. [Google Scholar]
- 111.Müller-Niklas G, Heissenberger A, Puskaric S, Herndl G. Ultraviolet-B radiation and bacterial metabolism in coastal waters. Aquat Microb Ecol. 1995;9:111–116. [Google Scholar]
- 112.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 113.Mur L R, Burger-Wiersma T. The order Prochlorales. In: Balows A, Trüper H P, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. 2nd ed. Vol. 2. New York, N.Y: Springer Verlag; 1992. pp. 2105–2110. [Google Scholar]
- 114.Neveux J, Lantoine F. Spectrofluorimetric assay of chlorophylls and pheopigments using the least squares approximation technique. Deep-Sea Res I. 1993;40:1747–1765. [Google Scholar]
- 115.Neveux, J., F. Lantoine, D. Vaulot, D. Marie, and J. Blanchot. Phycoerythins in the southern tropical and equatorial Pacific: evidence for new cyanobacterial types. J. Geophys. Res., in press.
- 116.Neveux J, Vaulot D, Courties C, Fukai E. Green photosynthetic bacteria associated with the deep chlorophyll maximum of the Sargasso Sea. C R Acad Sci (Paris) Ser III. 1989;308:9–14. [Google Scholar]
- 117.Olson, R. J. Unpublished data.
- 118.Olson R J, Chisholm S W, Zettler E R, Armbrust E V. Analysis of Synechococcus pigment types in the sea using single and dual beam flow cytometry. Deep-Sea Res I. 1988;35:425–440. [Google Scholar]
- 119.Olson R J, Zettler E R, Altabet M A, Dusenberry J A, Chisholm S W. Spatial and temporal distributions of prochlorophyte picoplankton in the North Atlantic Ocean. Deep-Sea Res I. 1990;37:1033–1051. [Google Scholar]
- 120.Olson R J, Zettler E R, Armbrust E V, Chisholm S W. Pigment, size and distribution of Synechococcus in the North Atlantic and Pacific oceans. Limnol Oceanogr. 1990;35:45–58. [Google Scholar]
- 121.Olson R J, Zettler E R, Chisholm S W, Dusenberry J A. Advances in Oceanography through flow cytometry. NATO ASI Ser Ser G. 1991;27:351–399. [Google Scholar]
- 122.Ong L J, Glazer A N. Phycoerythrins of the marine unicellular cyanobacteria. J Biol Chem. 1991;266:9519–9527. [PubMed] [Google Scholar]
- 123.Oquist G, Campbell D, Clarke A K, Gustafsson P. The cyanobacterium Synechococcus modulates Photosystem II function in response to excitation stress through D1 exchange. Photosynth Res. 1995;46:151–158. doi: 10.1007/BF00020425. [DOI] [PubMed] [Google Scholar]
- 124.Palenik B. Cyanobacterial community structure as seen from RNA polymerase gene sequence analysis. Appl Environ Microbiol. 1994;60:3212–3219. doi: 10.1128/aem.60.9.3212-3219.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Palenik B, Haselkorn R. Multiple evolutionary origins of prochlorophytes, the chlorophyll b-containing prokaryotes. Nature (London) 1992;355:265–267. doi: 10.1038/355265a0. [DOI] [PubMed] [Google Scholar]
- 126.Parham R, Rebeiz C A. Chloroplast biogenesis: (4-vinyl)chlorophyllide a reductase is a divinyl chlorophyllide a-specific, NADPH-dependent enzyme. Biochemistry. 1992;31:8460–8464. doi: 10.1021/bi00151a011. [DOI] [PubMed] [Google Scholar]
- 127.Parpais J, Marie D, Partensky F, Morin P, Vaulot D. Effect of phosphorus starvation on the cell cycle of the photosynthetic prokaryote Prochlorococcus spp. Mar Ecol Prog Ser. 1996;132:265–274. [Google Scholar]
- 128.Partensky, F. Unpublished data.
- 129.Partensky F, Blanchot J, Lantoine F, Neveux J, Marie D. Vertical structure of picophytoplankton at different trophic sites of the tropical northeastern Atlantic Ocean. Deep-Sea Res I. 1996;43:1191–1213. [Google Scholar]
- 130.Partensky, F., J. Blanchot, and D. Vaulot. Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. In L. Charpy and A. W. D. Larkum (ed.), Marine cyanobacteria, in press. Institut Océanographique, Monaco.
- 131.Partensky F, Hoepffner N, Li W K W, Ulloa O, Vaulot D. Photoacclimation of Prochlorococcus sp. (Prochlorophyta) strains isolated from the North Atlantic and the Mediterranean Sea. Plant Physiol. 1993;101:295–296. doi: 10.1104/pp.101.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Partensky F, LaRoche J, Wyman K, Falkowski P G. The divinyl-chlorophyll a/b-protein complexes of two strains of the oxyphototrophic marine prokaryote Prochlorococcus—characterization and response to changes in growth irradiance. Photosynth Res. 1997;51:209–222. [Google Scholar]
- 133.Pichard S L, Campbell L, Paul J H. Diversity of the ribulose bisphosphate carboxylase/oxygenase form I gene (rbcL) in natural phytoplankton communities. Appl Environ Microbiol. 1997;63:3600–3606. doi: 10.1128/aem.63.9.3600-3606.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pile A J, Patterson M R, Witman J D. In situ grazing on plankton <10 μm by the boreal sponge Mycale lingua. Mar Ecol Prog Ser. 1996;141:95–102. [Google Scholar]
- 135.Pinevich A V, Averina S G, Velichko N V. Another view on the role of photosynthetic pigments in taxonomy of oxygenic-phototrophic bacteria: proposed rejection of the order Prochlorales Florenzano, Balloni, and Materassi 1986 (Emend. Burger-Wiersma, Stal, and Mur 1989), the family Prochloraceae Florenzano, Balloni, and Materassi 1986, and the family Prochlorotrichaceae Burger-Wiersma, Stal, and Mur 1989. Int J Syst Bacteriol. 1997;47:1264–1267. [Google Scholar]
- 136.Raven J A. Why are there no picoplanktonic O2 evolvers with volumes less than 10−19 m3? J Plankton Res. 1994;16:565–580. [Google Scholar]
- 137.Reckermann M, Veldhuis M J W. Trophic interactions between picophytoplankton and micro- and nanozooplankton in the western Arabian Sea during the NE monsoon 1993. Aquat Microb Ecol. 1997;12:263–273. [Google Scholar]
- 138.Richter S, Hagemann M, Messer W. Transcriptional analysis and mutation of a dnaA-like gene in Synechocystis sp. strain PCC 6803. J Bacteriol. 1998;180:4946–4949. doi: 10.1128/jb.180.18.4946-4949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Richter S, Hess W R, Krause M, Messer W. Unique organization of the dnaA region from Prochlorococcus marinus CCMP1375, a marine cyanobacterium. Mol Gen Genet. 1998;257:534–541. doi: 10.1007/s004380050679. [DOI] [PubMed] [Google Scholar]
- 140.Richter S, Messer W. Genetic structure of the dnaA region of the cyanobacterium Synechocystis sp strain PCC 6803. J Bacteriol. 1995;177:4245–4251. doi: 10.1128/jb.177.15.4245-4251.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rippka R. Isolation and purification of cyanobacteria. Methods Enzymol. 1988;167:3–27. doi: 10.1016/0076-6879(88)67004-2. [DOI] [PubMed] [Google Scholar]
- 142.Rippka, R. Unpublished data.
- 143.Rippka R, Coursin T, Lichtlé C, Partensky F, Houmard J, Herdman M. Proceedings of the IXth International Symposium on Photosynthetic Prokaryotes. 1997. Prochlorococcus sp. PCC 9511, an axenic, marine, chlorophyll b-containing oxyphotobacterium; p. 189. [Google Scholar]
- 144.Rüdiger W, Schoch S. Chlorophylls. In: Goodwin T W, editor. Plant pigments. New York, N.Y: Academic Press, Inc.; 1988. pp. 1–59. [Google Scholar]
- 145.Scanlan D J, Hess W R, Partensky F, Newman J, Vaulot D. High degree of genetic variation in Prochlorococcus (Prochlorophyta) revealed by RFLP analysis. Eur J Phycol. 1996;31:1–9. [Google Scholar]
- 146.Scanlan D J, Silman N J, Donald K M, Wilson W H, Carr N G, Joint I, Mann N H. An immunological approach to detect phosphate stress in populations and single cells of photosynthetic picoplankton. Appl Environ Microbiol. 1997;63:2411–2420. doi: 10.1128/aem.63.6.2411-2420.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schluchter W M, Shen G H, Zhao J D, Bryant D A. Characterization of psaI and psaL mutants of Synechococcus sp. strain PCC 7002: a new model for state transitions in cyanobacteria. Photochem Photobiol. 1996;64:53–66. doi: 10.1111/j.1751-1097.1996.tb02421.x. [DOI] [PubMed] [Google Scholar]
- 148.Schmidt T M, DeLong E F, Pace N R. Analysis of marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schyns G, Rippka R, Namane A, Campbell D, Herdman M, Houmard J. Prochlorothrix hollandica PCC 9006: genomic properties of an axenic representative of the chlorophyll a/b-containing oxyphotobacteria. Res Microbiol. 1997;148:345–354. doi: 10.1016/S0923-2508(97)81590-2. [DOI] [PubMed] [Google Scholar]
- 150.Shalapyonok A, Olson R J, Shalapyonok L S. Ultradian growth in Prochlorococcus spp. Appl Environ Microbiol. 1998;64:1066–1069. doi: 10.1128/aem.64.3.1066-1069.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shapiro L P, Haugen E M. Seasonal distribution and temperature tolerance of Synechococcus in Boothbay Harbor, Maine. Estuarine Coastal Shelf Sci. 1988;26:517–525. [Google Scholar]
- 152.Sharp P M, Li W H. An evolutionary perspective on synonymous codon usage in unicellular organisms. Mol Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- 153.Sharp P M, Matassi G. Codon usage and genome evolution. Curr Opin Genet Develop. 1994;4:851–860. doi: 10.1016/0959-437x(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 154.Shimada A, Hasegawa T, Umeda I, Kadoya N, Maruyama T. Spatial mesoscale patterns of West Pacific picophytoplankton as analyzed by flow cytometry: their contribution to subsurface chlorophyll maxima. Mar Biol. 1993;115:209–215. [Google Scholar]
- 155.Shimada A, Kanai S, Maruyama T. Partial sequence of ribulose-1,5-bisphosphate carboxylase/oxygenase and the phylogeny of Prochloron and Prochlorococcus (Prochlorales) J Mol Evol. 1995;40:671–677. doi: 10.1007/BF00160516. [DOI] [PubMed] [Google Scholar]
- 156.Shimada A, Maruyama T, Miyachi S. Vertical distributions and photosynthetic action spectra of two oceanic picophytoplankers, Prochlorococcus marinus and Synechococcus sp. Mar Biol. 1996;127:15–23. [Google Scholar]
- 157.Shimada A, Nishijima M, Maruyama T. Seasonal abundance of Prochlorococcus in Suruga Bay, Japan in 1992–1993. J Oceanogr. 1995;51:289–300. [Google Scholar]
- 158.Sieracki M E, Haugen E M, Cucci T L. Overestimation of heterotrophic bacteria in the Sargasso Sea: direct evidence by flow and imaging cytometry. Deep-Sea Res I. 1995;42:1399. [Google Scholar]
- 159.Skarstad K, Boye E. The initiator protein DnaA: evolution, properties and function. Biochim Biophys Acta. 1994;1217:111–130. doi: 10.1016/0167-4781(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 160.Soitamo A J, Zhou G, Clarke A K, Oquist G, Gustafsson P, Aro E M. Over-production of the D1:2 protein makes Synechococcus cells more tolerant to photoinhibition of Photosystem II. Plant Mol Biol. 1996;30:467–478. doi: 10.1007/BF00049325. [DOI] [PubMed] [Google Scholar]
- 161.Sueoka N. Directional mutation pressure, selective constraints, and genetic equilibria. J Mol Evol. 1992;34:95–114. doi: 10.1007/BF00182387. [DOI] [PubMed] [Google Scholar]
- 162.Thingstad T F, Rassoulzadegan F. Nutrient limitations, microbial food webs, and ‘biological C-pumps’: suggested interactions in a P-limited Mediterranean. Mar Ecol Prog Ser. 1995;117:299–306. [Google Scholar]
- 163.Urbach E. Evolution and population genetics of Prochlorococcus marinus. Ph.D. thesis. Cambridge: Massachusetts Institute of Technology; 1995. [Google Scholar]
- 164.Urbach E, Chisholm S W. Genetic diversity in Prochlorococcus populations flow cytometrically sorted from the Sargasso Sea and Gulf Stream. Limnol Oceanogr. 1998;43:1615–1630. [Google Scholar]
- 165.Urbach E, Robertson D L, Chisholm S W. Multiple evolutionary origins of prochlorophytes within the cyanobacterial radiation. Nature (London) 1992;355:267–269. doi: 10.1038/355267a0. [DOI] [PubMed] [Google Scholar]
- 166.Urbach E, Scanlan D J, Distel D L, Waterbury J B, Chisholm S W. Rapid diversification of marine picophytoplankton with dissimilar light harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (cyanobacteria) J Mol Evol. 1998;46:188–201. doi: 10.1007/pl00006294. [DOI] [PubMed] [Google Scholar]
- 167.van der Staay, G. W. M., and F. Partensky. Unpublished data.
- 168.van der Staay G W M, van der Staay-Moon S Y, Garczarek L, Partensky F. Characterization of the photosystem I subunits PsaI and PsaL from the marine oxyphotrophic prokaryotes Prochlorococcus spp. Photosynth Res. 1998;57:183–191. [Google Scholar]
- 169.van der Staay G W M, Yurkova N, Green B R. The 38 kDa chlorophyll a/b protein of the prokaryote Prochlorothrix hollandica is encoded by a divergent pcb gene. Plant Mol Biol. 1998;36:709–716. doi: 10.1023/a:1005930210515. [DOI] [PubMed] [Google Scholar]
- 170.van Lenning K, Garrido J L, Aristegui J, Zapata M. Temperature-programmed high performance liquid chromatography separation of mono- and divinyl chlorophyll forms from marine phytoplankton. Chromatographia. 1995;41:539–543. [Google Scholar]
- 171.Vaulot D. The cell cycle of phytoplankton: coupling cell growth to population growth. NATO ASI Ser Ser G. 1995;38:303–322. [Google Scholar]
- 172.Vaulot D, Courties C, Partensky F. A simple method to preserve oceanic phytoplankton for flow cytometric analyses. Cytometry. 1989;10:629–635. doi: 10.1002/cyto.990100519. [DOI] [PubMed] [Google Scholar]
- 173.Vaulot, D., and D. Marie. Diel variability of photosynthetic picoplankton in the equatorial Pacific. J. Geophys. Res., in press.
- 174.Vaulot D, Marie D, Olson R J, Chisholm S W. Growth of Prochlorococcus, a photosynthetic prokaryote, in the equatorial Pacific Ocean. Science. 1995;268:1480–1482. doi: 10.1126/science.268.5216.1480. [DOI] [PubMed] [Google Scholar]
- 175.Vaulot, D., D. Marie, and F. Partensky. Unpublished data.
- 176.Vaulot D, Partensky F. Cell cycle distributions of prochlorophytes in the North Western Mediterranean Sea. Deep-Sea Res I. 1992;39:727–742. [Google Scholar]
- 177.Vaulot D, Partensky F, Neveux J, Mantoura R F C, Llewellyn C. Winter presence of prochlorophytes in surface waters of the northwestern Mediterranean Sea. Limnol Oceanogr. 1990;35:1156–1164. [Google Scholar]
- 178.Veldhuis M J W, Kraay G W. Vertical distribution and pigment composition of a picoplanktonic prochlorophyte in the subtropical North-Atlantic: a combined study of HPLC-analysis of pigments and flow cytometry. Mar Ecol Prog Ser. 1990;68:121–127. [Google Scholar]
- 179.Veldhuis M J W, Kraay G W. Cell abundance and fluorescence of picoplankton in relation to growth irradiance and nitrogen availability in the Red Sea. Neth J Sea Res. 1993;31:135–145. [Google Scholar]
- 180.Veldhuis M J W, Kraay G W, Gieskes W W C. Growth and fluorescence characteristics of ultraplankton on a north-south transect in the eastern North Atlantic. Deep-Sea Res I. 1993;40:609–626. [Google Scholar]
- 181.Vidussi F, Claustre H, Bustillos-Guzman J, Cailliau C, Marty J C. Determination of chlorophylls and carotenoids of marine phytoplankton: separation of chlorophyll a from divinyl-chlorophyll a and zeaxanthin from lutein. J Plankton Res. 1996;18:2377–2382. [Google Scholar]
- 182.Waterbury J B, Valois F W. Resistance to co-occuring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl Environ Microbiol. 1993;59:3393–3399. doi: 10.1128/aem.59.10.3393-3399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Waterbury J B, Watson S W, Guillard R R L, Brand L E. Wide-spread occurence of a unicellular, marine planktonic, cyanobacterium. Nature (London) 1979;277:293–294. [Google Scholar]
- 184.Waterbury J B, Watson S W, Valois F W, Franks D G. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. Can Bull Fish Aquat Sci. 1986;214:71–120. [Google Scholar]
- 185.Watson G M F, Tabita F R. Regulation, unique gene organization, and unusual primary structure of carbon fixation genes from a marine phycoerythrin-containing cyanobacterium. Plant Mol Biol. 1996;32:1103–1115. doi: 10.1007/BF00041394. [DOI] [PubMed] [Google Scholar]
- 186.Wei H, Ca Q, Rahn R, Zhang X, Wang Y, Lebwohl M. DNA structural integrity and base composition affect ultraviolet light-induced oxidative DNA damage. Biochemistry. 1998;37:6485–6490. doi: 10.1021/bi972702f. [DOI] [PubMed] [Google Scholar]
- 187.Wikner J, Hagström Å. Evidence for tightly coupled nanoplanktonic predator-prey link regulating the bacterivores in the marine environment. Mar Ecol Prog Ser. 1988;50:137–145. [Google Scholar]
- 188.Wilhelm S W, Trick C G. Iron-limited growth of cyanobacteria: multiple siderophore is a common response. Limnol Oceanogr. 1994;39:1979–1984. [Google Scholar]
- 189.Wyman M, Gregory R P F, Carr N G. Novel role for phycoerythrin in a marine cyanobacterium, Synechococcus Strain DC2. Science. 1985;230:818–820. doi: 10.1126/science.230.4727.818. [DOI] [PubMed] [Google Scholar]
- 190.Yahel G, Post A F, Fabricius K, Marie D, Vaulot D, Genin A. Phytoplankton distribution and grazing near coral reefs. Limnol Oceanogr. 1998;43:551–563. [Google Scholar]
- 191.Zettler E R, Olson R J, Binder B J, Chisholm S W, Fitzwater S E, Gordon R M. Iron-enrichment bottle experiments in the equatorial Pacific: responses of individual phytoplankton cells. Deep-Sea Res II. 1996;43:1017–1029. [Google Scholar]
- 192.Zohary T, Brenner S, Krom M D, Angel D L, Kress N, Li W K W, Neori A, Yacobi Y Z. Buildup of microbial biomass during deep winter mixing in a Mediterranean warm-core eddy. Mar Ecol Prog Ser. 1998;167:47–57. [Google Scholar]