Both Transcriptional and Posttranscriptional Mechanisms Regulate Human Telomerase Template RNA Levels (original) (raw)
Abstract
The human telomerase RNA component (hTR) is present in normal somatic cells at lower levels than in cancer-derived cell lines. To understand the mechanisms regulating hTR levels in different cell types, we have compared the steady-state hTR levels in three groups of cells: (i) normal telomerase-negative human diploid cells; (ii) normal cells transfected with the human telomerase catalytic subunit, hTERT; and (iii) cells immortalized in vitro and cancer cells expressing their own endogenous hTERT. To account for the differences in steady-state hTR levels observed in these cell types, we compared the transcription rate and half-life of hTR in a subset of these cells. The half-life of hTR in telomerase-negative cells is about 5 days and is increased 1.6-fold in the presence of hTERT. The transcription rate of hTR is essentially unchanged in cells expressing exogenous hTERT, and the increased steady-state hTR level appears to be due to the increased half-life. However, the transcription rate of hTR is greatly increased in cells expressing endogenous hTERT, suggesting some overlap in transcriptional regulatory control. We conclude that the higher hTR level in cells expressing an endogenous telomerase can be a result of both increased transcription and a longer half-life and that the longer half-life might be partially a result of protection or stabilization by the telomerase catalytic subunit. The 4-week half-life of hTR in H1299 tumor cells is the longest half-life yet reported for any RNA.
Telomerase is a ribonucleoprotein in which a catalytic reverse transcriptase protein subunit uses the RNA as a template for the addition of telomeric repeat sequences to the ends of chromosomes (11, 16, 17, 28, 34, 41). Telomerase is responsible for the complete replication of the ends of chromosomes (telomeres) in most eukaryotes (26). In most normal human somatic cells, telomerase activity is nondetectable and telomere length decreases after each cell division due to the “end replication problem” (25, 42, 53).
Genes encoding the RNA component of telomerase have been cloned for many organisms, including Kluyveromyces lactis (32), Tetrahymena (45), Saccharomyces cerevisiae (48), Stylonychia mytilis (27), mouse (3), Bos taurus (Genbank accession no. AF054814), and human (11). The catalytic subunit of telomerase has also been cloned for a variety of organisms, including Euplotes aediculatus (28), Tetrahymena thermophila (8), Oxytricha trifallax (6), yeast (41), mouse (15, 30), and human (5, 24, 34, 41), all of which contain characteristic reverse transcriptase motifs (19, 28, 41). In humans, progressive telomere loss is believed to be the basis for cellular senescence and the limited life span of normal cells. Previously, it was demonstrated that ectopic expression of telomerase could greatly extend the life span of (4, 51) and potentially immortalize (22, 37) normal human cells in culture. Although telomerase is absent in most human adult somatic cells, telomerase activity is detected in most cancer cells (25). The upregulation or reactivation of telomerase in cancer cells or another mechanism to maintain telomere stability is likely to be necessary for the unlimited growth potential of cancer cells. However, at present, very little is known about the regulation of either the telomerase catalytic subunit or the telomerase RNA component.
Normal human diploid cells contain the integral RNA component of telomerase (hTR) but generally lack the mRNA for the catalytic subunit (hTERT) (34, 41), although there are some cases in which alternative splicing forms of hTERT are present in telomerase-negative cells (24, 50). The catalytic subunit is thought to be the only missing component necessary for at least a minimally functional enzyme. This is based on two lines of evidence: transfection of plasmids directing the expression of hTERT into telomerase-negative cell types results in the appearance of telomerase activity, and in vitro-transcribed hTR mixed with in vitro-translated hTERT in a rabbit reticulocyte lysate results in detectable telomerase (2, 4, 9, 51, 54). A 3- to 10-fold increase in steady-state levels of hTR has been observed in a variety of immortal cells (1), indicating that some regulation of hTR is associated with the acquisition of telomerase activity. In situ hybridization of human biopsy specimens indicates that the relative levels of hTR in tumor cells are sufficiently different from those in adjacent tissues to be clinically useful in the diagnosis of cancer (36, 38, 57, 58). Evidence from expression studies have defined a minimal hTR promoter (59), but studies of the actual rates of transcription in normal versus telomerase-expressing cells have not been reported.
Two mechanisms determine the steady-state level of hTR in cells: the rate of synthesis (the transcription rate) and the rate of degradation (the half-life). Elevated hTR levels in tumor cells might be due to an increased transcription rate, an increased half-life as a result of the association of hTR with the catalytic protein subunit or other regulatory modifications, or a combination of these factors. In the present study, we address this question by examining the steady-state levels, rates of transcription, and half-lives of hTR in three groups of human cells: (i) normal diploid telomerase-negative fibroblasts and epithelial cells, (ii) telomerase-negative cells converted to telomerase positivity by the forced expression of an exogenous hTERT cDNA, and (iii) telomerase-positive cells which have activated their endogenous hTERT gene during the process of immortalization, driven by the expression of the simian virus 40 (SV40) large T-antigen (T-Ag) or human papillomavirus E6/E7 proteins in vitro or by tumor formation in vivo. We report here that the differences in the steady-state levels of hTR can result from changes in both the rates of transcription of the template RNA and an increased half-life in the presence of the catalytic subunit of telomerase. To our knowledge, the half-life of hTR is the longest human RNA half-life yet described.
MATERIALS AND METHODS
Cell culture.
Human telomerase-positive non-small-cell lung cancer cell line H1299 (ATCC CRL-5803), telomerase-negative normal human diploid foreskin fibroblast BJ cells, BJ hTERT cells expressing a transfected hTERT (4), normal human diploid embryonic lung fibroblast strain IMR90 (ATCC CCL-186), and murine packaging cell lines PE501 and PA317 were cultured at 37°C under 5% CO2 in a 4:1 mixture of Dulbecco’s modified Eagle’s medium and medium 199 supplemented with 10% fortified bovine calf serum (HyClone, Logan, Utah) and 50 μg of gentamicin (Sigma, St. Louis, Mo.) per ml. IDH4 cells (47, 56) (immortal telomerase-positive cells derived from IMR90 fibroblasts stably transfected with SV40 T-Ag under the control of a dexamethasone-inducible promoter) are referred to here as IMR90 T-Ag. They were cultured in the same medium supplemented with 1 μg of dexamethasone per ml. Normal human mammary epithelial HME31 cells and all HME31-derived lines were grown in serum-free medium consisting of modified basal medium MCDB 170 (GIBCO BRL, Gaithersburg, Md.) supplemented with 0.4% bovine pituitary extract (Hammond Cell Tech, Alameda, Calif.), 5 μg of insulin (Sigma) per ml, 0.5 μg of hydrocortisone (Sigma) per ml, 50 μg of gentamicin per ml, 5 μg of transferrin per ml, and 10 ng of epidermal growth factor (GIBCO BRL) per ml. The medium used for HME31 cell growth was changed every other day.
Retroviral vector construction and infection.
The _Eco_RI fragment from pGRN145, containing the hTERT coding sequence (4) with a consensus Kozak sequence, was subcloned into the retroviral vector pBabe puro (39), in which the puromycin resistance gene is under the control of the SV40 promoter (Fig. 1A). Recombinant viruses were generated by first transfecting the ecotropic packaging cell line PE501 by electroporation and selecting with 4 μg of puromycin per ml. Viral supernatant derived from these cells was used to infect the amphotrophic PA317 packaging cell line (35) to generate clones containing unrearranged proviral copies of pBabe puro and pBabe puro hTERT. Medium containing released viruses produced from confluent dishes of selected populations of clones was filtered (pore size, 0.45 μm) and used to infect HME31 and IMR90 cells. IMR90 cells were selected on 750 ng of puromycin per ml, while HME31 cells, which are more sensitive to puromycin, were selected on 150 ng/ml.
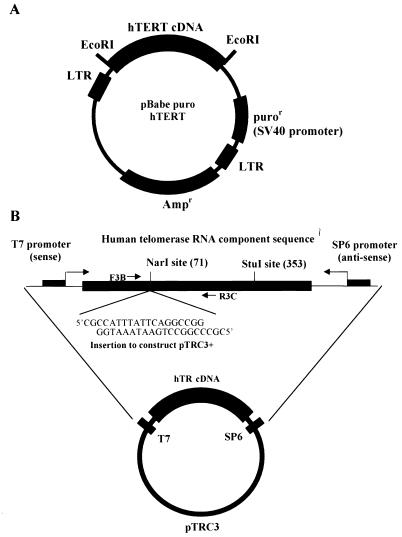
FIG. 1.
(A) Diagram of the construction of pBabe puro hTERT. An _Eco_RI fragment from pGRN145, containing the consensus Kozak sequence and the full-length human telomerase catalytic subunit cDNA, was cloned in the _Eco_RI site of the pBabe puro retroviral construct to form pBabe puro hTERT. (B) Diagram of plasmid construction for the generation of competitor RNA for quantitative RT-PCR (pTRC3+). The original plasmid pTRC3 is pGEM-5zf(+) with hTR cDNA cloned in its _Sac_I site, and the orientation is such that the T7 transcript of this construct is the sense strand. The _Nar_I and _Stu_I sites shown are unique sites in this plasmid. The location of the PCR primers used for RT-PCR assays in this study (F3B and R3C [Table 1]) are shown. A 20-bp fragment was inserted into pTRC3 by ligating the annealed oligonucleotides with pTRC3 linearized with _Nar_I. The oligonucleotides contained an equal number of G · C and A · T base pairs, so that the insertion did not change the G+C content of the PCR product compared to the RT-PCR product of hTR. The resulting plasmid, pTRC3+, was linearized with _Stu_I, and competitor RNA was generated with the T7 MAXIscript in vitro transcription kit.
Northern blot analysis of hTR expression.
Total RNA from these cells was prepared by a modification of the guanidium isothiocyanate-cesium chloride technique (7) with TriPure isolation reagent (Boehringer Mannheim Corp./Roche Molecular Biochemicals, Indianapolis, Ind.). The DNA probe for hTR was made by random priming with [α-32P]dCTP (GIBCO BRL), using a _Not_I-_Nsi_I fragment from pTRC3 [pGEM-5Zf(+) with an hTR insert cloned in its _Sac_I site] as the template (11). After stripping, the same blot was reprobed for 18S rRNA to normalize loading variations between RNA samples. Data were analyzed with ImageQuant version 3.3 software (Molecular Dynamics, Inc., Sunnyvale, Calif.).
Nuclear transcription runoff assay.
Nuclei from cultured BJ, BJ hTERT, IMR90, IMR90 T-Ag, and H1299 cells were prepared by the basic method described by Greenberg and coworkers (13, 14). Nuclei from 108 cells were resuspended in 500 μl of nucleus storage buffer (50 mM Tris-HCl [pH 8.3], 25% glycerol, 5 mM magnesium acetate, 0.1 mM EDTA, 5 mM dithiothreitol), quickly frozen in liquid nitrogen in 200-μl aliquots, and stored at −80°C. Transcription runoff assays were performed as described previously (31). Briefly, 200 μl of 2× reaction buffer containing 25% glycerol, 10 mM MgCl2, and 0.2 M KCl with three unlabeled nucleotides (1.2 mM ATP, 0.6 mM CTP, and 0.6 mM GTP) was added to 2 × 107 to 5 × 107 nuclei in 200 μl of nucleus storage buffer in an RNase-free 1.5-ml tube. After addition of 5 μl of 20-mCi/ml [α-32P]UTP (800 Ci/mmol; Amersham Co., Arlington Heights, Ill.) and incubation at room temperature for 45 min with occasional shaking, 50 U of RNase-free DNase I and 40 μl of 10 mM CaCl2 were added, and the mixture was incubated for 15 min at 37°C. Nuclear RNA was extracted, precipitated twice with ethanol and 5 M ammonium acetate, and resuspended in 500 μl of hybridization buffer containing 50% formamide, 0.5 M NaCl, 50 mM HEPES (pH 7.0), 0.4% sodium dodecyl sulfate (SDS), and 2 mM EDTA.
Target DNA (2 μg per slot) was linearized, denatured in 0.1 M NaOH at 37°C for 10 min, incubated on ice for 5 min, diluted in 10 volumes of ice-cold 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and bound in triplicate to Hybond-N+ (Amersham Co.) with a slot-blot manifold (Schleicher & Schuell, Keene, N.H.). The target DNA plasmids included (i) pTRC3 [pGEM-5Zf(+) with the hTR insert cloned in its _Sac_I site] (11) and its vector-only control [pGEM-5Zf(+); Promega, Madison, Wis.] and (ii) LK221 [human β-actin cDNA cloned in pBluescript SK(+)] (43) and its vector-only control [pBluescript SK(+); Stratagene, La Jolla, Calif.]. Each membrane was hybridized with an equal number of counts per minute (cpm). After prehybridization of membranes with 100 μg of yeast tRNA per ml for at least 4 h at 47°C, nuclear runoff transcripts were denatured (95°C for 5 min) and added to each membrane at a final concentration of at least 106 cpm/ml for 48 h at 47°C. The hybridization membranes were washed with 2× SSC at room temperature for 30 min followed by 2× SSC with 10 μg of RNase A per ml at 37°C for 30 min and then with 0.1× SSC–0.1% SDS at 50°C for 30 min. The hybridization membranes were exposed to PhosphorImager storage screens overnight. Hybridization bands were quantified with ImageQuant version 3.3 software (Molecular Dynamics, Inc.). The intensities of all hybridization bands were analyzed with a manually set local background. The signals obtained from the pGEM-5zf(+) and pBluescript SK(+) vectors were subtracted from the corresponding signals obtained from pTRC3 and LK221, respectively, and the data obtained from triplicate determinations were averaged. Human telomerase RNA component transcription rates were then normalized as a percentage of the human β-actin transcription rate in each cell line or strain (21).
Isolation of thiouridine-containing RNA by phenylmercury affinity chromatography for half-life determinations.
Mercurated agarose (equivalent to Bio-Rad Affi-Gel 501) was prepared from Affi-Gel 10 (Bio-Rad Laboratories, Hercules, Calif.) by a procedure specified by Bio-Rad Laboratories, Inc. In brief, Affi-Gel 10 was incubated with _p_-aminophenylmercuric acetate (Sigma) in dimethylformamide for 4 h, and then the unreacted succinimide groups on Affi-Gel 10 were blocked with ethanolamine. After a final solvent wash, Affi-Gel 501 was resuspended in anhydrous isopropanol and kept at −20°C in a dark bottle.
To keep the cells in log-phase growth during the time course of the pulse-chase experiments, cultured cells were passaged at 1:8 split ratios. After 24 h, the cells were pulse-labeled with 4-thiouridine (Sigma) at a final concentration of 100 μM for 5 h. To monitor the efficiency of the procedure, 5 μCi of [5,6-3H]uridine (Amersham Co.) per ml was included. At the end of the 5-h period, the cells were rinsed once in phosphate-buffered saline and fresh medium was added. The cells were collected in cold phosphate-buffered saline (pH 7.4) at each chase time point by being treated in trypsin-EDTA (GIBCO BRL) for 5 min at 37°C.
Total RNA from these cells was prepared with TriPure isolation reagent as described above. 4-Thiouridine-labeled RNA was separated with mercurated agarose (equivalent to Affi-Gel 501) as described previously (55). In brief, total RNA was dissolved in buffer A (50 mM sodium acetate [pH 5.5], 0.1% SDS, 0.15 M NaCl, 4 mM EDTA), heated to 95°C for 3 min, cooled rapidly in an ice-water bath, and batch-absorbed at 300 μg of RNA per ml (packed volume) of mercurated agarose resin in the dark for 2 h at 4°C with shaking. The resin was then packed into a PolyPrep column (Bio-Rad Laboratories) and washed with 2.5 column volumes of buffer A. Nonspecifically bound RNA was washed off with buffer B (buffer A containing 0.5 M NaCl instead of 0.15 M NaCl). Thiouridine-labeled RNA was eluted with buffer C (buffer A containing 10 mM β-mercaptoethanol). The recovery of thiouridine-labeled RNA was determined by counting 3H in aliquots of each fraction. Fractions eluted with buffer C were isopropanol precipitated, redissolved in 0.5 M ammonium acetate, pooled, and reprecipitated with ethanol.
The reliability of thiouridine labeling for analyzing the half-life was confirmed by determining the half-life of 18S rRNA in H1299 cells. Thiouridine-labeled RNA recovered on mercurated columns after increasing periods of growth in the absence of thiouridine was analyzed on Northern blots. The half-life (determined as described below) of 3.8 days is consistent with the half-life of 3 to 7 days previously reported for 18S rRNA in mammalian cells (12, 18).
After 10 to 12 days of continuous log-phase growth, 4-thiouridine-labeled RNA was a very small fraction of the total RNA. Control experiments with nonthiolated [3H]uridine-labeled RNA showed that two cycles of phenylmercury affinity chromatography were sufficient to eliminate background binding (10).
RNase protection assay.
RNase protection assays to quantify hTR were performed with the HybSpeed RPA kit (Ambion, Austin, Tex.) as described by the manufacturer. The antisense riboprobe was generated with the MAXIscript SP6 in vitro transcription kit (Ambion), using pTRC3 (Fig. 1B) linearized with _Stu_I as a template. For each RNase protection assay, 2 × 105 cpm of DNase I-treated probe was annealed to target RNA at 68°C for 18 h. The RNase-protected fragment (∼100 bp) was analyzed on a nondenaturing 6% polyacrylamide gel in 0.5× Tris-borate-EDTA (TBE). To compensate for experimental variations in individual RNase protection assays, relative quantitation of hTR levels in H1299, BJ hTERT, and BJ total RNA was performed simultaneously with the same antisense riboprobe. Three assays were performed with each RNA sample, and the average intensity of the protected band per microgram of total RNA of each sample was determined by plotting the intensity against input RNA amount. Quantitation was performed with ImageQuant version 3.3 software.
Quantitative RT-PCR.
Quantitative reverse transcription-PCR (RT-PCR) was based on the method originally described by Wang et al. (52) and modified by Nagano and Kelly (40). The plasmid used to synthesize the internal control RNA (competitor RNA) was constructed by inserting a 20-bp fragment into the unique _Nar_I site in pTRC3 (Fig. 1B). The RT-PCR product derived from the internal control is thus 20 bp longer (146 bp) than that of the RT-PCR fragment derived from the human telomerase RNA component (126 bp). The new plasmid (pTRC3+) was linearized with _Stu_I and transcribed with a MAXIscript T7 in vitro transcription kit (Ambion). The RNA was treated with DNase I to digest template plasmid DNA and ethanol precipitated. The concentration of this competitor RNA was calculated based on measurements of the optical density at 260 nm, and serial dilutions were made for the RT reactions. Control experiments established that the efficiency of the RT step for the competitor RNA is the same as the efficiency for hTR.
The quantitative analysis of human telomerase RNA component was accomplished in two steps. (i) An initial titration assay was performed to estimate the number of human telomerase RNA molecules (hTR) in each sample. 4-Thiouridine-labeled RNA purified from the progeny of two 150-mm culture plates of cells labeled at t = 0 was treated with 10 U of DNase I in a final volume of 20 μl. After heat inactivation of DNase I (5 min at 75°C), 2-μl aliquots of this RNA and 1-μl aliquots of competitor RNA at various dilutions were mixed and then reverse transcribed in 20 μl by using decamers and 10× alternate buffer supplied in the RETROscript kit (500 mM Tris-HCl [pH 8.3], 750 mM KCl, 30 mM MgCl2, 50 mM dithiothreitol) as specified by the manufacturer (Ambion). Then 2 μl of each RT mixture was PCR amplified with primers F3B and R3C (41) (Table 1) (25 cycles of 94°C for 20 s, 55°C for 30 s, and 72°C for 40 s) with 2 U of Taq DNA polymerase (GIBCO BRL) and the RT-PCR buffer included in the RETROscript kit. Under these experimental conditions, the PCR efficiency for both the competitor RNA and hTR was 88% in the linear amplification range (up to 25 cycles). The number of hTR molecules per microliter of RNA was estimated from this initial titration assay. (ii) In the quantitative step, equal numbers of competitor RNA molecules, calculated from the above titration, were mixed with 2 μl of thiouridine-labeled RNA to ensure consistent and accurate quantitative RT-PCR (49). The RT and PCR amplification were performed as described above.
TABLE 1.
Sequences of the oligonucleotides used in this study
| Name | Purpose | Sequence |
|---|---|---|
| F3B | Forward primer for hTR | 5′TCTAACCCTAACTGAGAAGG GCGTAG |
| R3C | Reverse primer for hTR | 5′GTTTGCTCTAGAATGAACGG TGGAAG |
| RT1 | Top strand to construct competitor for hTR | 5′CGCCATTTATTCAGGCCGGG |
| RT2 | Bottom strand to construct competitor for hTR | 5′CGCCCGGCCTGAATAAATGG |
In both the titration and quantitation steps, one of the PCR primers was labeled at the 5′ end with [γ-32P]ATP, using T4 polynucleotide kinase (Promega). A 10-μl aliquot of each PCR mixture (50 μl) was electrophoresed in a 6% nondenaturing polyacrylamide gel in 0.5× TBE buffer, along with a 5′-end-labeled 10-bp ladder (GIBCO BRL) as a molecular size marker. The gel was dried and exposed to a PhosphorImager storage screen. The number of hTR molecules in each sample was calculated based on the relative amounts of PCR product corresponding to hTR and competitor RNA.
Determination of the half-life of hTR.
Newly synthesized RNA was pulse-labeled with 4-thiouridine, chased for 0 to 10 days (experiment 1) or 0 to 12 days (experiment 2), and purified with mercurated agarose. The number of hTR molecules at each time point was analyzed by quantitative RT-PCR. Data from two independent experiments were combined for four of the five cell types analyzed. Based on first-order kinetics, ln(hTR) has a linear relationship with the chase time, and the half-life of hTR (_t_1/2) is inversely related to the slope of this line (k). The relationship between _t_1/2 and k is t_1/2 = (ln 2)/−_k. Linear regression was performed for each plot to estimate the slope of each line.
RESULTS
Comparison of steady-state hTR levels.
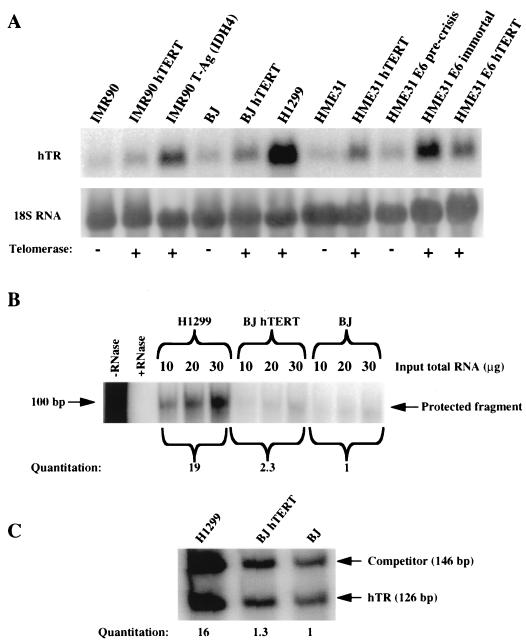
The relative hTR levels were quantitated by three different methods: Northern hybridization (Fig. 2A), RNase protection assay (Fig. 2B), and quantitative RT-PCR (Fig. 2C). The results of these analyses are summarized in Table 2. The Northern hybridization results showed that cells expressing an exogenous hTERT (BJ hTERT, IMR90 hTERT, HME31 hTERT, and HME31 E6 hTERT) contained approximately twice as much hTR as did their corresponding telomerase-negative cells (BJ, IMR90, HME31, and HME31 E6 precrisis). However, tumor and oncogene-immortalized lines expressing their endogenous hTERT (H1299, IMR90 T-Ag, and HME31 E6 immortal) had the largest amounts of hTR, ranging from 6- to 16-fold over those for the telomerase-negative cells. Quantitative RT-PCR and RNase protection assays confirmed these results from Northern hybridization (Table 2 and Fig. 2). These results also showed that the quantitative RT-PCR method we used was consistent with other methods.
FIG. 2.
Relative quantitation of steady-state hTR levels by Northern hybridization, RNase protection assay, and quantitative RT-PCR. (A) Northern hybridization of hTR and 18S rRNA. The intensities of the hTR and 18S hybridization bands were quantitated, and the intensities of the 18S bands were used as a loading control to normalize the hTR hybridization bands. (B) Comparison of steady-state hTR levels in H1299, BJ, and BJ hTERT cells by the RNase protection assay. The intensity of the protected band in each assay was plotted against the amount of input total RNA for each cell type, and the average intensity per 10 μg of input total RNA was calculated by linear regression based on individual graphs. (C) Comparison of steady-state hTR levels in H1299, BJ, and BJ hTERT cells by quantitative RT-PCR. The number of hTR molecules in 2 μg of total RNA from H1299, BJ, or BJ hTERT cells was calculated by measuring the amount of added competitor RNA and the ratio of intensities of the bands corresponding to competitor RNA and hTR.
TABLE 2.
Comparison of relative steady-state hTR levels
| Cell line | Description | Relative hTR levela quantitated by: | ||
|---|---|---|---|---|
| Northern hybridizationb | RNase protection assay | Quantitative RT-PCR | ||
| Telomerase-negative cells | ||||
| BJ | Normal foreskin fibroblasts | 1 | 1 | 1 |
| IMR90 | Normal lung fibroblasts | 1.0 ± 0.6 | ||
| HME31 | Normal mammary epithelial cells | 0.91 ± 0.06 | ||
| HME31 E6 precrisis | Precrisis HME31 cells expressing E6 | 1.6 ± 0.2 | ||
| Cells expressing exogenous hTERT | ||||
| BJ hTERT | Derived from BJ | 2.6 ± 0.3 | 2.3 | 1.3 |
| IMR90 hTERT | Derived from IMR90 | 1.4 ± 0.4 | ||
| HME31 hTERT | Derived from HME31 | 2.7 ± 0.2 | ||
| HME31 E6 hTERT | Derived from HME31 E6 precrisis | 3.4 ± 0.7 | ||
| Cells expressing endogenous hTERT | ||||
| HME31 E6 immortal | Postcrisis HME31 cells expressing E6 | 6.0 ± 0.3 | ||
| IMR90 T-Ag | SV40 T-Ag-immortalized IMR90 cells | 5.0 ± 0.9 | ||
| H1299 | Non-small-lung carcinoma cells | 14 ± 1.0 | 19 | 16 |
Transcription rates of hTR.
We measured the transcription rate of hTR to determine the fraction of the steady-state levels that were due to differences in the rates of synthesis. Transcription of hTR was determined by transcription runoff assays and normalized to human β-actin. Figure 3 shows an example of one set of slot blots, each of which was run in triplicate during the transcription runoff assays. The relative intensities of hybridization bands were quantitated as described in Materials and Methods. The averaged hTR transcription rates of three independent sets of transcription runoff assays are shown in Table 3. The transcription rate of hTR in H1299 cells was increased sixfold compared to that in BJ cells, and the transcription rate of hTR in IMR90 T-Ag cells was increased sevenfold compared to that in its parental IMR90 cells, while BJ hTERT showed a statistically insignificant increase in hTR transcription rate compared to its parental BJ cells. In all cases, the differences in the transcription rates of hTR in these cell types did not fully account for the differences in the steady-state hTR levels.
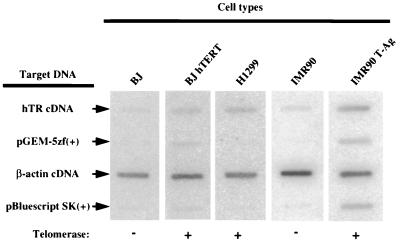
FIG. 3.
Transcription runoff analysis of hTR. Transcription runoff assays were performed with nuclei isolated from BJ, BJ hTERT, H1299, IMR90, and IMR90 T-Ag cells. Runoff transcripts were hybridized to a nylon membrane bearing the target DNA plasmids indicated. Shown is one set of slot blots that were done in triplicate for each cell type, from one of the nuclear runoff assays. Because BJ hTERT and IMR90 T-Ag contain stably integrated plasmid DNA, nuclear runoff transcripts from these cells also hybridized to pGEM-5zf(+) and pBluescript SK(+) on the membrane. After correction for the background hybridization of vector-only plasmids, the transcription rates of hTR were normalized to the human β-actin signal (see Materials and Methods) and expressed relative to the rate obtained in BJ fibroblasts (Table 3).
TABLE 3.
hTR transcription rate, half-life, and steady-state levela
| Cell type | Telomerase present | Relative half-lifeb | Relative transcription ratec | Expected relative steady-state hTR leveld | Actual relative steady-state hTR levele |
|---|---|---|---|---|---|
| BJ | − | 1.0 | 1.0 ± 0 | 1.0 | 1.0 |
| BJ hTERT | + | 1.6 | 1.3 ± 0.3 | 2.1 | 2.1 |
| IMR90 | − | 1.5 | 0.60 ± 0.2 | 0.9 | 1.0 |
| IMR90 T-Ag | + | 1.1 | 4.2 ± 0.5 | 4.6 | 5.0 |
| H1299 | + | 7.2 | 5.7 ± 0.4 | 41 | 16 |
Determination of the half-lives of hTR.
One method to measure the RNA half-life is to monitor the time course of the loss of the RNA while treating the cells with transcriptional inhibitors such as actinomycin D (20, 29). This technique works well for most mRNAs, since their half-lives are less than 17 h (46). Initial attempts to estimate the half-life of human telomerase RNA with actinomycin D were not successful. After 48 h of actinomycin D treatment (10 μg/ml), roughly half of the cells in culture had died due to the toxic effects of actinomycin D (44) but the hTR levels remained almost unchanged (data not shown). The half-life of the human telomerase RNA component is therefore too long to be measured by this method. Instead, an alternative approach involving 4-thiouridine to label newly synthesized RNA was used. The half-life of the labeled RNA was measured by purifying thiolated RNA on mercurated agarose (10, 23, 33, 55). Although other thiol-containing nucleotides or nucleosides can be used, we chose 4-thiouridine for its lack of toxicity (33), since the cells were pulse-labeled with 4-thiouridine and then cultured for up to 12 days for an accurate determination of the long half-life of hTR.
Control experiments established the following. (i) When cells were simultaneously labeled with 4-thiouridine and [3H]uridine (see Materials and Methods), 70% of the 3H label was consistently bound to the mercurated column and could be specifically eluted with β-mercaptoethanol. This indicates a high and reproducible efficiency of recovery of labeled RNA. (ii) Almost none (less than 0.01%) of the control [3H]uridine labeled RNAs that did not contain 4-thiouridine bound to the mercurated column material, demonstrating a low level of background binding. (iii) [3H]RNA did not bind to the column when [3H]uridine was added to the medium for 1 h immediately after 4-thiouridine was removed. This shows that the 4-thiouridine label was incorporated into newly synthesized RNA only during the pulse-labeling period and was not reutilized. This established the basis for estimating the half-life of hTR by this pulse-chase method.
Equal fractions of the cells originally labeled with 4-thiouridine were used for each chase time point. Since the cells were kept in continuous growth, long chase periods generated large numbers of cells in which the thiouridine-labeled RNA was a progressively smaller fraction of the total RNA. For example, after 10 days, two dishes of labeled cells generated at least 16 near-confluent dishes for normal cells such as BJ and IMR90 fibroblasts and 32 or 64 dishes for immortal cells such as BJ hTERT, IMR90 T-Ag, and H1299. We found that it was necessary to perform two cycles of mercurated affinity chromatography to reduce the background binding to satisfactory levels (see Materials and Methods) (10). When total RNA from 32 dishes of cells that were not labeled with 4-thiouridine (the equivalent number of H1299 cells to that in the 7-day time point of the chase experiment) was twice purified on the mercurated column, the hTR content was less than 10% of that found in the 7-day thiouridine-labeled sample, indicating a very low level of contamination of unlabeled RNA after two rounds of mercurated-column purification.
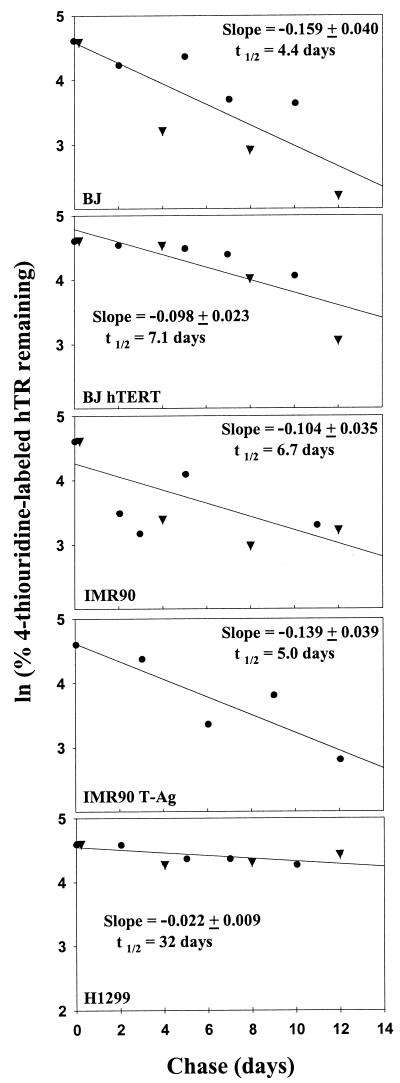
The amount of hTR remaining in the purified thiouridine-labeled RNA samples was determined by a two-step quantitative RT-PCR with a competitor for hTR that contained a 20-nucleotide insert. Because of plateau effects in PCRs, accurate quantitation was obtained only if the amount of competitor was equal to the amount of target RNA, so that RT-PCR products of target RNA and competitor RNA could be quantitated while they were both in the log range (49). An initial titration step of quantitative RT-PCR assays was performed with three threefold serial dilutions of competitor RNA to get a rough estimate of the amount of hTR present in the sample. In the quantitation step, the adjusted concentration of competitor RNA molecules was added to each thiouridine-labeled RNA sample for the RT reaction. The number of hTR molecules was calculated based on the number of molecules of competitor RNA divided by the ratio of the intensities of the 146-bp band (corresponding to the competitor RNA) and the 126-bp band (corresponding to hTR). Figure 4 shows a representative example of the second step of quantitative RT-PCR for BJ hTERT cells. Two independent determinations were performed for BJ, BJ hTERT, IMR90, and H1299, and one determination was performed for IMR90 T-Ag. The results for these five different cell types are shown in Fig. 5. The slopes of these plots were used to calculate the half-life of hTR, which ranged from 4.4 to 32 days. The results are also summarized in Table 3.
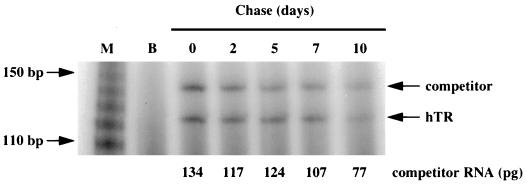
FIG. 4.
Half-life of hTR as determined by 4-thiouridine labeling and quantitative RT-PCR. The cells were labeled with 4-thiouridine for 5 h (experiment 1) or 12 h (experiment 2). After 0, 2, 5, 7, and 10 days of chase (experiment 1) and 0, 4, 8, and 12 days of chase (experiment 2), cells derived from the same original numbers of labeled cells were collected. RNA was extracted, thiouridine-labeled RNA was purified, and two-step quantitative RT-PCR was performed to quantitate the number of hTR molecules in 1/10 of each purified RNA sample. An example of the final quantitation step of 4-thiouridine-labeled RNA from BJ hTERT cells is shown. Lane M contains the 5′-32P-end-labeled 10-bp marker (GIBCO BRL), and lane B is the blank (no template in the PCR step). The amount of competitor RNA (in picograms) in each lane is also indicated.
FIG. 5.
Half-life of hTR. Plots of ln (% 4-thiouridine-labeled hTR remaining) versus chase time for each cell type. The numbers of hTR molecules determined by quantitative RT-PCR for BJ, BJ hTERT, IMR90, IMR90 T-Ag, and H1299 at each chase time point were expressed as percentages of their initial values, and these percentages were used to prepare the logarithmic plots. The slopes (−k ± standard error) of these lines were estimated by linear regression and used to calculate the half-life of hTR in each cell strain or cell line, using the formula t_1/2 = (ln 2)/−_k. The half-lives of hTR in BJ, BJ hTERT, IMR90, IMR90 T-Ag, and H1299 cells were determined to be 4.4, 7.1, 6.7, 5.0, and 32 days, respectively (Table 3). The different symbols represent data from two independent experiments (●, experiment 1; ▾, experiment 2) performed for all cell types other than IMR90 T-Ag.
DISCUSSION
The rather modest (3- to 10-fold) increase in hTR levels that has been found in cultured normal versus tumor cells is consistent with the hypothesis that stabilization of the integral RNA by its association with the hTERT protein might be sufficient to account for this increase. Our present study clearly indicates that this is not the case. Although some stabilization can occur, the major factor determining the increased steady-state hTR levels is the upregulation of hTR transcription. Furthermore, we show that this increased transcription is not a result of feedback regulation due to the presence of hTERT protein, since the increased transcription is not present in cells expressing an exogenous hTERT but occurs only in cells in which the endogenous gene has been activated by the process of in vitro immortalization by viral oncogenes or in vivo tumor formation. This result provides the first evidence that at some level there is a coordinated program for the derepression of telomerase during tumor formation (involving the regulation of both hTR and hTERT) rather than just a focal activation of hTERT (for example, by deletion of repressive sequences in the hTERT gene, activation of hTERT expression by translocation next to strong promoters, or any other mechanism that would activate only hTERT).
The steady-state hTR level is increased in telomerase-positive cells by both transcriptional and posttranscriptional mechanisms.
The steady-state hTR levels are higher in telomerase-positive cells than in telomerase-negative cells (Fig. 2 and Table 2). Mathematically, the steady-state RNA level is directly related to the transcription rate and half-life of the RNA, so that the steady-state level equals the product of the transcription rate and the half-life (Table 3). The present study reveals that both the half-life and the transcription rate of hTR can contribute to steady-state levels of hTR in the telomerase-positive cells.
The cell types rendered telomerase positive by expression of hTERT cDNA (exogenous hTERT) have slightly (two- to threefold) increased hTR levels compared to their telomerase-negative counterparts (HME31 hTERT versus HME31, BJ hTERT versus BJ, IMR90 hTERT versus IMR90, and HME31 E6 hTERT versus HME31 E6 precrisis). For the one pair in which both the transcription and half-life of hTR were measured (BJ hTERT and BJ), the very small increase in the transcription rate in BJ hTERT cells (1.3- ± 0.3-fold) is not significantly different from that in BJ parental cells, while the 1.6-fold increase in half-life is sufficient to explain the 2.1-fold increase in its steady-state hTR level. The observation that the expression of exogenous hTERT in BJ cells has little effect on the transcription rate of hTR suggests that the expression of hTERT alone is not sufficient to upregulate hTR transcription significantly.
The cell types that became telomerase positive by activating their endogenous hTERT, such as the non-small-lung carcinoma line H1299, IMR90 T-Ag (IMR90 immortalized by SV40 T-Ag), and HME31 E6 immortal (immortalized by E6 of human papillomavirus type 16) have 5- to 15-fold-higher hTR levels than do normal diploid cell types. The transcription rates of hTR in both IMR90 T-Ag and H1299 cells are increased approximately fivefold compared to those in normal IMR90 and BJ fibroblasts. In BJ hTERT cells, although expression of hTERT is sufficient to restore telomerase activity and maintain the telomere length in BJ fibroblasts (4), it did not result in an increased rate of hTR transcription. The increased transcription rate of hTR, seen only in immortal cell types expressing their endogenous hTERT, suggests that the changes or mutations that permit the reactivation of the endogenous hTERT during the immortalization process affect a coordinated telomerase reactivation program that may also upregulate hTR expression. In H1299 cells, increased hTR levels are partially accounted for by a sevenfold increase in the hTR half-life. Steady-state hTR levels in these immortal cell types can thus be higher because of the combined effects on both half-life and transcription rate.
In summary, expression of the telomerase catalytic subunit alone may cause a moderate increase in the steady-state level of hTR by increasing its half-life without affecting its transcription rate. Much higher steady-state hTR levels were observed in immortal cell types that expressed their endogenous hTERT, in which hTR transcription was also increased. In fact, in both cases examined, increases in hTR transcription made a major contribution to the elevated steady-state hTR levels in cells expressing their endogenous hTERT.
hTR RNA component has a very long half-life.
hTR was detected in all the cell types used in this study, including telomerase-negative cells such as BJ foreskin fibroblasts, IMR90 lung fibroblasts, and HME31 mammary epithelial cells. The half-life of hTR ranged from 4.4 to 32 days for the cell types examined. Even the shortest half-life of hTR measured (4.4 days in BJ cells and 5 days in IDH4 cells) is very long with respect to the half-lives of other RNAs that have been studied (46). The half-lives of a few structural RNAs have been described. The half-life of rRNA in cultured rat fibroblast cells is 7.5 ± 1.5 days (18), and the half-life of rRNA in human fibroblasts is roughly estimated to be less than 3 days (12). The half-life of 18S rRNA in H1299 cells, as determined by thiouridine labeling, is 3.8 days (data not shown), which is consistent with these previous reports. To our knowledge, hTR in H1299 cells has the longest half-life reported for any eukaryotic RNA.
An increased half-life for hTR was observed in some telomerase-positive cells. The half-life of hTR in BJ hTERT cells (7.1 days) was increased compared to that in the parental BJ cells (4.4 days). The longest hTR half-life was observed in H1299 cells (32 days), which have the highest telomerase activity of all the cell types examined in this study. However, the half-life of hTR was increased in the presence of hTERT in only two of the three cases studied. The exception is IMR90 T-Ag (telomerase positive), in which hTR had a half-life of 5.0 days, which is essentially the same as that of its parental IMR90 cells (6.7 days). Stabilization of hTR by hTERT does not seem to play a significant role in the increased hTR level in IMR90 T-Ag cells. Although some protection or stabilization of hTR may be provided by the presence of hTERT in BJ hTERT and H1299 cells, the unchanged hTR half-life in the telomerase-positive IMR90 T-Ag cells suggests that other factors affecting hTR turnover are likely to be involved as well and that they could counteract the stabilizing effect of hTERT protein and shorten the half-life of hTR.
In situ hybridization experiments have shown dramatically elevated hTR levels in a wide variety of human tumors (36, 38, 57, 58). In the present study, we have determined that the association of telomerase RNA with the telomerase catalytic subunit hTERT may contribute to an increase in the half-life and steady-state level of hTR, but other factors clearly affect the hTR half-life as well. Transcriptional upregulation of hTR is observed in immortalized cell types that express their endogenous hTERT. Understanding the transcriptional regulation of hTR that contributes to these higher levels may provide additional diagnostic and therapeutic opportunities for the treatment of cancer.
ACKNOWLEDGMENTS
BJ neonatal foreskin fibroblasts were kindly provided by James Smith, Baylor University Medical Center, Houston, Tex. The hTR and hTERT cDNAs were provided by Geron Corp., Menlo Park, Calif.
This work was supported by NIA grant AG07992. Jerry W. Shay is an Ellison Medical Foundation senior scholar.
REFERENCES
- 1.Avilion A A, Piatyszek M A, Gupta J, Shay J W, Bacchetti S, Greider C W. Human telomerase RNA and telomerase activity in immortal cell lines and tumor tissues. Cancer Res. 1996;56:645–650. [PubMed] [Google Scholar]
- 2.Beattie T L, Zhou W, Robinson M O, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 3.Blasco M A, Lee H W, Hande M P, Samper E, Lansdorp P M, DePinho R A, Greider C W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 4.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 5.Bryan T M, Marusic L, Bacchetti S, Namba M, Reddel R R. The telomere lengthening mechanism in telomerase-negative immortal human cells does not involve the telomerase RNA subunit. Hum Mol Genet. 1997;6:921–926. doi: 10.1093/hmg/6.6.921. [DOI] [PubMed] [Google Scholar]
- 6.Bryan T M, Sperger J M, Chapman K B, Cech T R. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc Natl Acad Sci USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc Natl Acad Sci USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Counter C M, Meyerson M, Eaton E N, Ellisen L W, Caddle S D, Haber D A, Weinberg R A. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene. 1998;16:1217–1222. doi: 10.1038/sj.onc.1201882. [DOI] [PubMed] [Google Scholar]
- 10.Cramer C L, Ryder T B, Bell J N, Lamb C J. Rapid switching of plant gene expression induced by fungal elicitor. Science. 1985;227:1240–1243. doi: 10.1126/science.227.4691.1240. [DOI] [PubMed] [Google Scholar]
- 11.Feng J, Funk W D, Wang S S, Weinrich S L, Avilion A A, Chiu C P, Adams R R, Chang E, Allsopp R C, Yu J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 12.Gillery P, Georges N, Wegrowski J, Randoux A, Borel J P. Protein synthesis in collagen lattice-cultured fibroblasts is controlled at the ribosomal level. FEBS Lett. 1995;357:287–289. doi: 10.1016/0014-5793(94)01375-b. [DOI] [PubMed] [Google Scholar]
- 13.Greenberg M E, Bender T P. Identification of newly transcribed RNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1997. pp. 4.10.1–4.10.11. [DOI] [PubMed] [Google Scholar]
- 14.Greenberg M E, Ziff E B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg R A, Allsopp R C, Chin L, Morin G B, DePinho R A. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- 16.Greider C W, Blackburn E H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 17.Greider C W, Blackburn E H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 18.Halle J P, Muller S, Simm A, Adam G. Copy number, epigenetic state and expression of the rRNA genes in young and senescent rat embryo fibroblasts. Eur J Cell Biol. 1997;74:281–288. [PubMed] [Google Scholar]
- 19.Harrington L, Zhou W, McPhail T, Oulton R, Yeung D S, Mar V, Bass M B, Robinson M O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrold S, Genovese C, Kobrin B, Morrison S L, Milcarek C. A comparison of apparent mRNA half-life using kinetic labeling techniques vs. decaying following administration of transcriptional inhibitors. Anal Biochem. 1991;198:19–29. doi: 10.1016/0003-2697(91)90500-s. [DOI] [PubMed] [Google Scholar]
- 21.Horikoshi T, Danenberg K D, Stadlbauer T H, Volkenandt M, Shea L C, Aigner K, Gustavsson B, Leichman L, Frosing R, Ray M, Gibson N W, Spears P, Danenberg P V. Quantitation of thymidylate synthase, dihydrofolate reductase and DT-diaphorase gene expression in human tumors using the polymerase chain reaction. Cancer Res. 1992;52:108–116. [PubMed] [Google Scholar]
- 22.Jiang X R, Jimenez G, Chang E, Frolkis M, Kusler B, Sage M, Beeche M, Bodnar A G, Wahl G M, Tlsty T D, Chiu C P. Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nat Genet. 1999;21:111–114. doi: 10.1038/5056. [DOI] [PubMed] [Google Scholar]
- 23.Johnson T R, Rudin S D, Blossey B K, Ilan J. Newly synthesized RNA: simultaneous measurement in intact cells of transcription rates and RNA stability of insulin-like growth factor I, actin, and albumin in growth hormone-stimulated hepatocytes. Proc Natl Acad Sci USA. 1991;88:5287–5291. doi: 10.1073/pnas.88.12.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilian A, Bowtell D D, Abud H E, Hime G R, Venter D J, Keese P K, Duncan E L, Reddel R R, Jefferson R A. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 25.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L, Coviello G M, Wright W E, Weinrich S L, Shay J W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 26.Lingner J, Cech T R. Telomerase and chromosome end maintenance. Curr Opin Genet Dev. 1998;8:226–232. doi: 10.1016/s0959-437x(98)80145-7. [DOI] [PubMed] [Google Scholar]
- 27.Lingner J, Hendrick L L, Cech T R. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- 28.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 29.Maity A, McKenna W G, Muschel R J. Evidence for posttranscriptional regulation of cyclin B1 mRNA in the cell cycle and following irradiation in HeLa cells. EMBO J. 1995;14:603–609. doi: 10.1002/j.1460-2075.1995.tb07036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin-Rivera L, Herrera E, Albar J P, Blasco M A. Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc Natl Acad Sci USA. 1998;95:10471–10476. doi: 10.1073/pnas.95.18.10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marzluff W F, Huang R C C. Transcription of RNA in isolated nuclei. In: Hames B D, Higgins S J, editors. Transcription and translation: a practical approach. Washington, D.C: IRL Press; 1984. pp. 89–128. [Google Scholar]
- 32.McEachern M J, Blackburn E H. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 33.Melvin W T, Milne H B, Silver A A, Allen H J, Kier H M. Incorporation of 6-thioguanosine and 4-thiouridine into RNA—application to isolation of newly synthesised RNA by affinity chromatography. Eur J Biochem. 1978;92:373–379. doi: 10.1111/j.1432-1033.1978.tb12756.x. [DOI] [PubMed] [Google Scholar]
- 34.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, Bacchetti S, Haber D A, Weinberg R A. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 35.Miller A D, Rosman G. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 36.Morales C P, Burdick J S, Saboorian M H, Wright W E, Shay J W. In situ hybridization for telomerase RNA in routine cytologic brushings for the diagnosis of pancreaticobiliary malignancies. Gastrointest Endosc. 1998;48:402–405. doi: 10.1016/s0016-5107(98)70011-2. [DOI] [PubMed] [Google Scholar]
- 37.Morales C P, Holt S E, Ouellette M, Kaur K J, Yan Y, Wilson K S, White M A, Wright W E, Shay J W. Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat Genet. 1999;21:115–118. doi: 10.1038/5063. [DOI] [PubMed] [Google Scholar]
- 38.Morales C P, Lee E L, Shay J W. In situ hybridization for the detection of telomerase RNA in the progression from Barrett’s esophagus to esophageal adenocarcinoma. Cancer. 1998;83:652–659. [PubMed] [Google Scholar]
- 39.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagano M, Kelly P A. Tissue distribution and regulation of rat prolactin receptor gene expression. J Biol Chem. 1994;269:13337–13345. [PubMed] [Google Scholar]
- 41.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 42.Olovnikov A M. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 43.Ponte P, Ng S Y, Engel J, Gunning P, Kedes L. Evolutionary conservation in the untranslated regions of actin mRNAs: DNA sequence of a human beta-actin cDNA. Nucleic Acids Res. 1984;12:1687–1696. doi: 10.1093/nar/12.3.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puckett L, Darnell J E. Essential factors in the kinetic analysis of RNA synthesis in HeLa cells. J Cell Physiol. 1977;90:521–534. doi: 10.1002/jcp.1040900315. [DOI] [PubMed] [Google Scholar]
- 45.Romero D P, Blackburn E H. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 46.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shay J W, West M D, Wright W E. Re-expression of senescent markers in deinduced reversibly immortalized cells. Exp Gerontol. 1992;27:477–492. doi: 10.1016/0531-5565(92)90003-i. [DOI] [PubMed] [Google Scholar]
- 48.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 49.Souaze F, Ntodou-Thome A, Tran C Y, Rostene W, Forgez P. Quantitative RT-PCR: limits and accuracy. BioTechniques. 1996;21:280–285. doi: 10.2144/96212rr01. [DOI] [PubMed] [Google Scholar]
- 50.Ulaner G A, Hu J F, Vu T H, Giudice L C, Hoffman A R. Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res. 1998;58:4168–4172. [PubMed] [Google Scholar]
- 51.Vaziri H, Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 52.Wang A M, Doyle M V, Mark D F. Quantitation of mRNA by the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson J D. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 54.Weinrich S L, Pruzan R, Ma L, Ouellette M, Tesmer V M, Holt S E, Bodnar A G, Lichtsteiner S, Kim N W, Trager J B, Taylor R D, Carlos R, Andrews W H, Wright W E, Shay J W, Harley C B, Morin G B. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 55.Woodford T A, Schlegel R, Pardee A B. Selective isolation of newly synthesized mammalian mRNA after in vivo labeling with 4-thiouridine or 6-thioguanosine. Anal Biochem. 1988;171:166–172. doi: 10.1016/0003-2697(88)90138-8. [DOI] [PubMed] [Google Scholar]
- 56.Wright W E, Pereira-Smith O M, Shay J W. Reversible cellular senescence: implication for a two-stage model for the immortalization of normal human diploid fibroblasts. Mol Cell Biol. 1989;9:3088–3092. doi: 10.1128/mcb.9.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yashima K, Litzky L A, Kaiser L, Rogers T, Lam S, Wistuba I I, Milchgrub S, Srivastava S, Piatyszek M A, Shay J W, Gazdar A F. Telomerase expression in respiratory epithelium during the multistage pathogenesis of lung carcinomas. Cancer Res. 1997;57:2373–2377. [PubMed] [Google Scholar]
- 58.Yashima K, Piatyszek M A, Saboorian H M, Virmani A K, Brown D, Shay J W, Gazdar A F. Telomerase activity and in situ telomerase RNA expression in malignant and non-malignant lymph nodes. J Clin Pathol. 1997;50:110–117. doi: 10.1136/jcp.50.2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao J Q, Hoare S F, McFarlane R, Muir S, Parkinson E K, Black D M, Keith W N. Cloning and characterization of human and mouse telomerase RNA gene promoter sequences. Oncogene. 1998;16:1345–1350. doi: 10.1038/sj.onc.1201892. [DOI] [PubMed] [Google Scholar]